Proteomic Characterization of Host-Pathogen Interactions during Bovine Trophoblast Cell Line Infection by Neospora caninum
Abstract
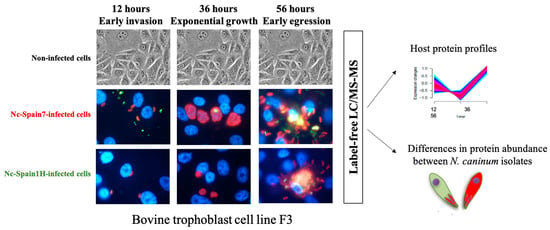
1. Introduction
2. Results and Discussion
2.1. Neospora caninum and Bovine Trophoblast Cell Proteomes
2.2. Host Proteome Remodelling during N. caninum Infection
2.3. Host Immune Response during N. caninum Infection
2.4. Proteomic Differences between High- and Low-Virulence N. caninum Isolates at Different Stages of the Lytic Cycle
3. Materials and Methods
3.1. Parasites and Cell Cultures
3.2. Experimental Design and Sample Production for Proteome Analyses
3.3. LC-MS/MS Analyses
3.4. Data Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dubey, J.P.; Schares, G.; Ortega-Mora, L.M. Epidemiology and control of neosporosis and Neospora caninum. Clin. Microbiol. Rev. 2007, 20, 323–367. [Google Scholar] [CrossRef]
- Reichel, M.P.; Alejandra Ayanegui-Alcérreca, M.; Gondim, L.F.P.; Ellis, J.T. What is the global economic impact of Neospora caninum in cattle—The billion dollar question. Int. J. Parasitol. 2013, 43, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Almería, S.; Serrano-Pérez, B.; López-Gatius, F. Immune response in bovine neosporosis: Protection or contribution to the pathogenesis of abortion. Microb. Pathog. 2017, 109, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Pelayo, L.; García-Sánchez, M.; Regidor-Cerrillo, J.; Horcajo, P.; Collantes-Fernández, E.; Gómez-Bautista, M.; Hambruch, N.; Pfarrer, C.; Ortega-Mora, L.M. Differential susceptibility of bovine caruncular and trophoblast cell lines to infection with high and low virulence isolates of Neospora caninum. Parasites Vectors 2017, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Horcajo, P.; Jiménez-Pelayo, L.; García-Sánchez, M.; Regidor-Cerrillo, J.; Collantes-Fernández, E.; Rozas, D.; Hambruch, N.; Pfarrer, C.; Ortega-Mora, L.M. Transcriptome modulation of bovine trophoblast cells in vitro by Neospora caninum. Int. J. Parasitol. 2017, 47, 791–799. [Google Scholar] [CrossRef]
- Jiménez-Pelayo, L.; García-Sánchez, M.; Regidor-Cerrillo, J.; Horcajo, P.; Collantes-Fernández, E.; Gómez-Bautista, M.; Hambruch, N.; Pfarrer, C.; Ortega-Mora, L. Immune response profile of caruncular and trophoblast cell lines infected by high- (Nc-Spain7) and low virulente (Nc-Spain1H) isolates of Neospora caninum. Parasites Vectors 2019, 12, 218. [Google Scholar] [CrossRef]
- Rojo-Montejo, S.; Collantes-Fernández, E.; Blanco-Murcia, J.; Rodríguez-Bertos, A.; Risco-Castillo, V.; Ortega-Mora, L.M. Experimental infection with a low virulence isolate of Neospora caninum at 70 days gestation in cattle did not result in foetopathy. Vet. Res. 2009, 40, 49. [Google Scholar] [CrossRef]
- Caspe, S.G.; Moore, D.P.; Leunda, M.R.; Cano, D.B.; Lischinsky, L.; Regidor-Cerrillo, J.; Álvarez-García, G.; Echaide, I.G.; Bacigalupe, D.; Ortega-Mora, L.M.; et al. The Neospora caninum-Spain 7 isolate induces placental damage, fetal death and abortion in cattle when inoculated in early gestation. Vet. Parasitol. 2012, 189, 171–181. [Google Scholar] [CrossRef]
- Regidor-Cerrillo, J.; Arranz-Solis, D.; Benavides, J.; Gomez-Bautista, M.; Castro-Hermida, J.A.; Mezo, M.; Perez, V.; Ortega-Mora, L.M.; Gonzalez-Warleta, M. Neospora caninum infection during early pregnancy in cattle: How the isolate influences infection dynamics, clinical outcome and peripheral and local immune responses. Vet. Res. 2014, 45, 10. [Google Scholar] [CrossRef]
- Chryssafidis, A.L.; Canton, G.; Chianini, F.; Innes, E.A.; Madureira, E.H.; Gennari, S.M. Pathogenicity of Nc-Bahia and Nc-1 strains of Neospora caninum in experimentally infected cows and buffaloes in early pregnancy. Parasitol. Res. 2014, 113, 1521–1528. [Google Scholar] [CrossRef]
- Jiménez-Pelayo, L.; García-Sánchez, M.; Vázquez, P.; Regidor-Cerrillo, J.; Horcajo, P.; Collantes-Fernández, E.; Blanco-Murcia, J.; Gutiérrez-Expósito, D.; Román-Trufero, A.; Osoro, K. Early Neospora caninum infection dynamics in cattle after inoculation at mid-gestation with high (Nc-Spain7)-or low (Nc-Spain1H)-virulence isolates. Vet. Res. 2019, 50, 72. [Google Scholar] [CrossRef] [PubMed]
- Szklanna, P.B.; Wynne, K.; Nolan, M.; Egan, K.; Ainle, F.N.; Maguire, P.B. Comparative proteomic analysis of trophoblast cell models reveals their differential phenotypes, potential uses, and limitations. Proteomics 2017, 17, e1700037. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.G.; Kim, J.H.; Shin, Y.S.; Shin, G.W.; Kim, Y.R.; Palaksha, K.J.; Kim, D.Y.; Yamane, I.; Kim, Y.H.; Kim, G.S.; et al. Application of proteomics for comparison of proteome of Neospora caninum and Toxoplasma gondii tachyzoites. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005, 815, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.S.; Shin, G.W.; Kim, Y.R.; Lee, E.Y.; Yang, H.H.; Palaksha, K.J.; Youn, H.J.; Kim, J.H.; Kim, D.Y.; Marsh, A.E.; et al. Comparison of proteome and antigenic proteome between two Neospora caninum isolates. Vet. Parasitol. 2005, 134, 41–52. [Google Scholar] [CrossRef]
- Regidor-Cerrillo, J.; Álvarez-García, G.; Pastor-Fernández, I.; Marugán-Hernández, V.; Gómez-Bautista, M.; Ortega-Mora, L.M. Proteome expression changes among virulent and attenuated Neospora caninum isolates. J. Proteom. 2012, 75, 2306–2318. [Google Scholar] [CrossRef]
- Ramaprasad, A.; Mourier, T.; Naeem, R.; Malas, T.B.; Moussa, E.; Panigrahi, A.; Vermont, S.J.; Otto, T.D.; Wastling, J.; Pain, A. Comprehensive evaluation of Toxoplasma gondii VEG and Neospora caninum LIV genomes with tachyzoite stage transcriptome and proteome defines novel transcript features. PLoS ONE 2015, 10, e0124473. [Google Scholar] [CrossRef]
- Horcajo, P.; Xia, D.; Randle, N.; Collantes-Fernández, E.; Wastling, J.; Ortega-Mora, L.; Regidor-Cerrillo, J. Integrative transcriptome and proteome analyses define marked differences between Neospora caninum isolates throughout the tachyzoite lytic cycle. J. Proteom. 2018, 180, 108–119. [Google Scholar] [CrossRef]
- Xu, T.; Ping, J.; Yu, Y.; Yu, F.; Yu, Y.; Hao, P.; Li, X. Revealing parasite influence in metabolic pathways in Apicomplexa infected patients. BMC Bioinform. 2010, 11, S13. [Google Scholar] [CrossRef]
- Remmerie, A.; Scott, C.L. Macrophages and lipid metabolism. Cell Immunol. 2018, 330, 27–42. [Google Scholar] [CrossRef]
- Hargrave, K.E.; Woods, S.; Millington, O.; Chalmers, S.; Westrop, G.D.; Roberts, C.W. Multi-Omics Studies Demonstrate Toxoplasma gondii-Induced Metabolic Reprogramming of Murine Dendritic Cells. Front. Cell Infect. Microbiol. 2019, 9, 309. [Google Scholar] [CrossRef]
- García-Sánchez, M. Interactions between high and low virulence isolates of Neospora caninum and target cells of the bovine innate immune response. Ph.D. Thesis, Complutense University of Madrid, Madrid, Spain, November 2019. [Google Scholar]
- Jiménez-Pelayo, L.; García-Sánchez, M.; Collantes-Fernández, E.; Regidor-Cerrillo, J.; Horcajo, P.; Gutiérrez-Expósito, D.; Espinosa, J.; Benavides, J.; Osoro, K.; Pfarrer, C.; et al. The cross-talk at the placenta between Neospora caninum and the bovine host determines the outcome of the infection. Vet. Res. 2020. submitted. [Google Scholar]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Sinai, A.P.; Webster, P.; Joiner, K.A. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: A high affinity interaction. J. Cell Sci. 1997, 110 Pt 17, 2117–2128. [Google Scholar]
- Nelson, M.M.; Jones, A.R.; Carmen, J.C.; Sinai, A.P.; Burchmore, R.; Wastling, J.M. Modulation of the host cell proteome by the intracellular apicomplexan parasite Toxoplasma gondii. Infect. Immun. 2008, 76, 828–844. [Google Scholar] [CrossRef]
- Pernas, L.; Adomako-Ankomah, Y.; Shastri, A.J.; Ewald, S.E.; Treeck, M.; Boyle, J.P.; Boothroyd, J.C. Toxoplasma effector MAF1 mediates recruitment of host mitochondria and impacts the host response. PLoS Biol. 2014, 12, e1001845. [Google Scholar] [CrossRef] [PubMed]
- Nolan, S.J.; Romano, J.D.; Luechtefeld, T.; Coppens, I. Neospora caninum Recruits Host Cell Structures to Its Parasitophorous Vacuole and Salvages Lipids from Organelles. Eukaryot. Cell 2015, 14, 454–473. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, X.; He, L.; Zhang, A.; Li, Q.; Hu, W.; Chen, H.; Du, J.; Shen, J. Toxoplasma gondii isolate with genotype Chinese 1 triggers trophoblast apoptosis through oxidative stress and mitochondrial dysfunction in mice. Exp. Parasitol. 2015, 154, 51–61. [Google Scholar] [CrossRef]
- Syn, G.; Anderson, D.; Blackwell, J.M.; Jamieson, S.E. Toxoplasma gondii infection is Associated with mitochondrial dysfunction in-vitro. Front. Cell Infect. Microbiol. 2017, 7, 512. [Google Scholar] [CrossRef]
- García-Sánchez, M.; Jiménez-Pelayo, L.; Horcajo, P.; Regidor-Cerrillo, J.; Ólafsson, E.B.; Bhandage, A.K.; Barragan, A.; Werling, D.; Ortega-Mora, L.M.; Collantes-Fernández, E. Differential responses of bovine monocyte-derived macrophages to infection by Neospora caninum isolates of high and low virulence. Front. Immunol. 2019, 10, 915. [Google Scholar] [CrossRef]
- Zhou, D.H.; Yuan, Z.G.; Zhao, F.R.; Li, H.L.; Zhou, Y.; Lin, R.Q.; Zou, F.C.; Song, H.Q.; Xu, M.J.; Zhu, X.Q. Modulation of mouse macrophage proteome induced by Toxoplasma gondii tachyzoites in vivo. Parasitol. Res. 2011, 109, 1637–1646. [Google Scholar] [CrossRef]
- Guleria, I.; Pollard, J.W. The trophoblast is a component of the innate immune system during pregnancy. Nat. Med. 2000, 6, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Wheelhouse, N.; Coyle, C.; Barlow, P.G.; Mitchell, S.; Greub, G.; Baszler, T.; Rae, M.T.; Longbottom, D. Waddlia chondrophila infects and multiplies in ovine trophoblast cells stimulating an inflammatory immune response. PLoS ONE 2014, 9, e102386. [Google Scholar] [CrossRef] [PubMed]
- Konno, A.; Ahn, J.-S.; Kitamura, H.; Hamilton, M.J.; Gebe, J.A.; Aruffo, A.; Davis, W.C. Tissue distribution of CD6 and CD6 ligand in cattle: Expression of the CD6 ligand (CD166) in the autonomic nervous system of cattle and the human. J. Leukoc. Biol. 2001, 69, 944–950. [Google Scholar] [PubMed]
- Zimmerman, A.W.; Joosten, B.; Torensma, R.; Parnes, J.R.; van Leeuwen, F.N.; Figdor, C.G. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood 2006, 107, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Montes, M.J.; Tortosa, C.G.; Borja, C.; Abadia, A.C.; González-Gómez, F.; Ruiz, C.; Olivares, E.G. Constitutive secretion of interleukin-6 by human decidual stromal cells in culture. Regulatory effect of progesterone. Am. J. Reprod. Immunol. 1995, 34, 188–194. [Google Scholar] [CrossRef]
- Steinborna, A.; Geisse, M.; Kaufmann, M. Expression of cytokine receptors in the placenta in term and preterm labour. Placenta 1998, 19, 165–170. [Google Scholar] [CrossRef]
- Steinborn, A.; Von Gall, C.; Hildenbrand, R.; Stutte, H.; Kaufmann, M. Identification of placental cytokine-producing cells in term and preterm labor. Obstet. Gynecol. 1998, 91, 329–335. [Google Scholar] [CrossRef]
- Sibley, L.D.; Qiu, W.; Fentress, S.; Taylor, S.J.; Khan, A.; Hui, R. Forward genetics in Toxoplasma gondii reveals a family of rhoptry kinases that mediates pathogenesis. Eukaryot. Cell 2009, 8, 1085–1093. [Google Scholar] [CrossRef]
- Hakimi, M.A.; Olias, P.; Sibley, L.D. Toxoplasma Effectors Targeting Host Signaling and Transcription. Clin. Microbiol. Rev. 2017, 30, 615–645. [Google Scholar] [CrossRef]
- Ma, L.; Liu, G.; Liu, J.; Li, M.; Zhang, H.; Tang, D.; Liu, Q. Neospora caninum ROP16 play an important role in the pathogenicity by phosphorylating host cell STAT3. Vet. Parasitol. 2017, 243, 135–147. [Google Scholar] [CrossRef]
- Ma, L.; Liu, J.; Li, M.; Fu, Y.; Zhang, X.; Liu, Q. Rhoptry protein 5 (ROP5) is a key virulence factor in Neospora caninum. Front. Microbiol. 2017, 8, 370. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rico-San Román, L.; Horcajo, P.; Regidor-Cerrillo, J.; Fernández-Escobar, M.; Collantes-Fernández, E.; Gutiérrez-Blázquez, D.; Hernaéz-Sánchez, M.L.; Saeij, J.; Ortega-Mora, L.M. Comparative tachyzoite proteome analyses among six Neospora caninum isolates with different virulence. Int. J. Parasitol. 2019. accepted. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, M.A.; Bougdour, A. Toxoplasma’s ways of manipulating the host transcriptome via secreted effectors. Curr. Opin. Microbiol. 2015, 26, 24–31. [Google Scholar] [CrossRef] [PubMed]
- García-Sánchez, M.; Jiménez-Pelayo, L.; Horcajo, P.; Regidor-Cerrillo, J.; Collantes-Fernández, E.; Ortega-Mora, L.M. Gene expression profiling of Neospora caninum in bovine macrophages reveals differences between isolates associated with key parasite functions. Front. Cell Infect. Microbiol. 2019, 9, 354. [Google Scholar] [CrossRef]
- Song, C.; Chiasson, M.A.; Nursimulu, N.; Hung, S.S.; Wasmuth, J.; Grigg, M.E.; Parkinson, J. Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii. Mol. Syst. Biol. 2013, 9, 708. [Google Scholar] [CrossRef]
- Croken, M.M.; Ma, Y.; Markillie, L.M.; Taylor, R.C.; Orr, G.; Weiss, L.M.; Kim, K. Distinct strains of Toxoplasma gondii feature divergent transcriptomes regardless of developmental stage. PLoS ONE 2014, 9, e111297. [Google Scholar] [CrossRef][Green Version]
- Regidor-Cerrillo, J.; Gómez-Bautista, M.; Sodupe, I.; Aduriz, G.; Álvarez-García, G.; Del Pozo, I.; Ortega-Mora, L.M. In vitro invasion efficiency and intracellular proliferation rate comprise virulence-related phenotypic traits of Neospora caninum. Vet. Res. 2011, 42, 41. [Google Scholar] [CrossRef]
- Hambruch, N.; Haeger, J.D.; Dilly, M.; Pfarrer, C. EGF stimulates proliferation in the bovine placental trophoblast cell line F3 via Ras and MAPK. Placenta 2010, 31, 67–74. [Google Scholar] [CrossRef]
- Bridger, P.; Haupt, S.; Klisch, K.; Leiser, R.; Tinneberg, H.; Pfarrer, C. Validation of primary epitheloid cell cultures isolated from bovine placental caruncles and cotyledons. Theriogenology 2007, 68, 592–603. [Google Scholar] [CrossRef]
- Rigbolt, K.T.; Vanselow, J.T.; Blagoev, B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell Proteom. 2011, 10, O110.007450. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Pérez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Sanderson, S.J.; Jones, A.R.; Prieto, J.H.; Yates, J.R.; Bromley, E.; Tomley, F.M.; Lal, K.; Sinden, R.E.; Brunk, B.P.; et al. The proteome of Toxoplasma gondii: Integration with the genome provides novel insights into gene expression and annotation. Genome Biol. 2008, 9. [Google Scholar] [CrossRef]
- Sheiner, L.; Demerly, J.L.; Poulsen, N.; Beatty, W.L.; Lucas, O.; Behnke, M.S.; White, M.W.; Striepen, B. A Systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011, 7, e1002392. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, Y.; Shimoda, N.; Fereig, R.M.; Moritaka, T.; Umeda, K.; Nishimura, M.; Ihara, F.; Kobayashi, K.; Himori, Y.; Suzuki, Y.; et al. Neospora caninum Dense Granule Protein 7 regulates the pathogenesis of neosporosis by modulating host immune response. Appl. Environ. Microbiol. 2018, 84, e01350-18. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regidor-Cerrillo, J.; Xia, D.; Jiménez-Pelayo, L.; García-Sánchez, M.; Collantes-Fernández, E.; Randle, N.; Wastling, J.; Ortega-Mora, L.-M.; Horcajo, P. Proteomic Characterization of Host-Pathogen Interactions during Bovine Trophoblast Cell Line Infection by Neospora caninum. Pathogens 2020, 9, 749. https://doi.org/10.3390/pathogens9090749
Regidor-Cerrillo J, Xia D, Jiménez-Pelayo L, García-Sánchez M, Collantes-Fernández E, Randle N, Wastling J, Ortega-Mora L-M, Horcajo P. Proteomic Characterization of Host-Pathogen Interactions during Bovine Trophoblast Cell Line Infection by Neospora caninum. Pathogens. 2020; 9(9):749. https://doi.org/10.3390/pathogens9090749
Chicago/Turabian StyleRegidor-Cerrillo, Javier, Dong Xia, Laura Jiménez-Pelayo, Marta García-Sánchez, Esther Collantes-Fernández, Nadine Randle, Jonathan Wastling, Luis-Miguel Ortega-Mora, and Pilar Horcajo. 2020. "Proteomic Characterization of Host-Pathogen Interactions during Bovine Trophoblast Cell Line Infection by Neospora caninum" Pathogens 9, no. 9: 749. https://doi.org/10.3390/pathogens9090749
APA StyleRegidor-Cerrillo, J., Xia, D., Jiménez-Pelayo, L., García-Sánchez, M., Collantes-Fernández, E., Randle, N., Wastling, J., Ortega-Mora, L.-M., & Horcajo, P. (2020). Proteomic Characterization of Host-Pathogen Interactions during Bovine Trophoblast Cell Line Infection by Neospora caninum. Pathogens, 9(9), 749. https://doi.org/10.3390/pathogens9090749