Biosurfactant Production by Bacillus amyloliquefaciens C11 and Streptomyces lavendulae C27 Isolated from a Biopurification System for Environmental Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Isolation and Selection
2.2. Selection of Biosurfactant-Producing Bacteria
2.3. Strains Identification
2.3.1. Bacterial Identification by Molecular Biology
2.3.2. Bacterial Identification via MALDI-TOF/TOF MS
2.4. Biosurfactant Production at Bench Scale
2.5. Biosurfactant Extract Preparation and Characterization
2.6. Analyses
2.6.1. Drop-Collapse
2.6.2. Oil Displacement Test
2.6.3. Superficial Tension
2.6.4. Biosurfactant Crude Extract Characterization
2.7. Statistical Analysis
3. Results
3.1. Bacteria Isolation and Selection
3.2. Biosurfactant Production
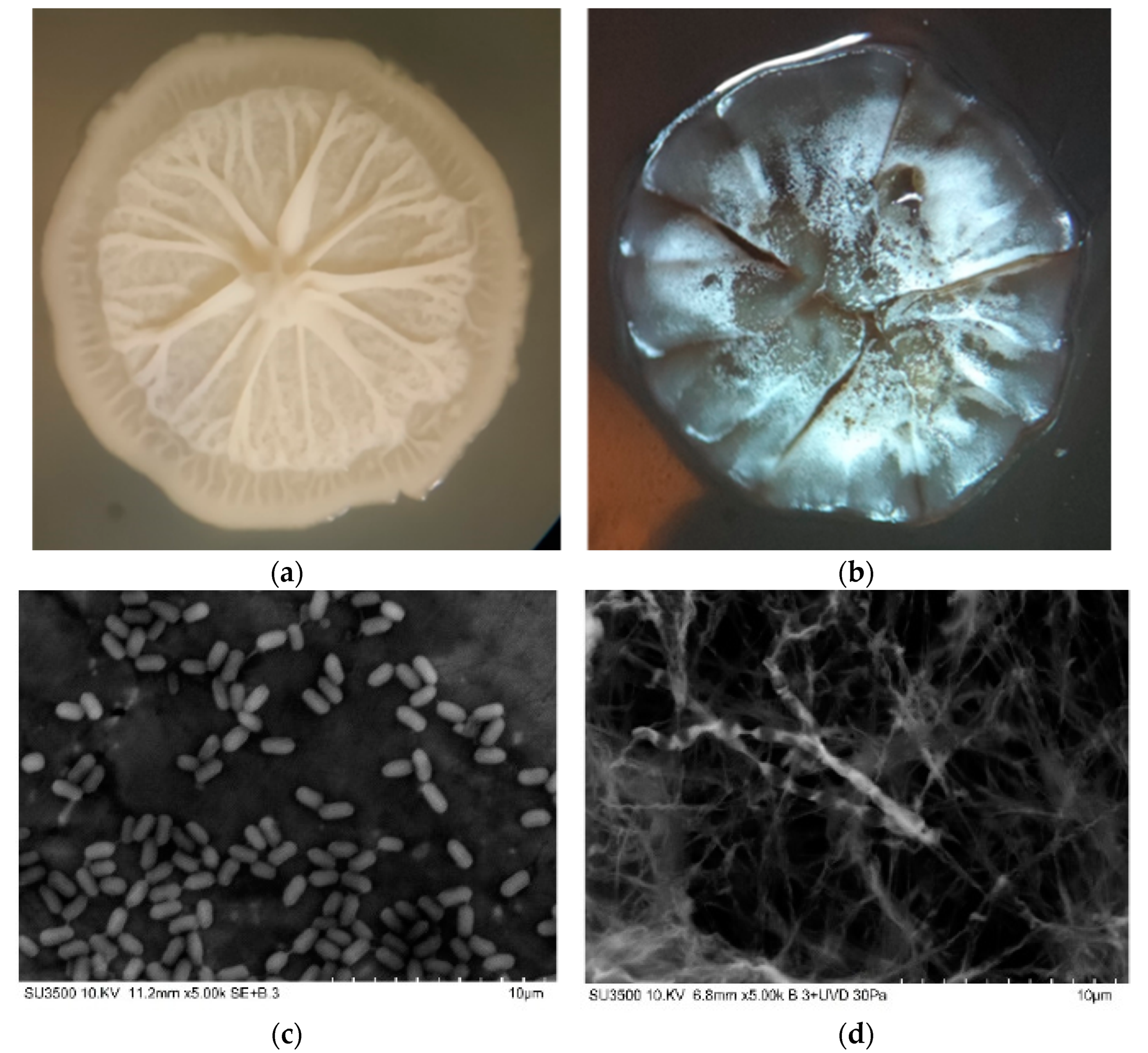
3.3. Strains Identification and Characterization
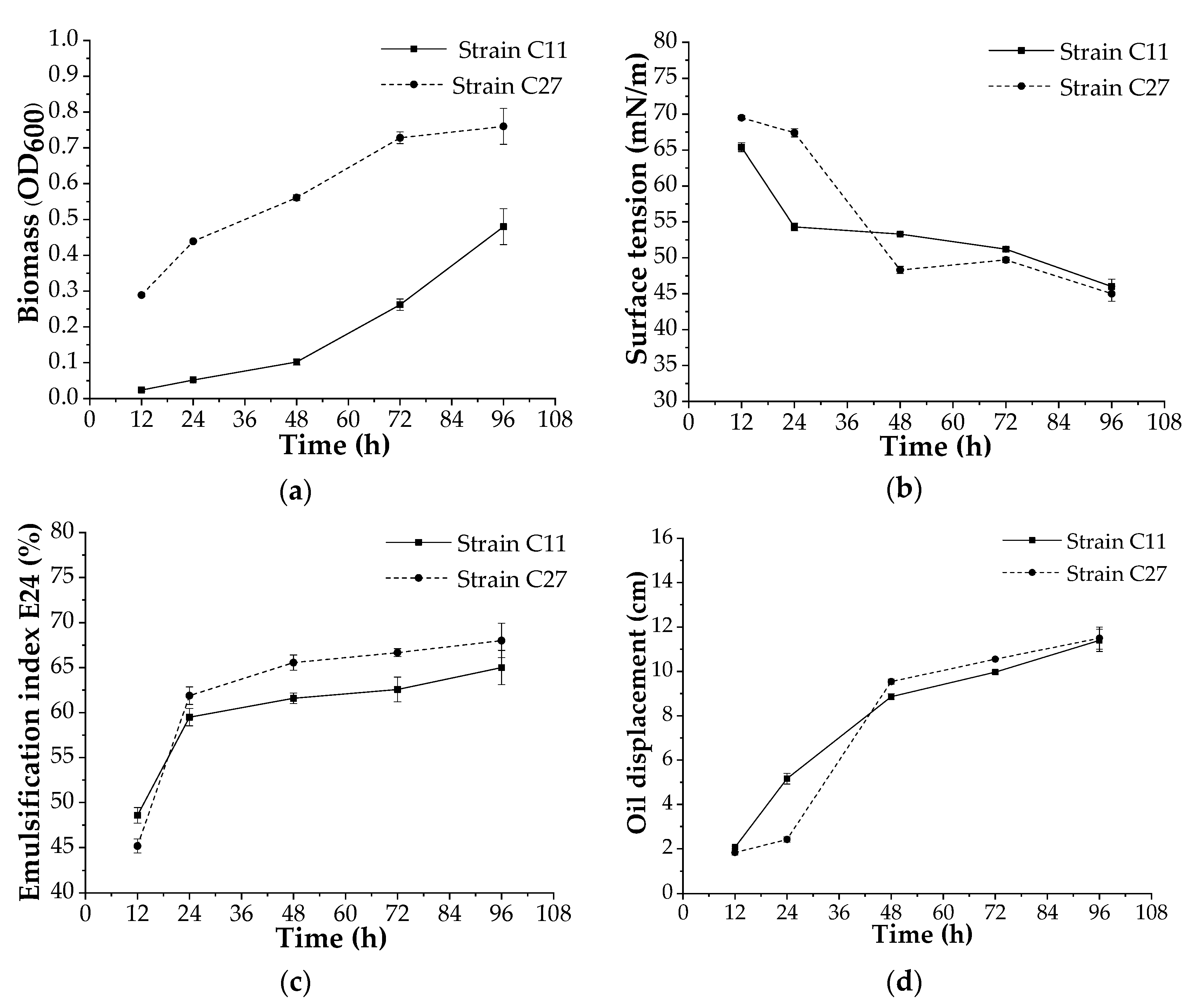
3.4. Biosurfactant Production at Bench Scale
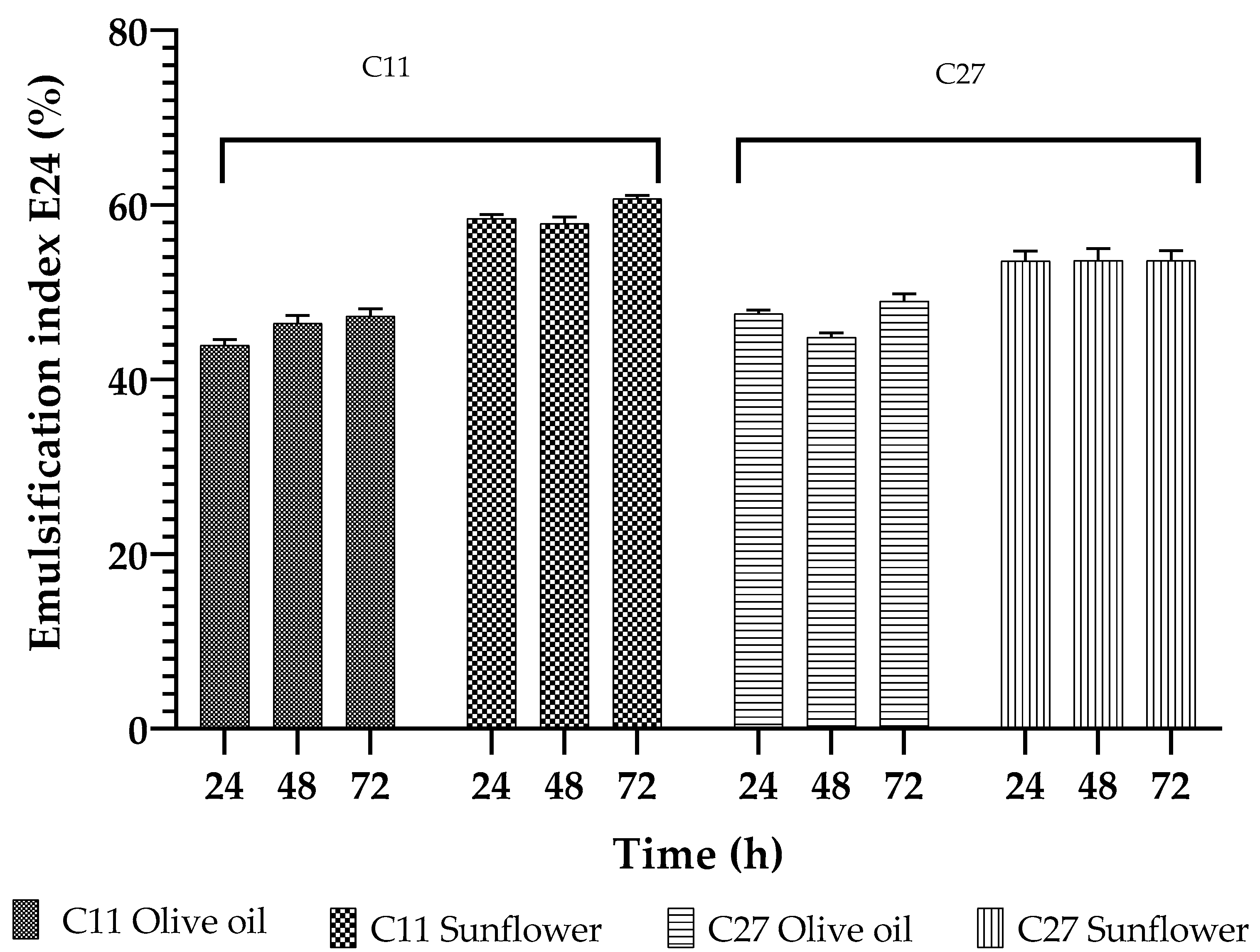
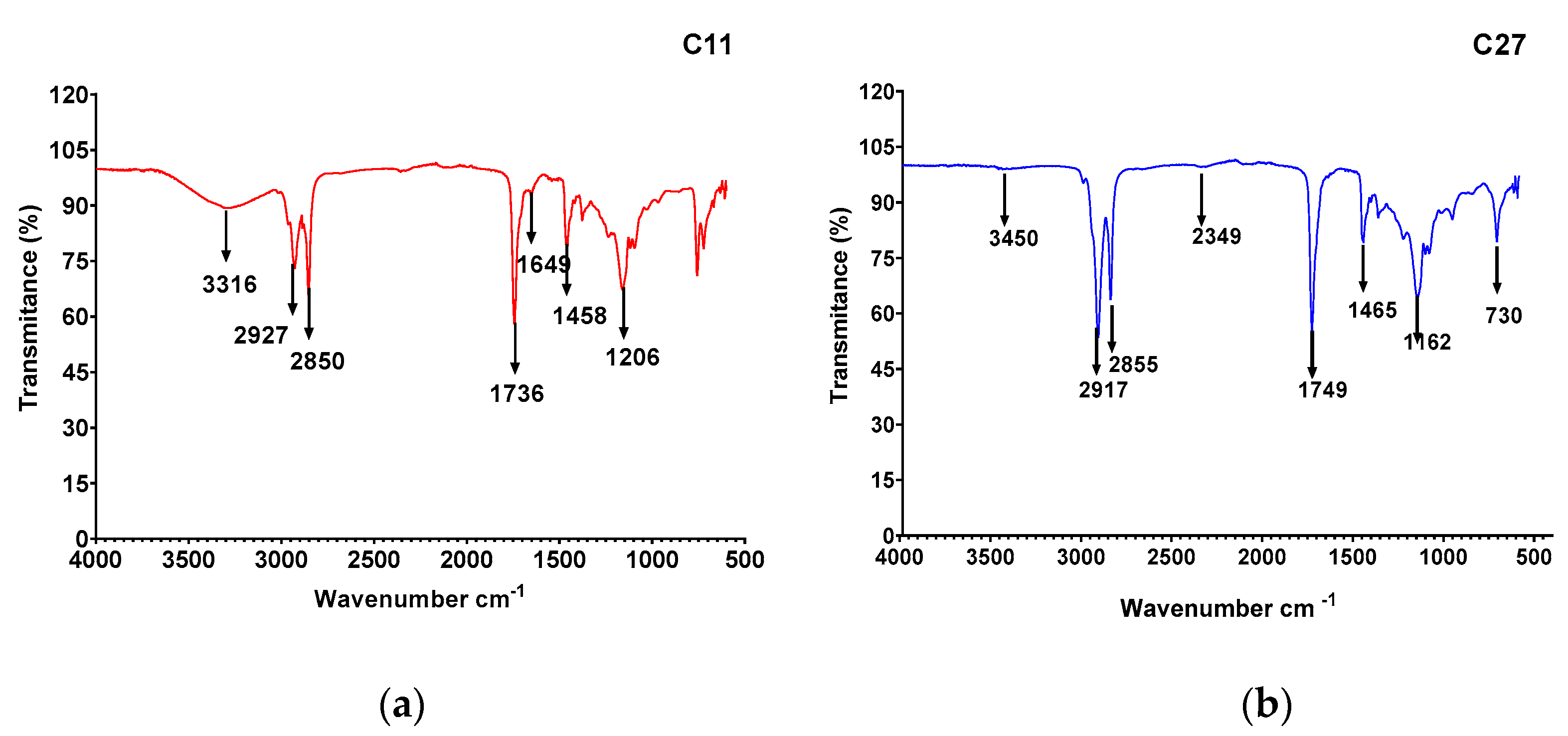
3.5. Biosurfactant Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shoeb, E.; Akhlaq, F.; Badar, U.; Akhter, J.; Imtiaz, S. Classification and industrial applications of biosurfactants. Acad. Res. Int. 2013, 4, 243. [Google Scholar]
- Saharan, B.S.; Sahu, R.K.; Sharma, D. A Review on biosurfactants: Fermentation, current developments and perspectives. Genet. Eng. Biotechnol. J. 2011, 2011, 1–42. [Google Scholar]
- Kosaric, N.; Vardar-Sukan, F. Biosurfactants: Production and Utilization--Processes, Technologies, and Economics; Surfactant Science Series; CRC Press: Boca Raton, FL, USA, 2015; ISBN 9781523120499. [Google Scholar]
- Kumar, A.; Singh, S.K.; Kant, C.; Verma, H.; Kumar, D.; Singh, P.P.; Modi, A.; Droby, S.; Kesawat, M.S.; Alavilli, H.; et al. Microbial Biosurfactant: A new frontier for sustainable agriculture and pharmaceutical industries. Antioxidants 2021, 10, 1472. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, C.U. Surface activity of extracellular products of a Pseudomonas aeruginosa isolated from petroleum—Contaminated soil. Int. J. Environ. Sci. 2010, 1, 225–235. [Google Scholar]
- Yin, H.; Qiang, J.; Jia, Y.; Ye, J.; Peng, H.; Qin, H.; Zhang, N.; He, B. Characteristics of biosurfactant produced by Pseudomonas aeruginosa S6 isolated from oil-containing wastewater. Proc. Biochem. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, Y.; Bhatt, P.; Chen, S. Biosurfactant is a powerful tool for the bioremediation of heavy metals from contaminated soils. J. Hazard. Mater. 2021, 418, 126253. [Google Scholar] [CrossRef]
- Delegan, Y.; Petrikov, K.; Frantsuzova, E.; Rudenko, N.; Solomentsev, V.; Suzina, N.; Travkin, V.; Solyanikova, I.P. Complete Genome Analysis of Rhodococcus opacus S8 capable of degrading alkanes and producing biosurfactant reveals Its genetic adaptation for crude oil decomposition. Microorganism 2022, 10, 1172. [Google Scholar] [CrossRef]
- Bhatt, P.; Verma, A.; Gangola, S.; Bhandari, G.; Chen, S. Microbial glycoconjugates in organic pollutant bioremediation: Recent advances and applications. Microb. Cell Factories 2021, 20, 1–18. [Google Scholar] [CrossRef]
- García-Reyes, S.; Yañez Ocampo, G.; Wong Villarreal, A.; Rajesh Kannan, R.; Citarasu, T.; Patiño, R.; Ortiz-Hernandez, L. Partial characterization of a biosurfactant extracted from Pseudomonas sp. B0406 that enhances the solubility of pesticides. Environ. Technol. 2017, 39, 1–16. [Google Scholar] [CrossRef]
- Calvo, C.; Manzanera, M.; Silva-Castro, G.A.; Uad, I.; Gonzalez-Lopez, J. Application of bioemulsifiers in soil oil bioremediation processes. Future prospects. Sci. Total Environ. 2009, 407, 3634–3640. [Google Scholar] [CrossRef]
- De la Rosa Cruz, N.L.; Sánchez-Salinas, E.; Ortiz-Hernández, M.L. Biosurfactantes y su papel en la biorremediación de suelos contaminados con plaguicidas. Rev. Latinoam. Biotecnol. Ambient. Algal. 2014, 5, 4. [Google Scholar] [CrossRef]
- Dadrasnia, A.; Ismail, S. Biosurfactant production by Bacillus salmalaya for lubricating oil solubilization and biodegradation. Int. J. Environ. Res. Public Health 2015, 12, 9848–9863. [Google Scholar] [CrossRef]
- Singh, P.; Tiwary, B.N. Isolation and characterization of glycolipid biosurfactant produced by a Pseudomonas otitidis strain isolated from Chirimiri coal mines, India. Bioresour. Bioproc. 2016, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Awashti, N.; Kumar, A.; Makkar, R.C.S. A chlorinated pesticide in presence of biosurfactant. Environ. Sci. Health B 1999, 34, 793–803. [Google Scholar]
- Bagheri Lotfabad, T.; Ebadipour, N.; Roostaazad, R.; Partovi, M.; Bahmaei, M. Two schemes for production of biosurfactant from Pseudomonas aeruginosa MR01: Applying residues from soybean oil industry and silica sol–gel immobilized cells. Colloids Surf. B Biointerfaces 2017, 152, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Atakpa, E.O.; Zhou, H.; Jiang, L.; Ma, Y.; Liang, Y.; Li, Y.; Zhang, D.; Zhang, C. Improved degradation of petroleum hydrocarbons by co-culture of fungi and biosurfactant-producing bacteria. Chemosphere 2022, 290, 133337. [Google Scholar] [CrossRef] [PubMed]
- Luna, J.M.; Rufino, R.D.; Sarubbo, L.A.; Campos-Takaki, G.M. Characterisation, surface properties and biological activity of a biosurfactant produced from industrial waste by Candida sphaerica UCP0995 for application in the petroleum industry. Colloids Surf. B Biointerfaces 2013, 102, 202–209. [Google Scholar] [CrossRef]
- Santos, A.P.P.; Silva, M.D.S.; Costa, E.V.L.; Rufino, R.D.; Santos, V.A.; Ramos, C.S.; Sarubbo, L.A.; Porto, A.L.F. Production and characterization of a biosurfactant produced by Streptomyces sp. 1559 isolated from lichens of the Amazon region. Braz. J. Med. Biol. Res. 2017, 51, e6657. [Google Scholar] [CrossRef]
- Silva, E.J.; Rocha e Silva, N.M.P.; Rufino, R.D.; Luna, J.M.; Silva, R.O.; Sarubbo, L.A. Characterization of a biosurfactant produced by Pseudomonas cepacia CCT6659 in the presence of industrial wastes and its application in the biodegradation of hydrophobic compounds in soil. Colloids Surf. B Biointerfaces 2014, 117, 36–41. [Google Scholar] [CrossRef]
- Zambry, N.S.; Ayoib, A.; Md Noh, N.A.; Yahya, A.R.M. Production and partial characterization of biosurfactant produced by Streptomyces sp. R1. Bioprocess Biosyst. Eng. 2017, 40, 1007–1016. [Google Scholar] [CrossRef]
- Lamilla, C.; Braga, D.; Castro, R.; Guimar, C.; De, L.V.A.; Freire, D.M.G.; Barrientos, L. Streptomyces luridus So3. 2 from Antarctic soil as a novel producer of compounds with bioemulsification potential. PLoS ONE 2018, 13, e0196054. [Google Scholar]
- Singh, P.B.; Sharma, S.; Saini, H.S.; Chadha, B.S. Biosurfactant production by Pseudomonas sp. and its role in aqueous phase partitioning and biodegradation of chlorpyrifos. Lett. Appl. Microbiol. 2009, 49, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Wattanaphon, H.T.; Kerdsin, A.; Thammacharoen, C.; Sangvanich, P.; Vangnai, A.S. A biosurfactant from Burkholderia cenocepacia BSP3 and its enhancement of pesticide solubilization. J. Appl. Microbiol. 2008, 105, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Lamilla, C.; Schalchli, H.; Briceño, G.; Leiva, B.; Donoso-Piñol, P.; Barrientos, L.; Rocha, V.A.L.; Freire, D.M.G.; Diez, M.C. A Pesticide Biopurification System: A source of biosurfactant-producing bacteria with environmental biotechnology applications. Agronomy 2021, 11, 624. [Google Scholar] [CrossRef]
- Al Disi, Z.; Al-Ghouti, M.A.; Zouari, N. Investigating the simultaneous removal of hydrocarbons and heavy metals by highly adapted Bacillus and Pseudomonas strains. Environ. Technol. Innov. 2022, 27, 102513. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmail, G.A.; Valan Arasu, M. Enhanced production of biosurfactant from Bacillus subtilis Strain Al-Dhabi-130 under solid-state fermentation using date molasses from saudi arabia for bioremediation of crude-oil-contaminated soils. Int. J. Curr. Microbiol. Appl. Sci. 2020, 17, 8446. [Google Scholar] [CrossRef]
- Maldonado Desena, F.; De la Cruz Ceferino, N.; Gómez Cornelio, S.; Alvarez Villagomez, C.; Herrera Candelario, J.L.; De la Rosa García, S. Bacteria Halotolerant from karst sinkholes as a source of biosurfactants and bioemulsifiers. Microorganisms 2022, 10, 1264. [Google Scholar] [CrossRef]
- Lotfabad, T.B.; Shourian, M.; Roostaazad, R.; Najafabadi, A.R.; Adelzadeh, M.R.; Noghabi, K.A. An efficient biosurfactant-producing bacterium Pseudomonas aeruginosa MR01, isolated from oil excavation areas in south of Iran. Colloids Surf. B Biointerfaces 2009, 69, 183–193. [Google Scholar] [CrossRef]
- Deepika, K.V.; Raghuram, M.; Bramhachari, P.V. Rhamnolipid biosurfactant production by Pseudomonas aeruginosa strain KVD-HR42 isolated from oil contaminated mangrove sediments. Afr. J. Microbiol. Res. 2017, 11, 218–231. [Google Scholar] [CrossRef]
- Adamczak, M.; Bednarski, W. Influence of medium composition and aeration on the synthesis of biosurfactants produced by Candida antarctica. Biotechnol. Lett. 2000, 22, 313–316. [Google Scholar] [CrossRef]
- Abouseoud, M.; Maachi, R.; Amrane, A.; Boudergua, S.; Nabi, A. Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens. Desalination 2008, 223, 143–151. [Google Scholar] [CrossRef]
- Maass, D.; Moya Ramírez, I.; García Román, M.; Jurado Alameda, E.; Ulson de Souza, A.A.; Borges Valle, J.A.; Altmajer Vaz, D. Two-phase olive mill waste (alpeorujo) as carbon source for biosurfactant production. J. Chem. Technol. Biotechnol. 2016, 91, 1990–1997. [Google Scholar] [CrossRef]
- Haba, E.; Espuny, M.J.; Busquets, M.; Manresa, A. Screening and production of rhamnolipids by Pseudomonas aeruginosa 47T2 NCIB 40044 from waste frying oils. J. Appl. Microbiol. 2000, 88, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, D.P.; Cameotra, S.S. Biosurfactants in agriculture. Appl. Microbiol. Biotechnol. 2013, 97, 1005–1016. [Google Scholar] [CrossRef]
- FAOSTAT 2018. Food and Agriculture Organization (FAO). Available online: https://www.fao.org/faostat/en/#data/RP/visualize (accessed on 15 August 2022).
- Sequinatto, L.; Reichert, J.M.; dos Santos, D.R.; Reinert, D.J.; Copetti, A.C.C. Occurrence of agrochemicals in surface waters of shallow soils and steep slopes cropped to tobacco. Quim. Nova 2013, 36, 768–772. [Google Scholar] [CrossRef] [Green Version]
- Muñiz-González, A.-B.; Paoli, F.; Martínez-Guitarte, J.-L.; Lencioni, V. Molecular biomarkers as tool for early warning by chlorpyrifos exposure on Alpine chironomids. Environ. Pollut. 2021, 290, 118061. [Google Scholar] [CrossRef]
- SAG. Available online: https://www.sag.gob.cl/sites/default/files/declaracion_de_ventas_de_plaguicidas_ano_2019_0.pdf (accessed on 15 August 2022).
- Levio-Raiman, M.; Briceño, G.; Leiva, B.; López, S.; Schalchli, H.; Lamilla, C.; Bornhardt, C.; Diez, M.C. Treatment of pesticide-contaminated water using a selected fungal consortium: Study in a batch and packed-bed bioreactor. Agronomy 2021, 11, 743. [Google Scholar] [CrossRef]
- Diez, M.C.; Leiva, B.; Gallardo, F. Novel insights in biopurification system for dissipation of a pesticide mixture in repeated applications. Environ. Sci. Pollut. Res. 2018, 25, 21440–21450. [Google Scholar] [CrossRef]
- Nandhini, B.; Josephine, R.M. A study on bacterial and fungal diversity in potted soil. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 1–5. [Google Scholar]
- Ren, W.; Zhong, Y.; Ding, Y.; Wu, Y.; Xu, X.; Zhou, P. Mismatches in 16S rRNA gene primers: An area worth further exploring. Front. Microbiol. 2022, 13, 888803. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Cui, H.; Li, Y.; Xu, N.; Lu, T.; Chen, J.; Penuelas, J.; Hu, B.; Qian, H. Composition identification and functional verification of bacterial community in disease-suppressive soils by machine learning. Environ. Microbiol. 2022, 24, 3405–3419. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.G.; Goldenberg, B.G. Surface-active agents from two Bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, msw054. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, L.A.; Alberton Magina, M.D.; Tavares, L.B.B.; Guelli Ulson de Souza, S.M.A.; García Román, M.; Altmajer Vaz, D. Biosurfactant production by Trametes versicolor grown on two-phase olive mill waste in solid-state fermentation. Environ. Technol. 2018, 39, 3066–3076. [Google Scholar] [CrossRef]
- Jain, R.M.; Mody, K.; Joshi, N.; Mishra, A.; Jha, B. Production and structural characterization of biosurfactant produced by an alkaliphilic bacterium, Klebsiella sp.: Evaluation of different carbon sources. Colloids Surf. B Biointerfaces 2013, 108, 199–204. [Google Scholar] [CrossRef]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure–function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta Molec. Cell Biol. Lipids 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Ullmann, K.; Poggemann, L.; Nirschl, H.; Leneweit, G. Adsorption process for phospholipids of different chain lengths at a fluorocarbon/water interface studied by Du Noüy ring and spinning drop. Colloid Polym. Sci. 2020, 298, 407–417. [Google Scholar] [CrossRef]
- Jacques, P. Surfactin and Other Lipopeptides from Bacillus spp. BT Biosurfactants: From Genes to Applications; Soberón-Chávez, G., Ed.; Springer: Berlin/ Heidelberg, Germany, 2011; pp. 57–91. ISBN 978-3-642-14490-5. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.-K. Pharmaceutically active secondary metabolites of marine Actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Briceño, G.; Lamilla, C.; Leiva, B.; Levio, M.; Donoso-Piñol, P.; Schalchli, H.; Gallardo, F.; Diez, M.C. Pesticide-tolerant bacteria isolated from a biopurification system to remove commonly used pesticides to protect water resources. PLoS ONE 2020, 15, e0234865. [Google Scholar] [CrossRef]
- Ferreira, I.N.S.; Rodríguez, D.M.; Campos-Takaki, G.M.; Andrade, R.F. da S. Biosurfactant and bioemulsifier as promising molecules produced by Mucor hiemalis isolated from Caatinga soil. Electron. J. Biotechnol. 2020, 47, 51–58. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, J.; Verma, N. Production, characterization and environmental applications of biosurfactants from Bacillus amyloliquefaciens and Bacillus subtilis. Biocatal. Agric. Biotechnol. 2018, 16, 132–139. [Google Scholar] [CrossRef]
- Al-Wahaibi, Y.; Joshi, S.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A.; Shibulal, B. Biosurfactant production by Bacillus subtilis B30 and its application in enhancing oil recovery. Colloids Surf. B Biointerfaces 2014, 114, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Das, A.J.; Kumar, R. Production of biosurfactant from agro-industrial waste by Bacillus safensis J2 and exploring its oil recovery efficiency and role in restoration of diesel contaminated soil. Environ. Technol. Innov. 2019, 16, 100450. [Google Scholar] [CrossRef]
- Elkhawaga, M.A. Optimization and characterization of biosurfactant from Streptomyces griseoplanus NRRL-ISP5009 (MS1). J. Appl. Microbiol. 2018, 124, 691–707. [Google Scholar] [CrossRef]
- Liu, Z.; Zeng, G.; Zhong, H.; Fu, H.; Liu, X. Production and characterization of biosurfactant from Bacillus subtilis CCTCC AB93108. J. Cent. South Univ. Technol. 2010, 17, 516–521. [Google Scholar] [CrossRef]
- Li, X.; Li, B.; Cai, S.; Zhang, Y.; Xu, M.; Zhang, C.; Yuan, B.; Xing, K.; Qin, S. Identification of rhizospheric Actinomycete Streptomyces lavendulae SPS-33 and the inhibitory effect of its volatile organic compounds against Ceratocystis fimbriata in Postharvest Sweet Potato (Ipomoea batatas (L.) Lam.). Microorganisms 2020, 8, 319. [Google Scholar] [CrossRef]
- Kadaikunnan, S.; Rejiniemon, T.S.; Khaled, J.M.; Alharbi, N.S.; Mothana, R. In-vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 9. [Google Scholar] [CrossRef]
- Kakou, A.; Kambire, O.; Boli, Z.; Yoro, T.; Koffi, N.; Koussemon, M. Diversity and enzymatic characterization of Bacillus species isolated from traditional cassava starters used for attiéké production. Int. J. Biol. Chem. Sci. 2017, 11, 531. [Google Scholar] [CrossRef]
- Ananda, R.; Dharumadurai, D.; Manogaran, G.P. An Introduction to Actinobacteria. In Actinobacteria—Basics and Biotechnological Applications; Dharumadurai, D., Ed.; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Colin, V.L.; Bourguignon, N.; Gómez, J.S.; de Carvalho, K.G.; Ferrero, M.A.; Amoroso, M.J. Production of surface-active compounds by a hydrocarbon-degrading Actinobacterium: Presumptive relationship with lipase activity. Water Air Soil Pollut. 2017, 228, 454. [Google Scholar] [CrossRef]
- Santos, E.F.; Teixeira, M.F.S.; Converti, A.; Porto, A.L.F.; Sarubbo, L.A. Production of a new lipoprotein biosurfactant by Streptomyces sp. DPUA1566 isolated from lichens collected in the Brazilian Amazon using agroindustry wastes. Biocatal. Agric. Biotechnol. 2019, 17, 142–150. [Google Scholar] [CrossRef]
- Heryani, H.; Putra, M.D. Dataset on potential large scale production of biosurfactant using Bacillus sp. Data Br. 2017, 13, 196–201. [Google Scholar] [CrossRef]
- Joshi, S.; Bharucha, C.; Desai, A.J. Production of biosurfactant and antifungal compound by fermented food isolate Bacillus subtilis 20B. Bioresour. Technol. 2008, 99, 4603–4608. [Google Scholar] [CrossRef] [PubMed]

| Strain | Gram (+/−) | Shape of Cell | Enzymatic Activity (cm) | |||||
|---|---|---|---|---|---|---|---|---|
| Gelatinase | Protease | Amylase | Lipolytic | Cellulase | Hemolytic | |||
| C11 | + | Rods | 2.2 | 2 | <1 | 1.1 | <1 | ++++ β |
| C12 | + | Rods | 1.1 | - | - | - | - | γ |
| C13 | + | Rods | 1.3 | 1.5 | - | <1 | <1 | γ |
| C14 | + | Rods | <1 | - | - | <1 | <1 | γ |
| C15 | + | Rods | - | - | - | - | - | γ |
| C16 | + | Rods | 1.1 | 1.8 | - | 1 | - | + α |
| C17-1 | − | Cocci | 1.1 | <1 | - | - | <1 | γ |
| C17-2 | + | Rods | <1 | - | - | <1 | <1 | γ |
| C18 | + | Rods | 1.1 | 1.6 | - | <1 | <1 | γ |
| C19 | + | Rods | <1 | - | - | - | - | γ |
| C20 | − | Rods | - | - | - | - | - | γ |
| C21 | + | Rods | - | - | - | - | - | γ |
| C22 | + | Rods | - | - | - | - | - | γ |
| C23 | + | Rods | <1 | <1 | - | 1.2 | <1 | γ |
| C24 | + | Rods | 1.1 | 1.5 | - | - | <1 | γ |
| C25 | − | Rods | - | - | - | - | - | + α |
| C26 | − | Rods | <1 | 1.6 | - | <1 | 1.7 | γ |
| C27 | + | Rods | <1 | - | - | - | - | +++ β |
| Strain | Strain Identity | Accession Number (Ribotyping) | MALDI-TOF Score |
|---|---|---|---|
| C11 | Bacillus amyloliquefaciens | ON732850 | 2.520 |
| C27 | Streptomyces lavendulae | ON732851 | 2.000 |
| Enzyme | B. amyloliquefaciens C11 | S. lavendulae C27 |
|---|---|---|
| Alkaline phosphatase | + | + |
| Esterase (C 4) | + | + |
| Esterase lipase (C 8) | + | - |
| Lipase (C 14) | + | - |
| Leucine arylamidase | + | + |
| Valine arylamidase | - | + |
| Cystine arylamidase | - | - |
| Trypsin | - | + |
| α-quimotrypsyne | - | + |
| Acid phosphatase | + | + |
| Naftol-AS-BI-phosphohydrolase | + | + |
| α-galactosidase | - | - |
| β-galactosidase | - | - |
| β-glucuronidase | - | - |
| α-glucosidase | - | + |
| β-glucosidase | - | + |
| N-acétil-β-glucosaminidase | + | - |
| α-mannosidase | - | - |
| α-fucosidase | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diez, M.C.; Llafquen, C.; Fincheira, P.; Lamilla, C.; Briceño, G.; Schalchli, H. Biosurfactant Production by Bacillus amyloliquefaciens C11 and Streptomyces lavendulae C27 Isolated from a Biopurification System for Environmental Applications. Microorganisms 2022, 10, 1892. https://doi.org/10.3390/microorganisms10101892
Diez MC, Llafquen C, Fincheira P, Lamilla C, Briceño G, Schalchli H. Biosurfactant Production by Bacillus amyloliquefaciens C11 and Streptomyces lavendulae C27 Isolated from a Biopurification System for Environmental Applications. Microorganisms. 2022; 10(10):1892. https://doi.org/10.3390/microorganisms10101892
Chicago/Turabian StyleDiez, M. Cristina, Cesar Llafquen, Paola Fincheira, Claudio Lamilla, Gabriela Briceño, and Heidi Schalchli. 2022. "Biosurfactant Production by Bacillus amyloliquefaciens C11 and Streptomyces lavendulae C27 Isolated from a Biopurification System for Environmental Applications" Microorganisms 10, no. 10: 1892. https://doi.org/10.3390/microorganisms10101892
APA StyleDiez, M. C., Llafquen, C., Fincheira, P., Lamilla, C., Briceño, G., & Schalchli, H. (2022). Biosurfactant Production by Bacillus amyloliquefaciens C11 and Streptomyces lavendulae C27 Isolated from a Biopurification System for Environmental Applications. Microorganisms, 10(10), 1892. https://doi.org/10.3390/microorganisms10101892








