Infection by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) Suppresses the Immune System of Hybrid Tilapia
Abstract
1. Introduction
2. Materials and Methods
2.1. Evaluating the Infectious Potential of the Fish Pond Water
2.2. Detection of Waterborne Actinospores, Filtering and DNA Extraction
2.3. PCR Analysis
2.4. Evaluating Percentage of Infected Fish
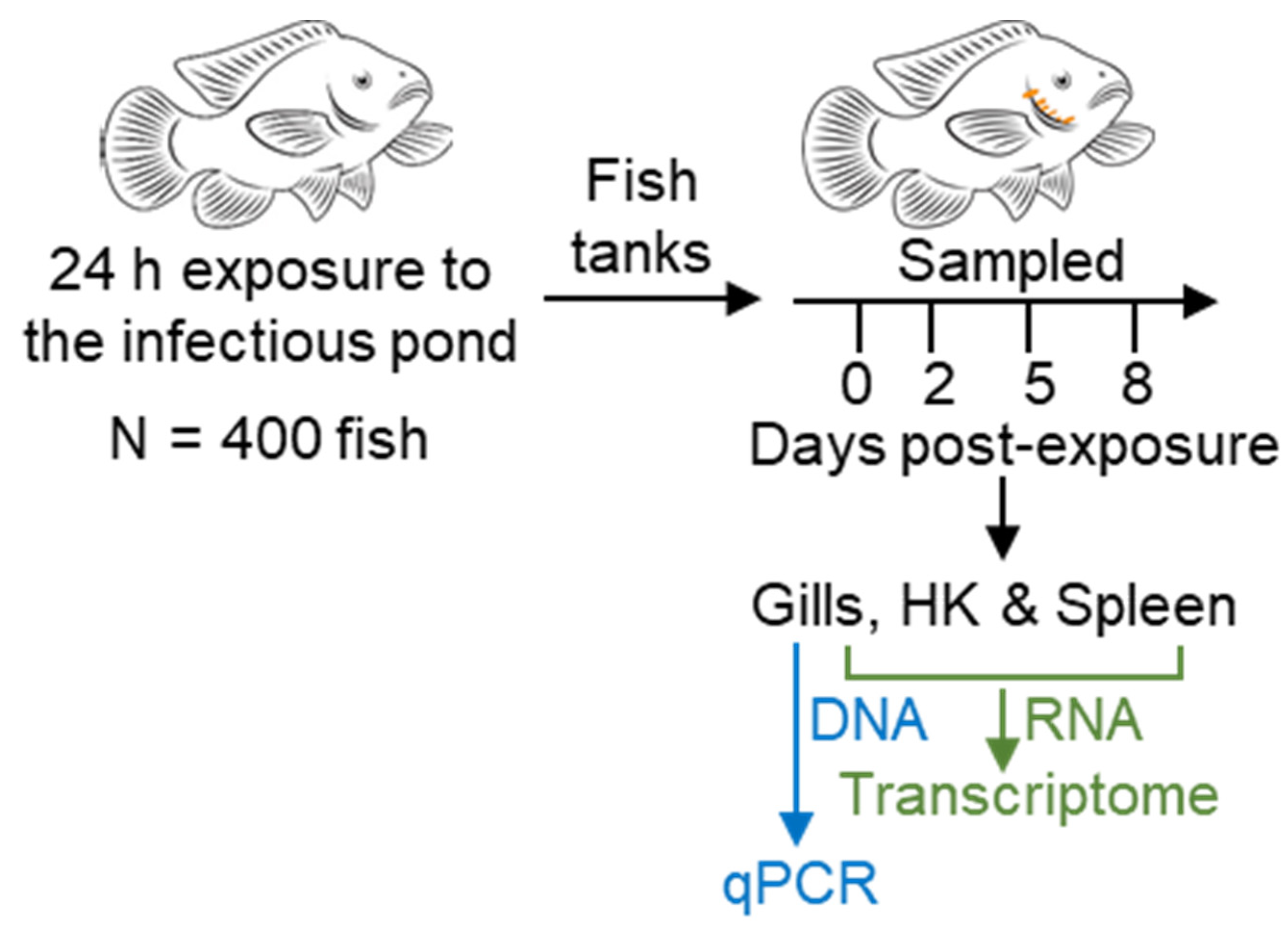
2.5. Transcriptome Experimental Design
2.6. RNA and DNA Extraction
2.7. Evaluation of Infection Severity by Quantitative RT-PCR
2.8. Library Preparation and Sequencing
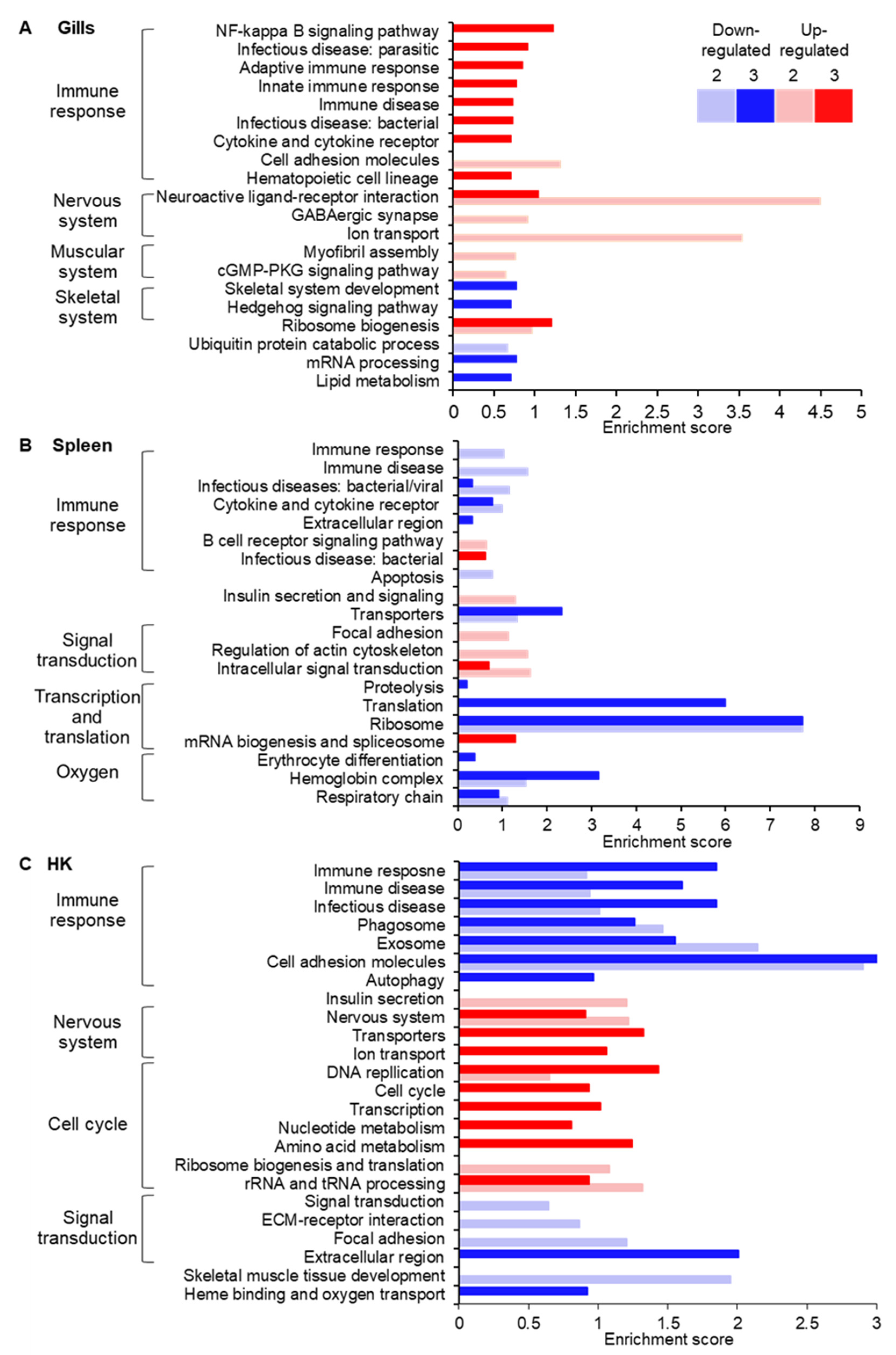
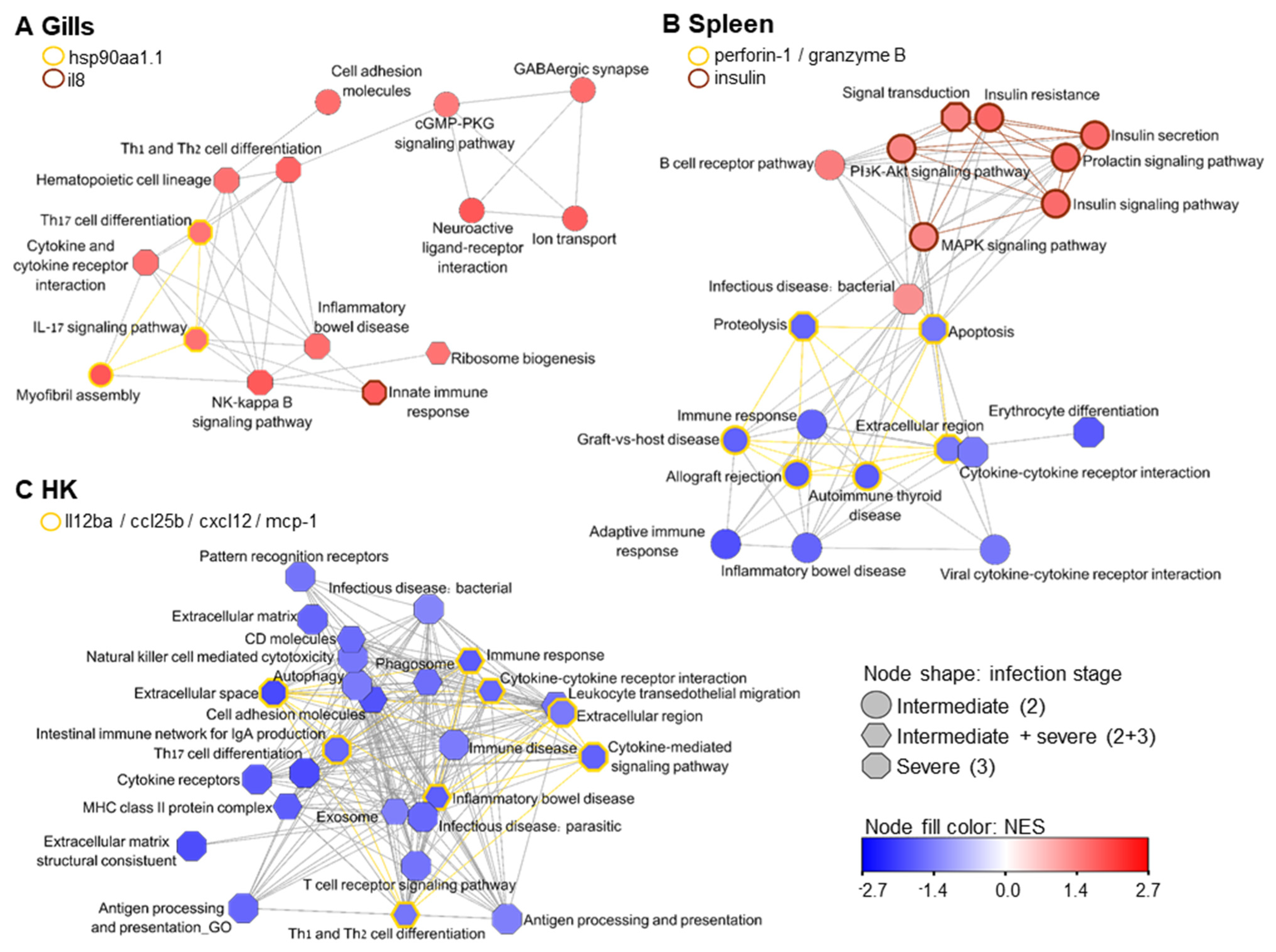
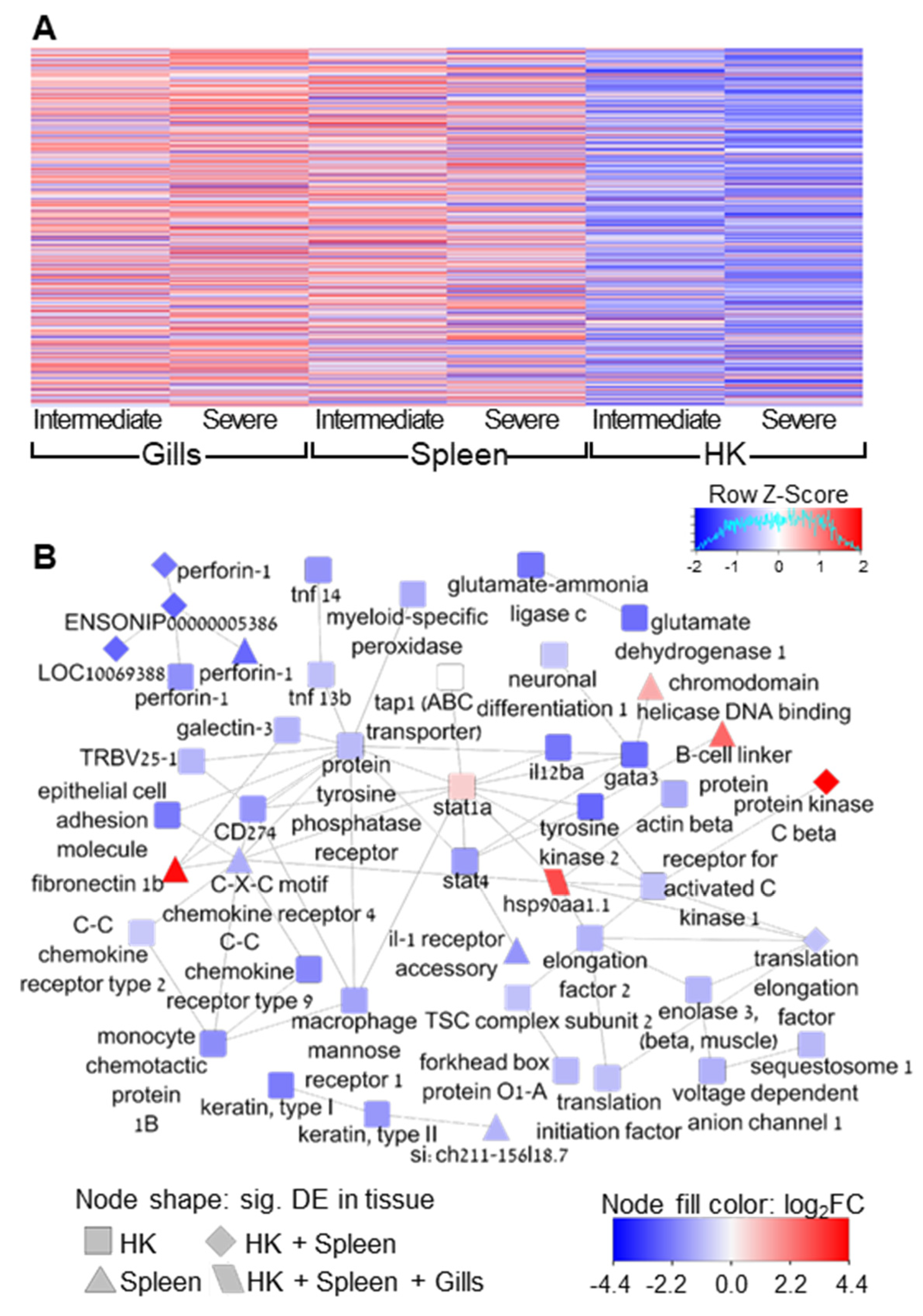
2.9. Quantification of Gene Expression and Enrichment Analysis
3. Results
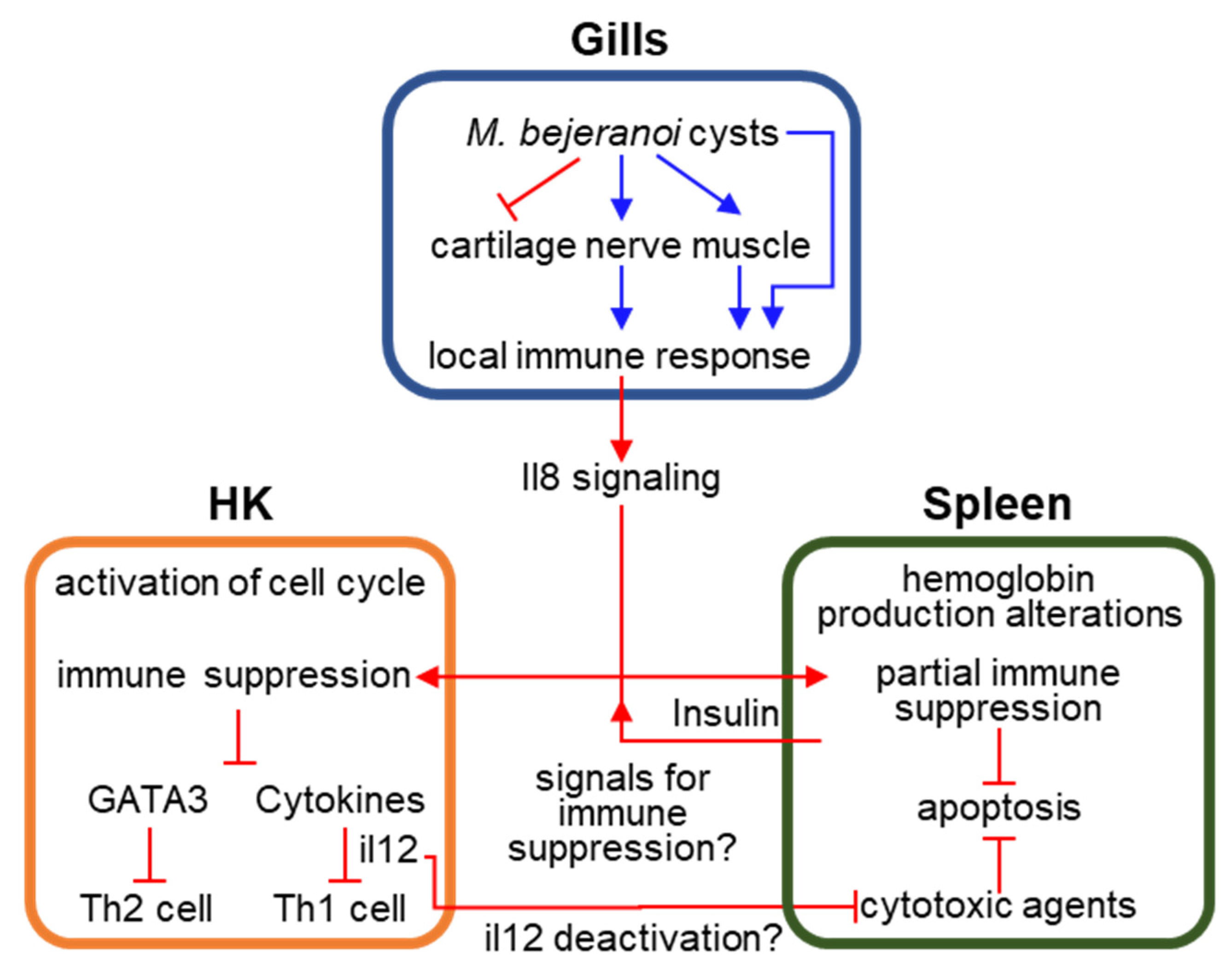
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Sayed, A.-F.M. (Ed.) Current state and future potential. In Tilapia Culture; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–20. [Google Scholar]
- Hulata, G. Tilapias-Biology and Culture; Ma’arechet Publishing House: Dalia, Israel, 2014. [Google Scholar]
- Eknath, A.E.; Hulata, G. Use and exchange of genetic resources of Nile tilapia (Oreochromis niloticus). Rev. Aquac. 2009, 1, 197–213. [Google Scholar] [CrossRef]
- Lövy, A.; Smirnov, M.; Brekhman, V.; Ofek, T.; Lotan, T. Morphological and molecular characterization of a novel myxosporean parasite Myxobolus bejeranoi n. sp. (Cnidaria: Myxosporea) from hybrid tilapia in Israel. Parasitol. Res. 2018, 117, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Okamura, B.; Gruhl, A.; Bartholomew, J.L. Myxozoan Evolution, Ecology and Development; Springer: Cham, Switzerland, 2015; Volume I, pp. 1–441. [Google Scholar]
- Atkinson, S.D.; Bartholomew, J.L.; Lotan, T. Myxozoans: Ancient metazoan parasites find a home in phylum Cnidaria. Zoology 2018, 129, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.S.; Neuhof, M.; Rubinstein, N.D.; Diamant, A.; Philippe, H.; Huchon, D.; Cartwright, P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. USA 2015, 112, 14912–14917. [Google Scholar] [CrossRef]
- Yang, Y.; Xiong, J.; Zhou, Z.; Huo, F.; Miao, W.; Ran, C.; Liu, Y.; Zhang, J.; Feng, J.; Wang, M.; et al. The Genome of the Myxosporean Thelohanellus kitauei Shows Adaptations to Nutrient Acquisition within Its Fish Host. Genome Biol. Evol. 2014, 6, 3182–3198. [Google Scholar] [CrossRef]
- Eszterbauer, E.; Atkinson, S.; Diamant, A.; Morris, D.; El-Matbouli, M.; Hartikainen, H. Myxozoan Life Cycles: Practical Approaches and Insights. In Myxozoan Evolution, Ecology and Development; Okamura, B., Gruhl, A.B.J., Eds.; Springer: Cham, Switzerland, 2015; pp. 175–198. [Google Scholar] [CrossRef]
- Holzer, A.S.; Bartošová-Sojková, P.; Born-Torrijos, A.; Lövy, A.; Hartigan, A.; Fiala, I. The joint evolution of the Myxozoa and their alternate hosts: A cnidarian recipe for success and vast biodiversity. Mol. Ecol. 2018, 27, 1651–1666. [Google Scholar] [CrossRef]
- Wolf, K.; Markiw, M.E. Biology Contravenes Taxonomy in the Myxozoa: New Discoveries Show Alternation of Invertebrate and Vertebrate Hosts. Science 1984, 225, 1449–1452. [Google Scholar] [CrossRef]
- Kent, M.L.; Margolis, L.; Corliss, J.O. The demise of a class of protists: Taxonomic and nomenclatural revisions proposed for the protist phylum Myxozoa Grassé, 1970. Can. J. Zool. 1994, 72, 932–937. [Google Scholar] [CrossRef]
- Holzer, A.S.; Piazzon, M.C.; Barrett, D.; Bartholomew, J.L.; Sitjà-Bobadilla, A. To React or Not to React: The Dilemma of Fish Immune Systems Facing Myxozoan Infections. Front. Immunol. 2021, 12, 734238. [Google Scholar] [CrossRef]
- Buchmann, K.; Secombes, C.J. Principles of Fish Immunology, From Host Cells and Molecules to Host Protection; Buchmann, K., Secombes, C.J., Eds.; Springer: Cham, Switzerland, 2021; ISBN 9783030854195. [Google Scholar]
- Tort, L.; Balasch, J.C.; Mackenzie, S. Fish immune system. A crossroads between innate and adaptive responses. Inmunología 2003, 22, 277–286. [Google Scholar]
- Zhu, L.-Y.; Nie, L.; Zhu, G.; Xiang, L.-X.; Shao, J.-Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Schmidt-posthaus, H.; Wahli, T.; Holland, J.W.; Secombes, C.J. Fish immune responses to Myxozoa. In Myxozoan Evolution, Ecology and Development; Okamura, B., Gruhl, A., Bartholome, J.L., Eds.; Springer: Cham, Switzerland, 2015; pp. 253–280. ISBN 9783319147536. [Google Scholar]
- Bjørgen, H.; Koppang, E.O. Anatomy of teleost fish immune structures and organs. In Principles of Fish Immunology, from Host Cells and Molecules to Host Protection; Buchmann, K., Secombes, C.J., Eds.; Springer: Cham, Switzerland, 2021; pp. 1–30. [Google Scholar]
- Estensoro, I.; Jung-Schroers, V.; Álvarez-Pellitero, P.; Steinhagen, D.; Sitjà-Bobadilla, A. Effects of Enteromyxum leei (Myxozoa) infection on gilthead sea bream (Sparus aurata) (Teleostei) intestinal mucus: Glycoprotein profile and bacterial adhesion. Parasitol. Res. 2013, 112, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-H.; Ma, A.-J.; Wang, X.-A. The immune response of turbot, Scophthalmus maximus (L.), skin to high water temperature. J. Fish Dis. 2011, 34, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Piazzon, M.C.; Galindo-Villegas, J.; Pereiro, P.; Estensoro, I.; Calduch-Giner, J.A.; Gómez-Casado, E.; Novoa, B.; Mulero, V.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. Differential Modulation of IgT and IgM upon Parasitic, Bacterial, Viral, and Dietary Challenges in a Perciform Fish. Front. Immunol. 2016, 7, 637. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Calduch-Giner, J.; Saera-Vila, A.; Palenzuela, O.; Álvarez-Pellitero, P.; Pérez-Sánchez, J. Chronic exposure to the parasite Enteromyxum leei (Myxozoa: Myxosporea) modulates the immune response and the expression of growth, redox and immune relevant genes in gilthead sea bream, Sparus aurata L. Fish Shellfish. Immunol. 2008, 24, 610–619. [Google Scholar] [CrossRef]
- Sitjà-Bobadilla, A.; Redondo, M.J.; Bermúdez, R.; Palenzuela, O.; Ferreiro, I.; Riaza, A.; Quiroga, I.; Nieto, J.M.; Alvarez-Pellitero, P. Innate and adaptive immune responses of turbot, Scophthalmus maximus (L.), following experimental infection with Enteromyxum scophthalmi (Myxosporea: Myxozoa). Fish Shellfish. Immunol. 2006, 21, 485–500. [Google Scholar] [CrossRef]
- Barrett, D.E.; Estensoro, I.; Sitjà-Bobadilla, A.; Bartholomew, J.L. Intestinal Transcriptomic and Histologic Profiling Reveals Tissue Repair Mechanisms Underlying Resistance to the Parasite Ceratonova shasta. Pathogens 2021, 10, 1179. [Google Scholar] [CrossRef]
- Cuesta, A.; Salinas, I.; Rodríguez, A.; Muñoz, P.; Sitjà-Bobadilla, A.; Álvarez-Pellitero, P.; Meseguer, J.; Esteban, M. Cell-mediated cytotoxicity is the main innate immune mechanism involved in the cellular defence of gilthead seabream (Teleostei: Sparidae) against Enteromyxum leei (Myxozoa). Parasite Immunol. 2006, 28, 657–665. [Google Scholar] [CrossRef]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef]
- Balouet, G.; Laurencin, F.B. Granulomatous nodules in fish: An experimental assessment in rainbow trout, Salmo gairdneri Richardson, and turbot, Scophthalmus maximus (L.). J. Fish Dis. 1986, 9, 417–429. [Google Scholar] [CrossRef]
- Massana, R.; Murray, A.E.; Preston, C.M.; DeLong, E.F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 1997, 63, 50–56. [Google Scholar] [CrossRef]
- Hadziavdic, K.; Lekang, K.; Lanzén, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA Gene for Designing Universal Eukaryote Specific Primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef]
- Holzer, A.S.; Sommerville, C.; Wootten, R. Molecular relationships and phylogeny in a community of myxosporeans and actinosporeans based on their 18S rDNA sequences. Int. J. Parasitol. 2004, 34, 1099–1111. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Xu, Q.; Yan, J.; Yu, F.; Wang, F.; Xiao, J.; Luo, Y.; Zhong, H. Nonadditive and allele-specific expression of insulin-like growth factor 1 in Nile tilapia (Oreochromis niloticus, ♀) × blue tilapia (O. aureus, ♂) hybrids. Comp. Biochem. Physiol. Part B—Biochem. Mol. Biol. 2019, 232, 93–100. [Google Scholar] [CrossRef]
- Hashimshony, T.; Senderovich, N.; Avital, G.; Klochendler, A.; de Leeuw, Y.; Anavy, L.; Gennert, D.; Li, S.; Livak, K.J.; Rozenblatt-Rosen, O.; et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016, 17, 77. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Khomtchouk, B.B.; Hennessy, J.R.; Wahlestedt, C. shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics. PLoS ONE 2017, 12, e0176334. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yazdanbakhsh, M.; Sacks, D.L. Why does immunity to parasites take so long to develop? Nat. Rev. Immunol. 2010, 10, 80–81. [Google Scholar] [CrossRef]
- Maizels, R.M. Parasite immunomodulation and polymorphisms of the immune system. J. Biol. 2009, 8, 62–64. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Immune defence, parasite evasion strategies and their relevance for ‘macroscopic phenomena’ such as virulence. Phil. Trans. R. Soc. B 2009, 364, 85–98. [Google Scholar] [CrossRef]
- Sudhagar, A.; Ertl, R.; Kumar, G.; El-Matbouli, M. Transcriptome profiling of posterior kidney of brown trout, Salmo trutta, during proliferative kidney disease. Parasites Vectors 2019, 12, 569. [Google Scholar] [CrossRef]
- Zhao, Y.; Weng, M.; Zhang, Q.; Li, A.; Zhang, J. Transcriptomics analysis of the infected tissue of gibel carp (Carassius auratus gibelio) with liver myxobolosis infers the underlying defense mechanisms from the perspective of immune-metabolic interactions. Aquaculture 2021, 542, 736867. [Google Scholar] [CrossRef]
- Chilmonczyk, S.; Monge, D.; De Kinkelin, P. Proliferative kidney disease: Cellular aspects of the rainbow trout, Oncorhynchus mykiss (Walbaum), response to parasitic infection. J. Fish Dis. 2002, 25, 217–226. [Google Scholar] [CrossRef]
- Davey, G.C.; Calduch-Giner, J.A.; Houeix, B.; Talbot, A.; Sitjà-Bobadilla, A.; Prunet, P.; Pérez-Sánchez, J.; Cairns, M.T. Molecular profiling of the gilthead sea bream (Sparus aurata L.) response to chronic exposure to the myxosporean parasite Enteromyxum leei. Mol. Immunol. 2011, 48, 2102–2112. [Google Scholar] [CrossRef] [PubMed]
- Gorgoglione, B.; Wang, T.; Secombes, C.J.; Holland, J.W. Immune gene expression profiling of Proliferative Kidney Disease in rainbow trout Oncorhynchus mykiss reveals a dominance of anti-inflammatory, antibody and T helper cell-like activities. Vet. Res. 2013, 44, 55. [Google Scholar] [CrossRef]
- Barrett, D.E.; Bartholomew, J.L. A tale of two fish: Comparative transcriptomics of resistant and susceptible steelhead following exposure to Ceratonova shasta highlights differences in parasite recognition. PLoS ONE 2020, 16, e0234837. [Google Scholar] [CrossRef]
- James, E.R.; Green, D.R. Manipulation of apoptosis in the host–parasite interaction. Trends Parasitol. 2004, 20, 280–287. [Google Scholar] [CrossRef]
- Robledo, D.; Ronza, P.; Harrison, P.W.; Losada, A.P.; Bermúdez, R.; Pardo, B.G.; Redondo, M.J.; Sitjà-Bobadilla, A.; Quiroga, M.I.; Martínez, P. RNA-seq analysis reveals significant transcriptome changes in turbot (Scophthalmus maximus) suffering severe enteromyxosis. BMC Genom. 2014, 15, 1149. [Google Scholar] [CrossRef]
- Wang, L.G.; Cheng, M.; Wen, X.; Xian, M.; Xia, H.; Nie, P. Molecular cloning, biological effect, and tissue distribution of interleukin-8 protein in mandarin fish (Siniperca chuasti) upon Flavobacterium columnare infection. Fish Shellfish. Immunol. 2017, 66, 112–119. [Google Scholar] [CrossRef]
- Wang, T.-T.; Song, X.-H.; Bao, G.-M.; Zhao, L.-X.; Yu, X.; Zhao, J. Molecular characterization, expression analysis, and biological effects of interleukin-8 in grass carp Ctenopharyngodon idellus. Fish Shellfish Immunol. 2013, 35, 1421–1432. [Google Scholar] [CrossRef]
- Chen, L.; He, C.; Baoprasertkul, P.; Xu, P.; Li, P.; Serapion, J.; Waldbieser, G.; Wolters, W.; Liu, Z. Analysis of a catfish gene resembling interleukin-8: cDNA cloning, gene structure, and expression after infection with Edwardsiella ictaluri. Dev. Comp. Immunol. 2005, 29, 135–142. [Google Scholar] [CrossRef]
- Caruso, M.A.; Sheridan, M.A. New insights into the signaling system and function of insulin in fish. Gen. Comp. Endocrinol. 2011, 173, 227–247. [Google Scholar] [CrossRef]
- Raadsen, S.A.; Yang, S.; Spaink, H. Hyperinsulinemia induces insulin resistance and immune suppression via Ptpn6 / Shp1 in zebrafish. J. Endocrinol. 2014, 222, 229–241. [Google Scholar]
- Yada, T.; Nakanishi, T. Interaction between endocrine and immune systems in fish. Int. Rev. Cytol. 2002, 220, 35–92. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, W.; Zhong, H.; Li, J.; Li, H.; He, F.; Zhou, J.; Tang, Y.; Yu, J.; Yu, F. Transcriptome analysis reveals that insulin is an immunomodulatory hormone in common carp. Fish Shellfish Immunol. 2016, 59, 213–219. [Google Scholar] [CrossRef]
- Vortkamp, A.; Lee, K.; Lanske, B.; Segre, G.V.; Kronenberg, H.M.; Tabin, C.J. Regulation of Rate of Cartilage Differentiation by Indian Hedgehog and PTH-Related Protein. Science 1996, 273, 613–622. [Google Scholar] [CrossRef]
- Lanske, B.; Karaplis, A.C.; Lee, K.; Luz, A.; Vortkamp, A.; Pirro, A.; Karperien, M.; Defize, L.H.K.; Ho, C.; Mulligan, R.C.; et al. PTH/PTHrP Receptor in Early Development and Indian Hedgehog—Regulated Bone Growth. Science 1996, 273, 663–666. [Google Scholar] [CrossRef]
- Koudijs, M.J.; Broeder, M.J.D.; Keijser, A.; Wienholds, E.; Houwing, S.; van Rooijen, E.M.H.C.; Geisler, R.; van Eeden, F.J.M. The Zebrafish Mutants dre, uki, and lep Encode Negative Regulators of the Hedgehog Signaling Pathway. PLoS Genet. 2005, 1, e19. [Google Scholar] [CrossRef]
- Cho, S.-W.; van Rijssel, J.C.; Witte, F.; de Bakker, M.A.; Richardson, M.K. The sonic hedgehog signaling pathway and the development of pharyngeal arch Derivatives in Haplochromis piceatus, a Lake Victoria cichlid. J. Oral Biosci. 2015, 57, 148–156. [Google Scholar] [CrossRef]
- Schwend, T.; Ahlgren, S.C. Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development. BMC Dev. Biol. 2009, 9, 59. [Google Scholar] [CrossRef]
- Cui, J.; Liu, S.; Zhang, B.; Wang, H.; Sun, H.; Song, S.; Qiu, X.; Liu, Y.; Wang, X.; Jiang, Z.; et al. Transciptome Analysis of the Gill and Swimbladder of Takifugu rubripes by RNA-Seq. PLoS ONE 2014, 9, e85505. [Google Scholar] [CrossRef]
- Zapata, A.; Diez, B.; Cejalvo, T.; Frías, C.G.-D.; Cortés, A. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 2006, 20, 126–136. [Google Scholar] [CrossRef]
- Tang, M.; Chen, Y.; Xian, H.; Tan, S.; Lian, Z.; Peng, X.; Hu, D. Circulating exosome level of indigenous fish may be a novel biomarker for the integrated ecotoxicity effect of water environment. Ecotoxicol. Environ. Saf. 2022, 229, 113084. [Google Scholar] [CrossRef]
- Iliev, D.B.; Jørgensen, S.M.; Rode, M.; Krasnov, A.; Harneshaug, I.; Jørgensen, J.B. CpG-induced secretion of MHCIIβ and exosomes from salmon (Salmo salar) APCs. Dev. Comp. Immunol. 2010, 34, 29–41. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Pan, J.; Zhu, M.; Liang, Z.; Shen, Z.; Dai, K.; Yan, B.; Dai, Y.; Xue, R.; et al. Proteomic analysis of the exosomes secreted from Ctenopharyngodon idellus kidney cells infected with grass carp reovirus reveals their involvement in the cellular responses to viral infection. Fish Physiol. Biochem. 2021, 47, 857–867. [Google Scholar] [CrossRef]
- He, J.; Chen, N.-N.; Li, Z.-M.; Wang, Y.-Y.; Weng, S.-P.; Guo, C.-J.; He, J.-G. Evidence for a Novel Antiviral Mechanism of Teleost Fish: Serum-Derived Exosomes Inhibit Virus Replication through Incorporating Mx1 Protein. Int. J. Mol. Sci. 2021, 22, 10346. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Mladineo, I.; Naya-Català, F.; Dirks, R.P.; Jong-Raadsen, S.; Vrbatović, A.; Hrabar, J.; Pérez-Sánchez, J.; Sitjà-Bobadilla, A. Acting locally-affecting globally: RNA sequencing of gilthead sea bream with a mild Sparicotyle chrysophrii infection reveals effects on apoptosis, immune and hypoxia related genes. BMC Genom. 2019, 20, 200. [Google Scholar] [CrossRef]
- Bailey, C.; von Siebenthal, E.W.; Rehberger, K.; Segner, H. Transcriptomic analysis of the impacts of ethinylestradiol (EE2) and its consequences for proliferative kidney disease outcome in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 222, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Secombes, C.J. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 2013, 35, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Araki, K.; Hayashi, K.; Takeuchi, Y.; Shiozaki, K.; Suetake, H.; Yamamoto, A. Adjuvant effect of recombinant interleukin-12 in the Nocardiosis formalin-killed vaccine of the amberjack Seriola dumerili. Fish Shellfish Immunol. 2017, 67, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Syahputra, K.; Kania, P.W.; Al-Jubury, A.; Jafaar, R.M.; Dirks, R.P.; Buchmann, K. Transcriptomic analysis of immunity in rainbow trout (Oncorhynchus mykiss) gills infected by Ichthyophthirius multifiliis. Fish Shellfish Immunol. 2019, 86, 486–496. [Google Scholar] [CrossRef]
- Martin, S.; Djordjevic, B.; Stanko, S. Modulation of splenic immune responses to bacterial lipopolysaccharide in rainbow trout (Oncorhynchus mykiss) fed lentinan, a beta-glucan from mushroom Lentinula edodes. Fish Shellfish Immunol. 2009, 26, 201–209. [Google Scholar]
- Galindo-villegas, J.; Mulero, I.; García-alcazar, A.; Muñoz, I.; Peñalver-mellado, M.; Streitenberger, S.; Scapigliati, G.; Meseguer, J.; Mulero, V. Recombinant TNF a as oral vaccine adjuvant protects European sea bass against vibriosis: Insights into the role of the CCL25 / CCR9 axis. Fish Shellfish Immunol. 2013, 35, 1260–1271. [Google Scholar] [CrossRef]
- Lu, I.-N.; Chiang, B.-L.; Lou, K.-L.; Huang, P.-T.; Yao, C.-C.J.; Wang, J.-S.; Lin, L.-D.; Jeng, J.-H.; Chang, B.-E. Cloning, expression and characterization of CCL21 and CCL25 chemokines in zebrafish. Dev. Comp. Immunol. 2012, 38, 203–214. [Google Scholar] [CrossRef]
- Aquilino, C.; Granja, A.G.; Castro, R.; Wang, T.; Abos, B.; Parra, D.; Secombes, C.J.; Tafalla, C. Rainbow trout CK9, a CCL25-like ancient chemokine that attracts and regulates B cells and macrophages, the main antigen presenting cells in fish. Oncotarget 2016, 7, 17547–17564. [Google Scholar] [CrossRef][Green Version]
- Zhou, S.; Liu, Y.; Dong, J.; Yang, Q.; Xu, N.; Yang, Y.; Gu, Z.; Ai, X. Transcriptome analysis of goldfish (Carassius auratus) in response to Gyrodactylus kobayashii infection. Parasit. Res. 2021, 120, 161–171. [Google Scholar] [CrossRef]
- Bai, H.; Zhou, T.; Zhao, J.; Chen, B.; Pu, F.; Bai, Y.; Wu, Y.; Chen, L.; Shi, Y.; Ke, Q.; et al. Transcriptome analysis reveals the temporal gene expression patterns in skin of large yellow croaker (Larimichthys Crocea) in response to Cryptocaryon irritans infection. Fish Shellfish Immunol. 2020, 99, 462–472. [Google Scholar] [CrossRef]
- Zhang, T.; Yuan, R.; Zhang, X.; Liu, B.; Zhou, Y.; Liang, Z.; Zhu, M. De novo RNA-seq assembly and di ff erential transcriptome analysis of Carassius auratus gibelio after Cyprinid herpesvirus 2 infection. Aquaculture 2020, 519, 734923. [Google Scholar] [CrossRef]
- Korytář, T. Functional and Molecular Investigations of the Disease Resistance in Rainbow Trout Using the Peritoneal Model of Inflammation. 2012. Available online: https://epub.ub.uni-greifswald.de/frontdoor/deliver/index/docId/1109/file/Dissertation_Tomas_Korytar_Functional_and_Molecular_Investigations_of_the_Disease_Resistance_in_Rainbow_Trout_Using_the_Peritoneal_Model_of_Inflammation.pdf (accessed on 10 April 2022).
- Ma, Q.; Jones, D.; Borghesani, P.R.; Segal, R.A.; Nagasawa, T.; Kishimoto, T.; Bronson, R.T.; Springer, T.A. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9448–9453. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.; Sato, H.; Zhang, Q.; Li, A.; Zhang, J. RNA-seq analysis of local tissue of arassius auratus gibelio with pharyngeal myxobolosis: Insights into the pharyngeal mucosal immune response in a fish-parasite dialogue. Fish Shellfish Immunol. 2019, 94, 99–112. [Google Scholar] [CrossRef]
- Yu, L.; Li, C.-H.; Chen, J. A novel CC chemokine ligand 2 like gene from ayu Plecoglossus altivelis is involved in the innate immune response against to Vibrio anguillarum. Fish Shellfish Immunol. 2019, 87, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Shilin, M.B.; Apalikova, O.V.; Lukina, J.N.; Golotin, V.A.; Li, J.; Zhang, J. The Anti-Infective Immune Mechanism of the CCL2 and CCL3 Chemokines in the Large Yellow Croaker (Larimichthys crocea). J. Appl. Ichthyol. 2021, 37, 916–924. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Estensoro, I.; Calduch-Giner, J.A.; Pozo, R.; Picard-Sánchez, A.; Pérez-Sánchez, J.; Sitjà-Bobadilla, A. Hints on T cell responses in a fish-parasite model: Enteromyxum leei induces differential expression of T cell signature molecules depending on the organ and the infection status. Parasites Vectors 2018, 11, 443. [Google Scholar] [CrossRef] [PubMed]
- Abós, B.; Bailey, C.; Tafalla, C. Adaptive Immunity. In Principles of Fish Immunology, from Cells and Molecules to Host Protection; Buchmann, K., Secombes, C.J., Eds.; Springer: Cham, Switzerland, 2021; pp. 105–140. [Google Scholar]
- Kobayashi, M.; Fitz, L.; Ryan, M.; Hewick, R.M.; Clark, S.C.; Chan, S.; Loudon, R.; Sherman, F.; Perussia, B.; Trinchieri, G. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 1989, 170, 827–845. [Google Scholar] [CrossRef]
- Walker, J.A.; McKenzie, A.N.J. TH2 cell development and function. Nat. Rev. Immunol. 2017, 18, 121–133. [Google Scholar] [CrossRef]
- Bailey, C.; Strepparava, N.; Wahli, T.; Segner, H. Exploring the immune response, tolerance and resistance in proliferative kidney disease of salmonids. Dev. Comp. Immunol. 2019, 90, 165–175. [Google Scholar] [CrossRef]
- Wang, T.; Holland, J.W.; Martin, S.A.M.; Secombes, C.J. Sequence and expression analysis of two T helper master transcription factors, T-bet and GATA3, in rainbow trout Oncorhynchus mykiss and analysis of their expression during bacterial and parasitic infection. Fish Shellfish Immunol. 2010, 29, 705–715. [Google Scholar] [CrossRef]
- Patil, H.J.; Gatica, J.; Zolti, A.; Benet-Perelberg, A.; Naor, A.; Dror, B.; Al Ashhab, A.; Marman, S.; Hasan, N.A.; Colwell, R.R.; et al. Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment. Microorganisms 2020, 8, 1984. [Google Scholar] [CrossRef]
- Patil, H.J.; Benet-Perelberg, A.; Naor, A.; Smirnov, M.; Ofek, T.; Nasser, A.; Minz, D.; Cytryn, E. Evidence of Increased Antibiotic Resistance in Phylogenetically-diverse Aeromonas Isolates from Semi-Intensive Fish Ponds Treated with Antibiotics. Front. Microbiol. 2016, 7, 1875. [Google Scholar] [CrossRef]
- Surachetpong, W.; Rajiv, S.; Roy, K.; Nicholson, P. Tilapia lake virus: The story so far. J. Fish Dis. 2020, 43, 1115–1132. [Google Scholar] [CrossRef]
- Gorgoglione, B.; Taylor, N.G.; Holland, J.W.; Feist, S.W.; Secombes, C.J. Immune response modulation upon sequential heterogeneous co-infection with Tetracapsuloides bryosalmonae and VHSV in brown trout (Salmo trutta). Fish Shellfish Immunol. 2019, 88, 375–390. [Google Scholar] [CrossRef]
- Kotob, M.H.; Menanteau-Ledouble, S.; Kumar, G.; Abdelzaher, M.; El-Matbouli, M. The impact of co-infections on fish: A review. Vet. Res. 2016, 47, 98. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maor-Landaw, K.; Smirnov, M.; Brekhman, V.; Ofek-Lalzar, M.; Yahav, T.; Lotan, T. Infection by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) Suppresses the Immune System of Hybrid Tilapia. Microorganisms 2022, 10, 1893. https://doi.org/10.3390/microorganisms10101893
Maor-Landaw K, Smirnov M, Brekhman V, Ofek-Lalzar M, Yahav T, Lotan T. Infection by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) Suppresses the Immune System of Hybrid Tilapia. Microorganisms. 2022; 10(10):1893. https://doi.org/10.3390/microorganisms10101893
Chicago/Turabian StyleMaor-Landaw, Keren, Margarita Smirnov, Vera Brekhman, Maya Ofek-Lalzar, Tal Yahav, and Tamar Lotan. 2022. "Infection by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) Suppresses the Immune System of Hybrid Tilapia" Microorganisms 10, no. 10: 1893. https://doi.org/10.3390/microorganisms10101893
APA StyleMaor-Landaw, K., Smirnov, M., Brekhman, V., Ofek-Lalzar, M., Yahav, T., & Lotan, T. (2022). Infection by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) Suppresses the Immune System of Hybrid Tilapia. Microorganisms, 10(10), 1893. https://doi.org/10.3390/microorganisms10101893





