Calmodulin in Paramecium: Focus on Genomic Data
Abstract
1. Introduction
2. Calmodulin
2.1. The EF-Hand Motif
3. Paramecium
3.1. Motility and Ion Channels
3.2. The Cortex and the Secretion Systems: The Trichocyst
4. Calmodulin in Paramecium
4.1. Cellular Localization
4.2. Isolation and Purification
4.3. Enzyme Targets
4.4. Other Protein Targets
4.5. Amino Acid Sequences
4.5.1. Mutant Versions
4.6. Methylation
4.7. Crystal Structures
4.8. Nucleotide Sequences
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carafoli, E.; Krebs, J. Why calcium? How calcium became the best communicator. J. Biol. Chem. 2016, 291, 20849–20857. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Villalobo, A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta 2014, 1843, 398–435. [Google Scholar] [CrossRef]
- Cheung, W.Y. Cyclic 3′,5′-nucleotide phosphodiesterase. Demonstration of an activator. Biochem. Biophys. Res. Commun. 1970, 38, 533–538. [Google Scholar] [CrossRef]
- Kakiuchi, S.; Yamazaki, R.; Nakajima, H. Properties of a heat-stable phosphodiesterase activating factor isolated from brain extract. Proc. Jpn. Acud. 1970, 46, 587–592. [Google Scholar] [CrossRef][Green Version]
- Cheung, W. Calmodulin plays a pivotal role in cellular regulation. Science 1980, 207, 19–27. [Google Scholar] [CrossRef]
- Teo, T.S.; Wang, J.H. Mechanism of activation of a cyclic adenosine 3′:5′-monophosphate phosphodiesterase from bovine heart by calcium ions. Identification of the protein activator as a Ca2+ binding protein. J. Biol. Chem. 1973, 248, 5950–5955. [Google Scholar] [CrossRef]
- LaPorte, D.C.; Wierman, B.M.; Storm, D.R. Calcium-induced exposure of a hydrophobic surface on calmodulin. Biochemistry 1980, 19, 3814–3819. [Google Scholar] [CrossRef]
- Shen, X.; Valencia, C.A.; Szostak, J.W.; Dong, B.; Liu, R. Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 5969–5974. [Google Scholar] [CrossRef] [PubMed]
- Brostrom, C.O.; Wolff, D.J. Properties and functions of calmodulin. Biochem. Pharmacol. 1981, 30, 1395–1405. [Google Scholar] [CrossRef]
- Villalobo, A.; Ishida, H.; Vogel, H.J.; Berchtold, M.W. Calmodulin as a protein linker and a regulator of adaptor/scaffold proteins. Biochim. Biophys Acta Mol. Cell. Res. 2018, 1865, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H.; Nockolds, C.E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar] [CrossRef]
- Strynadka, N.C.; James, M.N. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu. Rev. Biochem. 1989, 58, 951–998. [Google Scholar] [CrossRef] [PubMed]
- Onek, L.A.; Smith, R.J. Calmodulin and calcium mediated regulation in prokaryotes. J. Gen. Microbiol. 1992, 138, 1039–1049. [Google Scholar] [CrossRef][Green Version]
- Michiels, J.; Xi, C.; Verhaert, J.; Vanderleyden, J. The functions of Ca2+ in bacteria: A role for EF-hand proteins? Trends Microbiol. 2002, 10, 87–93. [Google Scholar] [CrossRef]
- Domínguez, D.C.; Guragain, M.; Patrauchan, M. Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 2015, 57, 151–165. [Google Scholar] [CrossRef]
- Zhou, Y.; Tzeng, W.P.; Yang, W.; Zhou, Y.; Ye, Y.; Lee, H.W.; Frey, T.K.; Yang, J. Identification of a Ca2+-binding domain in the rubella virus nonstructural protease. J. Virol. 2007, 81, 7517–7528. [Google Scholar] [CrossRef]
- Rahman, S.K.; Kerviel, A.; Mohl, B.P.; He, Y.; Zhou, Z.H.; Roy, P. A Calcium Sensor Discovered in Bluetongue Virus Nonstructural Protein 2 Is Critical for Virus Replication. J. Virol. 2020, 94, e01099-20. [Google Scholar] [CrossRef]
- Ishida, H.; Vogel, H.J. EF-Hand Proteins. In Encyclopedia of Metalloproteins; Kretsinger, R.H., Uversky, V.N., Permyakov, E.A., Eds.; Springer: New York, NY, USA, 2013; pp. 766–775. [Google Scholar] [CrossRef]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Kawasaki, H.; Kretsinger, R.H. Structural and functional diversity of EF-hand proteins: Evolutionary perspectives. Protein. Sci. 2017, 26, 1898–1920. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Ames, J.B.; Swindells, M.B.; Ikura, M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins 1999, 37, 499–507. [Google Scholar] [CrossRef]
- Nelson, M.R.; Chazin, W.J. Structures of EF-hand Ca2+-binding proteins: Diversity in the organization, packing and response to Ca2+ binding. Biometals 1998, 11, 297–318. [Google Scholar] [CrossRef]
- Grabarek, Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J. Mol. Biol. 2006, 359, 509–525. [Google Scholar] [CrossRef]
- Moncrief, N.D.; Kretsinger, R.H.; Goodman, M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J. Mol. Evol. 1990, 30, 522–562. [Google Scholar] [CrossRef]
- Allen, R.D. Cytology. In Paramecium; Görtz, H.-D., Ed.; Springer: Berlin, Germany, 1988; pp. 4–40. [Google Scholar]
- Lynn, D.H. Phylum CILIOPHORA–Conjugating, ciliated Protists with Nuclear Dualism. In The Ciliated Protozoa Characterization, Classification, and Guide to the Literature; Springer Science: Heidelberg, Germany, 2008; pp. 89–120. [Google Scholar]
- Hall, M.S.; Katz, L.A. On the nature of species: Insights from Paramecium and other ciliates. Genetica 2011, 139, 677–684. [Google Scholar] [CrossRef]
- Nyberg, D. The species concept and breeding systems. In Paramecium; Görtz, H.-D., Ed.; Springer: Berlin, Germany, 1988; pp. 41–58. [Google Scholar]
- Aury, J.M.; Jaillon, O.; Duret, L.; Noel, B.; Jubin, C.; Porcel, B.M.; Ségurens, B.; Daubin, V.; Anthouard, V.; Aiach, N.; et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006, 444, 171–178. [Google Scholar] [CrossRef]
- Arnaiz, O.; Meyer, E.; Sperling, L. ParameciumDB 2019: Integrating genomic data across the genus for functional and evolutionary biology. Nucleic Acids Res. 2020, 48, D599–D605. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive ree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tourancheau, A.B.; Villalobo, E.; Tsao, N.; Torres, A.; Pearlman, R.E. Protein coding gene trees in ciliates: Comparison with rRNA-based phylogenies. Mol. Phylogenet. Evol. 1998, 10, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Baroin-Tourancheau, A.; Delgado, P.; Perasso, R.; Adoutte, A. A broad molecular phylogeny of ciliates: Identification of major evolutionary trends and radiations within the phylum. Proc. Natl. Acad. Sci. USA 1992, 89, 9764–9768. [Google Scholar] [CrossRef] [PubMed]
- Strüder-Kypke, M.C.; Wright, A.D.; Fokin, S.I.; Lynn, D.H. Phylogenetic relationships of the genus Paramecium inferred from small subunit rRNA gene sequences. Mol. Phylogenet. Evol. 2000, 14, 122–130. [Google Scholar] [CrossRef]
- Hori, M.; Tomikawa, I.; Przyboś, E.; Fujishima, M. Comparison of the evolutionary distances among syngens and sibling species of Paramecium. Mol. Phylogenet. Evol. 2006, 38, 697–704. [Google Scholar] [CrossRef]
- Van Houten, J. Paramecium biology. In Evo-Devo: Non-Model Species in Cell and Developmental Biology. Results and Problems in Cell Differentiation; Tworzydlo, W., Bilinski, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 291–318. [Google Scholar] [CrossRef]
- Bouhouche, K.; Valentine, M.S.; Le Borgne, P.; Lemullois, M.; Yano, J.; Lodh, S.; Nabi, A.; Tassin, A.M.; Van Houten, J.L. Paramecium, a model to study ciliary beating and ciliogenesis: Insights from cutting-edge approaches. Front. Cell. Dev. Biol. 2022, 10, 847908. [Google Scholar] [CrossRef]
- Jennings, H.S. The basis for taxis and certain other terms in the behavior of infusoria. J. Comp. Neurol. Psychol. 1905, 15, 528–534. [Google Scholar] [CrossRef]
- Valentine, M.S.; Van Houten, J. Ion channels of cilia: Paramecium as a model. J. Eukaryot. Microbiol. 2022, 69, e12884. [Google Scholar] [CrossRef]
- Yano, J.; Wells, R.; Lam, Y.W.; Van Houten, J.L. Ciliary Ca2+ pumps regulate intraciliary Ca2+ from the action potential and may co-localize with ciliary voltage-gated Ca2+ channels. J. Exp. Biol. 2021, 224, jeb232074. [Google Scholar] [CrossRef]
- Saimi, Y.; Martinac, B.; Gustin, M.C.; Culbertson, M.R.; Adler, J.; Kung, C. Ion channels in Paramecium, yeast and Escherichia coli. Trends Biochem. 1988, 13, 304–309. [Google Scholar] [CrossRef]
- Kung, C.; Saimi, Y. The physiological basis of taxes in Paramecium. Annu. Rev. Physiol. 1982, 44, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Saimi, Y.; Kung, C. Behavioral genetics of Paramecium. Annu. Rev. Genet. 1987, 21, 47–65. [Google Scholar] [CrossRef]
- Lodh, S.; Yano, J.; Valentine, M.S.; Van Houten, J.L. Voltage-gated calcium channels of Paramecium cilia. J. Exp. Biol. 2016, 219, 3028–3038. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, J.; Chang, S.Y.; Kung, C. Genetic analyses of “paranoiac” mutants of Paramecium tetraurelia. Genetics 1977, 86, 113–120. [Google Scholar] [CrossRef]
- Cohen, J.; Beisson, J. The cytoskeleton. In Paramecium; Görtz, H.-D., Ed.; Springer: Berlin, Germany, 1988; pp. 363–392. [Google Scholar]
- Stelly, N.; Mauger, J.P.; Claret, M.; Adoutte, A. Cortical alveoli of Paramecium: A vast submembranous calcium storage compartment. J. Cell Biol. 1991, 113, 103–112. [Google Scholar] [CrossRef]
- Plattner, H. My favorite cell--Paramecium. Bioessays 2002, 24, 649–658. [Google Scholar] [CrossRef]
- Plattner, H. Trichocysts-Paramecium’s Projectile-like Secretory Organelles: Reappraisal of their Biogenesis, Composition, Intracellular Transport, and Possible Functions. J. Eukaryot. Microbiol. 2017, 64, 106–133. [Google Scholar] [CrossRef]
- Kerboeuf, D.; Le Berre, A.; Dedieu, J.C.; Cohen, J. Calmodulin is essential for assembling links necessary for exocytotic membrane fusion in Paramecium. EMBO J. 1993, 12, 3385–3390. [Google Scholar] [CrossRef]
- Vayssié, L.; Skouri, F.; Sperling, L.; Cohen, J. Molecular genetics of regulated secretion in Paramecium. Biochimie 2000, 82, 269–288. [Google Scholar] [CrossRef]
- Plattner, H. Calcium regulation in the protozoan model, Paramecium tetraurelia. J. Eukaryot. Microbiol. 2014, 61, 95–114. [Google Scholar] [CrossRef] [PubMed]
- Klauke, N.; Plattner, H. Imaging of Ca2+ transients induced in Paramecium cells by a polyamine secretagogue. J. Cell Sci. 1997, 110, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Kersken, H.; Tiggemann, R.; Westphal, C.; Plattner, H. The secretory contents of Paramecium tetraurelia trichocysts: Ultrastructural–cytochemical characterization. J. Histochem. Cytochem. 1984, 32, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Tiggemann, R.; Plattner, H. Possible involvement of a calmodulin regulated Ca2+-ATPase in exocytosis performance in Paramecium tetraurelia cells. FEBS Lett. 1982, 148, 226–230. [Google Scholar] [CrossRef]
- Maihle, N.J.; Satir, B.H. Indirect immunofluorescent localization of calmodulin in Paramecium tetraurelia. J. Protozool. 1979, 26, 10A. [Google Scholar] [CrossRef]
- Maihle, N.J.; Satir, B.H. Calmodulin in the ciliates Paramecium tetraurelia and Tetrahymena thermophila. Ann. N. Y. Acad. Sci. 1980, 356, 408–409. [Google Scholar] [CrossRef]
- Saimi, Y.; Kung, C. Calmodulin as an ion channel subunit. Annu. Rev. Physiol. 2002, 64, 289–311. [Google Scholar] [CrossRef]
- Kung, C.; Preston, R.R.; Maley, M.E.; Ling, K.Y.; Kanabrocki, J.A.; Seavey, B.R.; Saimi, Y. In vivo Paramecium mutants show that calmodulin orchestrates membrane responses to stimuli. Cell Calcium 1992, 13, 413–425. [Google Scholar] [CrossRef]
- Preston, R.R.; Kink, J.A.; Hinrichsen, R.D.; Saimi, Y.; Kung, C. Calmodulin mutants and Ca2+-dependent channels in Paramecium. Annu. Rev. Physiol. 1991, 53, 309–319. [Google Scholar] [CrossRef]
- Dedman, J.R.; Welsh, M.J.; Means, A.R. Ca2+-dependent regulator. Production and characterization of a monospecific antibody. J. Biol. Chem. 1978, 253, 7515–7521. [Google Scholar] [CrossRef]
- Maihle, N.J.; Dedman, J.R.; Means, A.R.; Chafouleas, J.G.; Satir, B.H. Presence and indirect immunofluorescent localization of calmodulin in Paramecium tetraurelia. J. Cell Biol. 1981, 89, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, S.; Steiner, A.L.; Schultz, J.E. Immunocytochemical localization of cyclic GMP, cGMP-dependent protein kinase, calmodulin and calcineurin in Paramecium tetraurelia. Eur. J. Cell Biol. 1983, 32, 164–170. [Google Scholar] [PubMed]
- Momayezi, M.; Kersken, H.; Gras, U.; Vilmart-Seuwen, J.; Plattner, H. Calmodulin in Paramecium tetraurelia: Localization from the in vivo to the ultrastructural level. J. Histochem. Cytochem. 1986, 34, 1621–1638. [Google Scholar] [CrossRef]
- Rauh, J.J.; Nelson, D.L. Calmodulin is a major component of extruded trichocysts from Paramecium tetraurelia. J. Cell Biol. 1981, 91, 860–865. [Google Scholar] [CrossRef]
- Levin, R.M.; Weiss, B. Binding of trifluoperazine to the calcium-dependent activator of cyclic nucleotide phosphodiesterase. Mol. Pharmacol. 1977, 13, 690–697. [Google Scholar]
- Walter, M.F.; Schultz, J.E. Calcium receptor protein calmodulin isolated from cilia and cells of Paramecium tetraurelia. Eur. J. Cell Biol. 1981, 24, 97–100. [Google Scholar] [PubMed]
- Fok, A.K.; Aihara, M.S.; Ishida, M.; Allen, R.D. Calmodulin localization and its effects on endocytic and phagocytic membrane trafficking in Paramecium multimicronucleatum. J. Eukaryot. Microbiol. 2008, 55, 481–491. [Google Scholar] [CrossRef]
- Burgess-Cassler, A.; Hinrichsen, R.D.; Maley, M.E.; Kung, C. Biochemical characterization of a genetically altered calmodulin in Paramecium. Biochim. Biophys. Acta 1987, 913, 321–328. [Google Scholar] [CrossRef]
- Suzuki, Y.; Ohnishi, K.; Hirabayashi, T.; Watanabe, Y. Tetrahymena calmodulin. Characterization of an anti-tetrahymena calmodulin and the immunofluorescent localization in Tetrahymena. Exp. Cell. Res. 1982, 137, 1–14. [Google Scholar] [CrossRef]
- Tindall, S.H.; DeVito, L.D.; Nelson, D.L. Biochemical characterization of the proteins of Paramecium secretory granules. J. Cell Sci. 1989, 92, 441–447. [Google Scholar] [CrossRef]
- Hinrichsen, R.D.; Burgess-Cassler, A.; Soltvedt, B.C.; Hennessey, T.; Kung, C. Restoration by calmodulin of a Ca2+-dependent K+ current missing in a mutant of Paramecium. Science 1986, 232, 503–506. [Google Scholar] [CrossRef]
- Burgess, W.H.; Jemiolo, D.K.; Kretsinger, R.H. Interaction of calcium and calmodulin in the presence of sodium dodecyl sulfate. Biochim. Biophys. Acta 1980, 623, 257–270. [Google Scholar] [CrossRef]
- Klee, C.B.; Vanaman, T.C. Calmodulin. Adv. Protein. Chem. 1982, 35, 213–321. [Google Scholar] [CrossRef] [PubMed]
- Satir, B.H.; Garofalo, R.S.; Gilligan, D.M.; Maihle, N.J. Possible functions of calmodulin in protozoa. Ann. N. Y. Acad. Sci. 1980, 356, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Momayezi, M.; Lumpert, C.J.; Kersken, H.; Gras, U.; Plattner, H.; Krinks, M.H.; Klee, C.B. Exocytosis induction in Paramecium tetraurelia cells by exogenous phosphoprotein phosphatase in vivo and in vitro: Possible involvement of calcineurin in exocytotic membrane fusion. J. Cell Biol. 1987, 105, 181–189. [Google Scholar] [CrossRef]
- Haeseleer, F.; Palczewski, K. Calmodulin and Ca2+-binding proteins (CaBPs): Variations on a theme. Adv. Exp. Med. Biol. 2002, 514, 303–317. [Google Scholar] [CrossRef]
- Jurado, L.A.; Chockalingam, P.S.; Jarrett, H.W. Apocalmodulin. Physiol. Rev. 1999, 79, 661–682. [Google Scholar] [CrossRef]
- Crivici, A.; Ikura, M. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 85–116. [Google Scholar] [CrossRef]
- Jamieson, G.A., Jr.; Vanaman, T.C.; Blum, J.J. Presence of calmodulin in Tetrahymena. Proc. Natl. Acad. Sci. USA 1979, 76, 6471–6475. [Google Scholar] [CrossRef]
- Ehrlich, B.E.; Jacobson, A.R.; Hinrichsen, R.; Sayre, L.M.; Forte, M.A. Paramecium calcium channels are blocked by a family of calmodulin antagonists. Proc. Natl. Acad. Sci. USA 1988, 85, 5718–5722. [Google Scholar] [CrossRef]
- Hennessey, T.M.; Kung, C. An anticalmodulin drug, W-7, inhibits the voltage-dependent calcium current in Paramecium caudatum. J. Exp. Biol. 1984, 110, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, R.S.; Gilligan, D.M.; Satir, B.H. Calmodulin antagonists inhibit secretion in Paramecium. J. Cell Biol. 1983, 96, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Burgess, W.H.; Watterson, D.M. Comparison of the NAD kinase and myosin light chain kinase activator properties of vertebrate, higher plant, and algal calmodulins. Plant Physiol. 1984, 75, 796–798. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.C.; Lukas, T.J.; Craig, T.A.; Wilson, E.; King, M.M.; Kwiatkowski, A.P.; Watterson, D.M. Computational and site-specific mutagenesis analyses of the asymmetric charge distribution on calmodulin. Proteins 1989, 6, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Lukas, T.J.; Wallen-Friedman, M.; Kung, C.; Watterson, D.M. In vivo mutations of calmodulin: A mutant Paramecium with altered ion current regulation has an isoleucine-to-threonine change at residue 136 and an altered methylation state at lysine residue 115. Proc. Natl. Acad. Sci. USA 1989, 86, 7331–7335. [Google Scholar] [CrossRef]
- Hinrichsen, R.; Wilson, E.; Lukas, T.; Craig, T.; Schultz, J.; Watterson, D.M. Analysis of the molecular basis of calmodulin defects that affect ion channel-mediated cellular responses: Site-specific mutagenesis and microinjection. J. Cell Biol. 1990, 111, 2537–2542. [Google Scholar] [CrossRef]
- Bemm, F.; Schwarz, R.; Förster, F.; Schultz, J. A kinome of 2600 in the ciliate Paramecium tetraurelia. FEBS Lett. 2009, 583, 3589–3592. [Google Scholar] [CrossRef]
- Schultz, J.E.; Klumpp, S. Guanylate cyclase in the excitable ciliary membrane of Paramecium. FEBS Lett. 1980, 122, 64–66. [Google Scholar] [CrossRef]
- Schultz, J.E.; Klumpp, S. Lanthanum dissociates calmodulin from the guanylate cyclase of the excitable ciliary membrane from Paramecium. FEMS Microbiol. Lett. 1982, 13, 303–306. [Google Scholar] [CrossRef]
- Klumpp, S.; Guerini, D.; Krebs, J.; Schultz, J.E. Effect of tryptic calmodulin fragments on guanylate cyclase activity from Paramecium tetraurelia. Biochem. Biophys. Res. Commun. 1987, 142, 857–864. [Google Scholar] [CrossRef]
- Klumpp, S.; Kleefeld, G.; Schultz, J.E. Calcium/calmodulin-regulated guanylate cyclase of the excitable ciliary membrane from Paramecium. Dissociation of calmodulin by La3+: Calmodulin specificity and properties of the reconstituted guanylate cyclase. J. Biol. Chem. 1983, 258, 12455–12459. [Google Scholar] [CrossRef]
- Potter, L.R. Guanylyl cyclase structure, function and regulation. Cell Signal. 2011, 23, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Linder, J.U.; Schultz, J.E. Guanylyl cyclases in unicellular organisms. Mol. Cell. Biochem. 2002, 230, 149–158. [Google Scholar] [CrossRef]
- Linder, J.U.; Engel, P.; Reimer, A.; Krüger, T.; Plattner, H.; Schultz, A.; Schultz, J.E. Guanylyl cyclases with the topology of mammalian adenylyl cyclases and an N-terminal P-type ATPase-like domain in Paramecium, Tetrahymena and Plasmodium. EMBO J. 1999, 18, 4222–4232. [Google Scholar] [CrossRef] [PubMed]
- Brini, M.; Calì, T.; Ottolini, D.; Carafoli, E. The plasma membrane calcium pump in health and disease. FEBS J. 2013, 280, 5385–5397. [Google Scholar] [CrossRef]
- Wright, M.V.; Van Houten, J.L. Characterization of a putative Ca2+-transporting Ca2+-ATPase in the pellicles of Paramecium tetraurelia. Biochim. Biophys. Acta 1990, 1029, 241–251. [Google Scholar] [CrossRef]
- Elwess, N.L.; Van Houten, J.L. Cloning and molecular analysis of the plasma membrane Ca2+-ATPase gene in Paramecium tetraurelia. J. Eukaryot. Microbiol. 1997, 44, 250–257. [Google Scholar] [CrossRef]
- Gietzen, K.; Wüthrich, A.; Bader, H. R 24571: A new powerful inhibitor of red blood cell Ca2+-transport ATPase and of calmodulin-regulated functions. Biochem. Biophys. Res. Commun. 1981, 101, 418–425. [Google Scholar] [CrossRef]
- Travis, S.M.; Nelson, D.L. Regulation of axonemal Mg2+-ATPase from Paramecium cilia: Effects of Ca2+ and cyclic nucleotides. Biochim. Biophys. Acta 1988, 966, 84–93. [Google Scholar] [CrossRef]
- Braun, A.P.; Schulman, H. The multifunctional calcium/calmodulin-dependent protein kinase: From form to function. Annu. Rev. Physiol. 1995, 57, 417–445. [Google Scholar] [CrossRef]
- Gundersen, R.E.; Nelson, D.L. A novel Ca2+-dependent protein kinase from Paramecium tetraurelia. J. Biol. Chem. 1987, 262, 4602–4609. [Google Scholar] [CrossRef]
- Son, M.; Gundersen, R.E.; Nelson, D.L. A second member of the novel Ca2+-dependent protein kinase family from Paramecium tetraurelia. Purification and characterization. J. Biol. Chem. 1993, 268, 5940–5948. [Google Scholar] [CrossRef]
- Kim, K.; Son, M.; Peterson, J.B.; Nelson, D.L. Ca2+-binding proteins of cilia and infraciliary lattice of Paramecium tetraurelia: Their phosphorylation by purified endogenous Ca2+-dependent protein kinases. J. Cell Sci. 2002, 115, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Kissmehl, R.; Treptau, T.; Hauser, K.; Plattner, H. A novel, calcium-inhibitable casein kinase in Paramecium cells. FEBS Lett. 1997, 402, 227–235. [Google Scholar] [CrossRef]
- Creamer, T.P. Calcineurin. Cell Commun. Signal. 2020, 18, 137. [Google Scholar] [CrossRef]
- Momayezi, M.; Kissmehl, R.; Plattner, H. Quantitative immunogold localization of protein phosphatase 2B (calcineurin) in Paramecium cells. J. Histochem. Cytochem. 2000, 48, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Fraga, D.; Sehring, I.M.; Kissmehl, R.; Reiss, M.; Gaines, R.; Hinrichsen, R.; Plattner, H. Protein phosphatase 2B (PP2B, calcineurin) in Paramecium: Partial characterization reveals that two members of the unusually large catalytic subunit family have distinct roles in calcium-dependent processes. Eukaryot. Cell 2010, 9, 1049–1063. [Google Scholar] [CrossRef]
- Westerlund, A.M.; Delemotte, L. Effect of Ca2+ on the promiscuous target-protein binding of calmodulin. PLoS Comput. Biol. 2018, 14, e1006072. [Google Scholar] [CrossRef]
- Saimi, Y.; Ling, K.Y. Calmodulin activation of calcium-dependent sodium channels in excised membrane patches of Paramecium. Science 1990, 249, 1441–1444. [Google Scholar] [CrossRef]
- Aderem, A. The MARCKS family of protein kinase-C substrates. Biochem. Soc. Trans. 1995, 23, 587–591. [Google Scholar] [CrossRef]
- Aderem, A. The MARCKS brothers: A family of protein kinase C substrates. Cell 1992, 71, 713–716. [Google Scholar] [CrossRef]
- Blackshear, P.J. The MARCKS family of cellular protein kinase C substrates. J. Biol. Chem. 1993, 268, 1501–1504. [Google Scholar] [CrossRef]
- Hinrichsen, R.D.; Blackshear, P.J. Regulation of peptide-calmodulin complexes by protein kinase C in vivo. Proc. Natl. Acad. Sci. USA 1993, 90, 1585–1589. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.C.; Nelson, D.L. The cilia of Paramecium tetraurelia contain both Ca2+-dependent and Ca2+-inhibitable calmodulin-binding proteins. Biochem. J. 1989, 259, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.W.; Saimi, Y.; Kung, C. A new multigene family encoding calcium-dependent calmodulin-binding membrane proteins of Paramecium tetraurelia. Gene 1999, 231, 21–32. [Google Scholar] [CrossRef]
- Park, H.Y.; Kim, S.A.; Korlach, J.; Rhoades, E.; Kwok, L.W.; Zipfel, W.R.; Waxham, M.N.; Webb, W.W.; Pollack, L. Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer. Proc. Natl. Acad. Sci. USA 2008, 105, 542–547. [Google Scholar] [CrossRef]
- Schaefer, W.H.; Lukas, T.J.; Blair, I.A.; Schultz, J.E.; Watterson, D.M. Amino acid sequence of a novel calmodulin from Paramecium tetraurelia that contains dimethyllysine in the first domain. J. Biol. Chem. 1987, 262, 1025–1029. [Google Scholar] [CrossRef]
- Yazawa, M.; Yagi, K.; Toda, H.; Kondo, K.; Narita, K.; Yamazaki, R.; Sobue, K.; Kakiuchi, S.; Nagao, S.; Nozawa, Y. The amino acid sequence of the Tetrahymena calmodulin which specifically interacts with guanylate cyclase. Biochem. Biophys. Res. Commun. 1981, 99, 1051–1057. [Google Scholar] [CrossRef]
- Kink, J.A.; Maley, M.E.; Preston, R.R.; Ling, K.Y.; Wallen-Friedman, M.A.; Saimi, Y.; Kung, C. Mutations in Paramecium calmodulin indicate functional differences between the C-terminal and N-terminal lobes in vivo. Cell 1990, 62, 165–174. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chagoyen, M.; García-Martín, J.A.; Pazos, F. Practical analysis of specificity-determining residues in protein families. Brief Bioinform. 2016, 17, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Rausell, A.; Juan, D.; Pazos, F.; Valencia, A. Protein interactions and ligand binding: From protein subfamilies to functional specificity. Proc. Natl. Acad. Sci. USA 2010, 107, 1995–2000. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Egli, R.; Rhyner, J.A.; Hameister, H.; Strehler, E.E. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics 1993, 16, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Berchtold, M.W. Structure and expression of the chicken calmodulin I gene. Gene 1997, 194, 63–68, A correction has appeared in Gene. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Komm, B.S.; McDonnell, D.P.; Pike, J.W.; Haussler, M.R. Immunoselection of cDNAs to avian intestinal calcium binding protein 28K and a novel calmodulin-like protein: Assessment of mRNA regulation by the vitamin D hormone. Biochemistry 1987, 26, 8332–8338. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Dutta, S.; Pal, A.; Sengupta, M.; Chattopadhyay, S. Calmodulin7: Recent insights into emerging roles in plant development and stress. Plant Mol. Biol. 2021, 107, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, W.H.; Hinrichsen, R.D.; Burgess-Cassler, A.; Kung, C.; Blair, I.A.; Watterson, D.M. A mutant Paramecium with a defective calcium-dependent potassium conductance has an altered calmodulin: A nonlethal selective alteration in calmodulin regulation. Proc. Natl. Acad. Sci. USA 1987, 84, 3931–3935. [Google Scholar] [CrossRef]
- Hinrichsen, R.D.; Pollock, M.; Hennessey, T.; Russell, C. An intragenic suppressor of a calmodulin mutation in Paramecium: Genetic and biochemical characterization. Genetics 1991, 129, 717–725. [Google Scholar] [CrossRef]
- Han, C.H.; Roberts, D.M. Altered methylation substrate kinetics and calcium binding of a calmodulin with a Val136-->Thr substitution. Eur. J. Biochem. 1997, 244, 904–912. [Google Scholar] [CrossRef]
- Kuriu, T.; Oosawa, Y.; Watanabe, Y.; Nakaoka, Y. Defect of cold-sensitive response in calmodulin mutants of Paramecium and the restoration by cyclic nucleotide. Cell Struct. Funct. 1997, 22, 493–500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Preston, R.R.; Wallen-Friedman, M.A.; Saimi, Y.; Kung, C. Calmodulin defects cause the loss of Ca2+-dependent K+ currents in two pantophobiac mutants of Paramecium tetraurelia. J. Membr. Biol. 1990, 115, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Saimi, Y.; Hinrichsen, R.D.; Forte, M.; Kung, C. Mutant analysis shows that the Ca2+-induced K+ current shuts off one type of excitation in Paramecium. Proc. Natl. Acad. Sci. USA 1983, 80, 5112–5116. [Google Scholar] [CrossRef] [PubMed]
- Hinrichsen, R.D.; Fraga, D.; Reed, M.W. 3’-modified antisense oligodeoxyribonucleotides complementary to calmodulin mRNA alter behavioral responses in Paramecium. Proc. Natl. Acad. Sci. USA 1992, 89, 8601–8605. [Google Scholar] [CrossRef] [PubMed]
- Linse, S.; Helmersson, A.; Forsén, S. Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 1991, 266, 8050–8054. [Google Scholar] [CrossRef]
- Martin, S.R.; Andersson Teleman, A.; Bayley, P.M.; Drakenberg, T.; Forsen, S. Kinetics of calcium dissociation from calmodulin and its tryptic fragments. A stopped-flow fluorescence study using Quin 2 reveals a two-domain structure. Eur. J. Biochem. 1985, 151, 543–550. [Google Scholar] [CrossRef]
- Oh, S.H.; Roberts, D.M. Analysis of the state of posttranslational calmodulin methylation in developing pea plants. Plant Physiol. 1990, 93, 880–887. [Google Scholar] [CrossRef]
- Magnani, R.; Dirk, L.M.; Trievel, R.C.; Houtz, R.L. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 2010, 1, 43. [Google Scholar] [CrossRef]
- Pech, L.L.; Nelson, D.L. Purification and characterization of calmodulin (lysine 115) N-methyltransferase from Paramecium tetraurelia. Biochim. Biophys. Acta 1994, 1199, 183–194. [Google Scholar] [CrossRef]
- Babu, Y.S.; Sack, J.S.; Greenhough, T.J.; Bugg, C.E.; Means, A.R.; Cook, W.J. Three-dimensional structure of calmodulin. Nature 1985, 315, 37–40. [Google Scholar] [CrossRef]
- Babu, Y.S.; Bugg, C.E.; Cook, W.J. Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 1988, 204, 191–204. [Google Scholar] [CrossRef]
- Taylor, D.A.; Sack, J.S.; Maune, J.F.; Beckingham, K.; Quiocho, F.A. Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-Å resolution. J. Biol. Chem. 1991, 26, 21375–21380. [Google Scholar] [CrossRef]
- Chattopadhyaya, R.; Meador, W.E.; Means, A.R.; Quiocho, F.A. Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 1992, 228, 1177–1192. [Google Scholar] [CrossRef]
- Rao, S.T.; Wu, S.; Satyshur, K.A.; Ling, K.Y.; Kung, C.; Sundaralingam, M. Structure of Paramecium tetraurelia calmodulin at 1.8 Å resolution. Protein Sci. 1993, 2, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Ban, C.; Ramakrishnan, B.; Ling, K.Y.; Kung, C.; Sundaralingam, M. Structure of the recombinant Paramecium tetraurelia calmodulin at 1.68 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 50–63. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Prot. Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Jaren, O.R.; Kranz, J.K.; Sorensen, B.R.; Wand, A.J.; Shea, M.A. Calcium-induced conformational switching of Paramecium calmodulin provides evidence for domain coupling. Biochemistry 2002, 41, 14158–14166. [Google Scholar] [CrossRef]
- VanScyoc, W.S.; Shea, M.A. Phenylalanine fluorescence studies of calcium binding to N-domain fragments of Paramecium calmodulin mutants show increased calcium affinity correlates with increased disorder. Protein Sci. 2001, 10, 1758–1768. [Google Scholar] [CrossRef]
- Prescott, D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994, 58, 233–267. [Google Scholar] [CrossRef]
- Kink, J.A.; Maley, M.E.; Ling, K.Y.; Kanabrocki, J.A.; Kung, C. Efficient expression of the Paramecium calmodulin gene in Escherichia coli after four TAA-to-CAA changes through a series of polymerase chain reactions. J. Protozool. 1991, 38, 441–447. [Google Scholar] [CrossRef]
- Katoh, M.; Hirono, M.; Takemasa, T.; Kimura, M.; Watanabe, Y. A micronucleus-specific sequence exists in the 5’-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 1993, 21, 2409–2414. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davis, T.N.; Urdea, M.S.; Masiarz, F.R.; Thorner, J. Isolation of the yeast calmodulin gene: Calmodulin is an essential protein. Cell 1986, 4, 423–431. [Google Scholar] [CrossRef]
- Takeda, T.; Yamamoto, M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 1987, 84, 3580–3584. [Google Scholar] [CrossRef] [PubMed]
- Robson, K.J.; Jennings, M.W. The structure of the calmodulin gene of Plasmodium falciparum. Mol. Biochem. Parasitol. 1991, 46, 19–34. [Google Scholar] [CrossRef]
- Zimmer, W.E.; Schloss, J.A.; Silflow, C.D.; Youngblom, J.; Watterson, D.M. Structural organization, DNA sequence, and expression of the calmodulin gene. J. Biol. Chem. 1988, 263, 19370–19383. [Google Scholar] [CrossRef]
- Chung, S.H.; Swindle, J. Linkage of the calmodulin and ubiquitin loci in Trypanosoma cruzi. Nucleic Acids Res. 1990, 18, 4561–4569. [Google Scholar] [CrossRef]
- Tschudi, C.; Young, A.S.; Ruben, L.; Patton, C.L.; Richards, F.F. Calmodulin genes in trypanosomes are tandemly repeated and produce multiple mRNAs with a common 5’ leader sequence. Proc. Natl. Acad. Sci. USA 1985, 82, 3998–4002. [Google Scholar] [CrossRef]
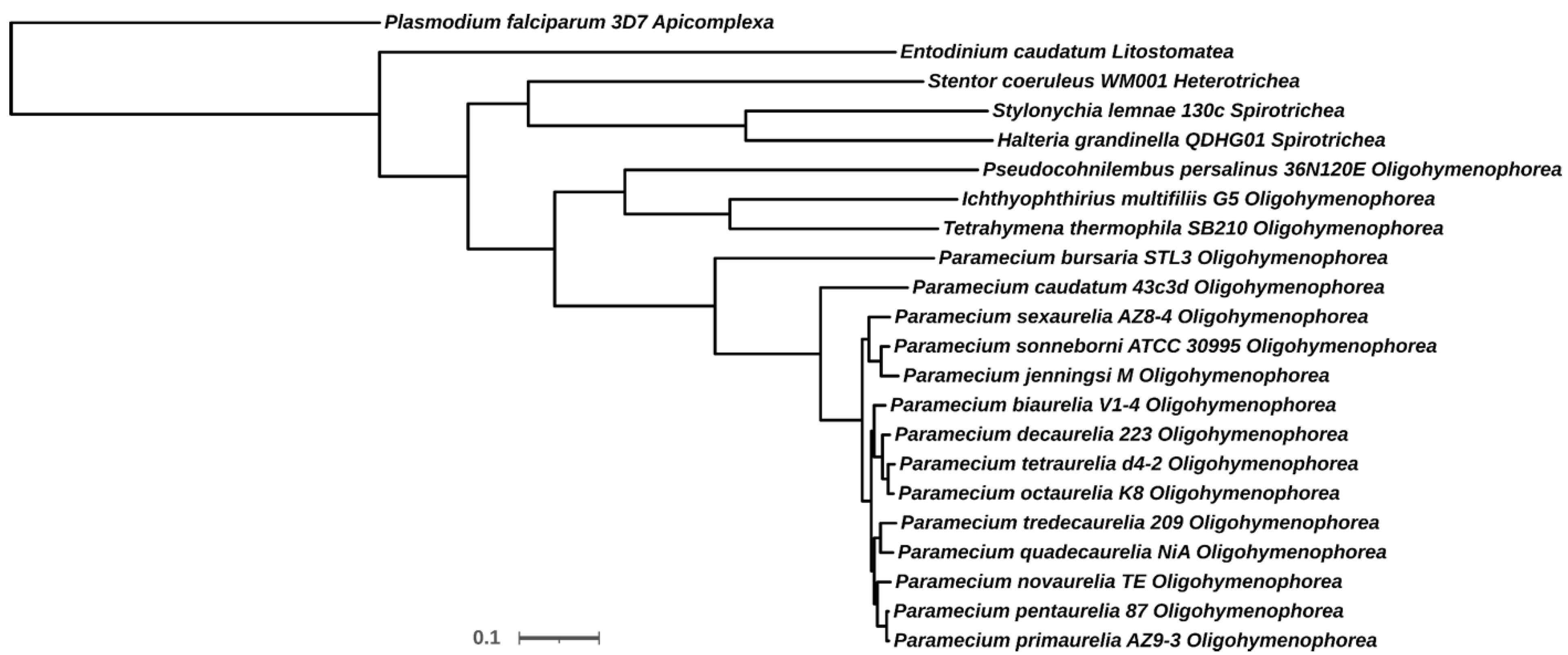

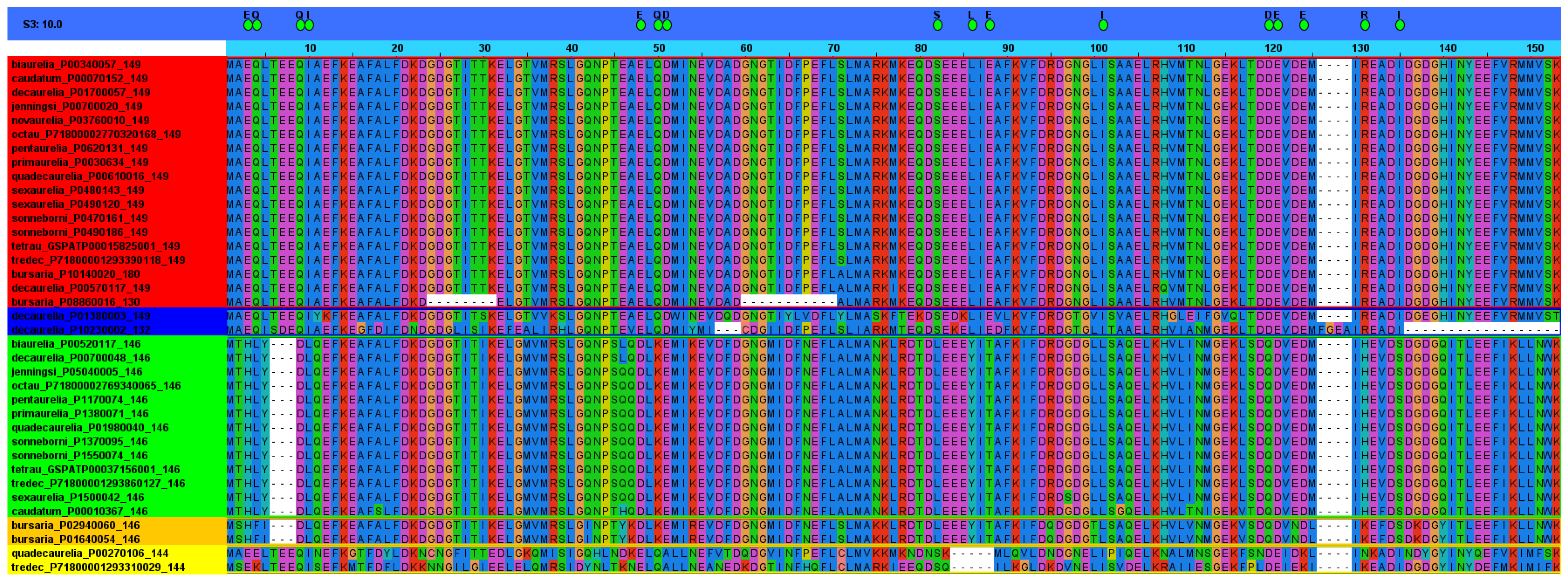

| Species, Class | Sequence IDs |
|---|---|
| Entodinium caudatum, Litostomatea | g10980, g11123, g16453, g28011, g28450, g40418, g47617, g55200 |
| Halteria grandinella QDHG01, Spirotrichea | TNV71997.1, TNV75227.1, TNV75230.1, TNV75245.1, TNV75536.1, TNV76174.1, TNV81842.1 |
| Ichthyophthirius multifiliis G5, Oligohymenophorea | EGR31776.1, EGR34608.1 |
| Paramecium biaurelia V1-4, Oligohymenophorea | P00340057, P00520117 |
| Paramecium bursaria STL3, Oligohymenophorea | P01640054, P02940060, P08860016, P10140020 |
| Paramecium caudatum 43c3d, Oligohymenophorea | P00010367, P00070152 |
| Paramecium decaurelia 223, Oligohymenophorea | P00570117, P00700048, P01380003, P01700057, P10230002 |
| Paramecium jenningsi M, Oligohymenophorea | P00700020, P05040005 |
| Paramecium novaurelia TE, Oligohymenophorea | P03760010 |
| Paramecium octaurelia K8, Oligohymenophorea | P71800002769340065, P71800002770320168 |
| Paramecium pentaurelia 87, Oligohymenophorea | P0620131, P1170074 |
| Paramecium primaurelia AZ9-3, Oligohymenophorea | P0030634, P1380071 |
| Paramecium quadecaurelia NiA, Oligohymenophorea | P00270106, P00610016, P01980040 |
| Paramecium sexaurelia AZ8-4, Oligohymenophorea | P0480143, P0490120, P1500042 |
| Paramecium sonneborni ATCC30995, Oligohymenophorea | P0470161, P0490186, P1370095, P1550074 |
| Paramecium tetraurelia d4-2, Oligohymenophorea | GSPATP00015825001, GSPATP00037156001 |
| Paramecium tredecaurelia 209, Oligohymenophorea | P71800001293310029, P71800001293390118, P71800001293860127 |
| Pseudocohnilembus persalinus 36N120E, Oligohymenophorea | KRX01518.1, KRX11001.1 |
| Stentor coeruleus WM001, Heterotrichea | OMJ67198.1, OMJ80243.1, OMJ88554.1 |
| Stylonychia lemnae 130c, Spirotrichea | CDW77243.1, CDW80161.1, CDW81015.1, CDW88706.1, CDW91107.1 |
| Tetrahymena thermophila SB210, Oligohymenophorea | EAS02529.2 |
| Plasmodium falciparum 3D7, Apicomplexa * | XP_001347585.1, XP_001348497.1 |
| Species, Class | TRUE | LIKE | TOTAL |
|---|---|---|---|
| Entodinium caudatum, Litostomatea | 0 | 8 | 8 |
| Halteria grandinella QDHG01, Spirotrichea | 1 | 6 | 7 |
| Ichthyophthirius multifiliis G5, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium biaurelia V1-4, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium bursaria STL3, Oligohymenophorea | 2 | 2 | 4 |
| Paramecium caudatum 43c3d, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium decaurelia 223, Oligohymenophorea | 4 | 1 | 5 |
| Paramecium jenningsi M, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium novaurelia TE, Oligohymenophorea | 1 | 0 | 1 |
| Paramecium octaurelia K8, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium pentaurelia 87, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium primaurelia AZ9-3, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium quadecaurelia NiA, Oligohymenophorea | 1 | 2 | 3 |
| Paramecium sexaurelia AZ8-4, Oligohymenophorea | 2 | 1 | 3 |
| Paramecium sonneborni ATCC30995, Oligohymenophorea | 2 | 2 | 4 |
| Paramecium tetraurelia d4-2, Oligohymenophorea | 1 | 1 | 2 |
| Paramecium tredecaurelia 209, Oligohymenophorea | 1 | 2 | 3 |
| Pseudocohnilembus persalinus 36N120E, Oligohymenophorea | 1 | 1 | 2 |
| Stentor coeruleus WM001, Heterotrichea | 2 | 1 | 3 |
| Stylonychia lemnae 130c, Spirotrichea | 1 | 4 | 5 |
| Tetrahymena thermophila SB210, Oligohymenophorea | 1 | 0 | 1 |
| Plasmodium falciparum 3D7, Apicomplexa * | 1 | 1 | 2 |
| Species | −4 | −3 | −2 | −1 | CALMODULIN | 1 | 2 | 3 |
|---|---|---|---|---|---|---|---|---|
| P. biaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P00340057 | OG0013227 | OG0001289 | OG0000584 |
| P. bursaria | OG0000460 | OG0034217 | OG0001284 | OG0022984 | P08860016 | OG0004706 | OG0034218 | OG0010485 |
| P. bursaria | OG0011926 | OG0034217 | OG0001284 | OG0022984 | P10140020 | OG0004706 | OG0034218 | OG0010485 |
| P. caudatum | OG0008990 | OG0004005 | OG0035853 | OG0035853 | P00070152 | OG0026817 | OG0005842 | OG0000048 |
| P. decaurelia | OG0005051 | OG0005051 | OG0006772 | OG0000494 | P00570117 | OG0017080 | OG0000016 | |
| P. decaurelia | OG0002384 | OG0018936 | OG0015720 | P01380003 | OG0000209 | OG0008678 | ||
| P. decaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P01700057 | OG0013227 | OG0001289 | OG0000584 |
| P. decaurelia | OG0003883 | OG0012438 | P10230002 | OG0013227 | OG0021818 | OG0000099 | ||
| P. jenningsi | OG0001678 | OG0007300 | P00700020 | OG0001289 | ||||
| P. novaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P03760010 | OG0013227 | OG0001289 | OG0000584 |
| P. octaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P71800002770320168 | OG0013227 | OG0001289 | |
| P. pentaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P0620131 | OG0013227 | OG0001289 | OG0000584 |
| P. primaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P0030634 | OG0013227 | OG0001289 | OG0000584 |
| P. quadecaurelia | OG0003656 | OG0001678 | OG0000201 | P00610016 | OG0013227 | OG0001289 | ||
| P. sexaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P0480143 | OG0000584 | OG0000014 | |
| P. sexaurelia | OG0014690 | OG0010650 | OG0011665 | OG0003706 | P0490120 | OG0013227 | OG0001289 | OG0000014 |
| P. sonneborni | OG0003656 | OG0001678 | OG0007300 | P0470161 | OG0001289 | OG0000584 | OG0000014 | |
| P. sonneborni | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P0490186 | OG0013227 | OG0001289 | OG0013227 |
| P. tetraurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | GSPATP00015825001 | OG0013227 | OG0001289 | OG0037287 |
| P. tredecaurelia | OG0003656 | OG0001678 | OG0007300 | OG0000201 | P71800001293390118 | OG0013227 | OG0001289 | OG0000584 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalobo, E.; Gutiérrez, G.; Villalobo, A. Calmodulin in Paramecium: Focus on Genomic Data. Microorganisms 2022, 10, 1915. https://doi.org/10.3390/microorganisms10101915
Villalobo E, Gutiérrez G, Villalobo A. Calmodulin in Paramecium: Focus on Genomic Data. Microorganisms. 2022; 10(10):1915. https://doi.org/10.3390/microorganisms10101915
Chicago/Turabian StyleVillalobo, Eduardo, Gabriel Gutiérrez, and Antonio Villalobo. 2022. "Calmodulin in Paramecium: Focus on Genomic Data" Microorganisms 10, no. 10: 1915. https://doi.org/10.3390/microorganisms10101915
APA StyleVillalobo, E., Gutiérrez, G., & Villalobo, A. (2022). Calmodulin in Paramecium: Focus on Genomic Data. Microorganisms, 10(10), 1915. https://doi.org/10.3390/microorganisms10101915





