Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans
Abstract
:1. Introduction
2. Discovery of UMI in Fungi and Cryptococcus
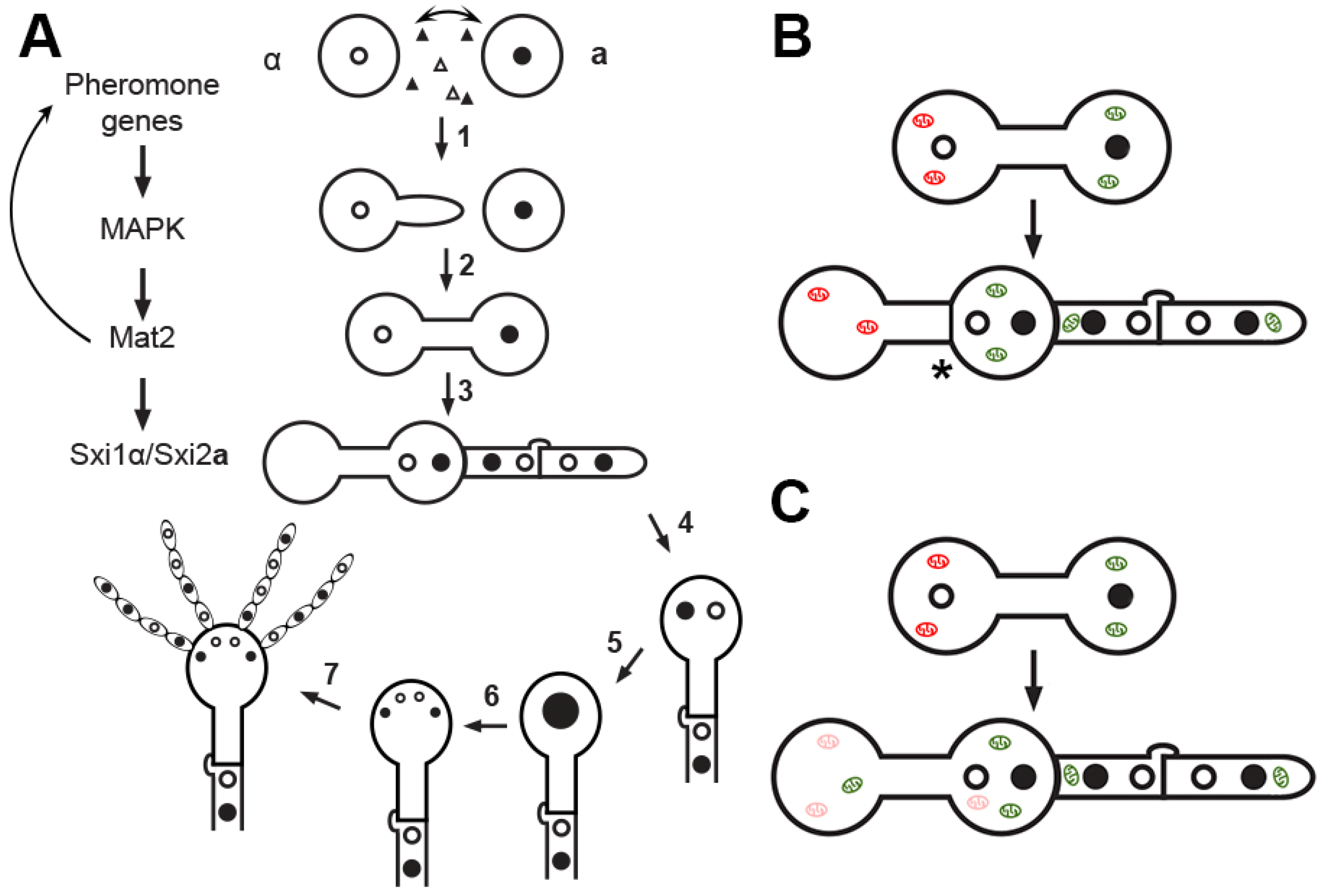
3. Cellular Features during Mating and Uniparental Mitochondrial Inheritance in C. neoformans
4. Factors Important for Uniparental Mitochondrial Inheritance in C. neoformans
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F.; Müller, M. The hydrogen hypothesis for the first eukaryote. Nature 1998, 392, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Mposhi, A.; Van Der Wijst, M.G.; Faber, K.N.; Rots, M.G. Regulation of mitochondrial gene expression the epigenetic enigma. Front. Biosci. 2017, 22, 1099–1113. [Google Scholar] [CrossRef] [Green Version]
- Birky, C.W. Uniparental inheritance of organelle genes. Curr. Biol. 2008, 18. [Google Scholar] [CrossRef] [Green Version]
- Kuroiwa, T. 100 years since the discovery of non-Mendelian plastid phenotypes. J. Plant Res. 2010, 123, 125–129. [Google Scholar] [CrossRef]
- Giles, R.E.; Blanc, H.; Cann, H.M.; Wallace, D.C. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 6715–6719. [Google Scholar] [CrossRef] [Green Version]
- Ankel-Simons, F.; Cummins, J.M. Misconceptions about mitochondria and mammalian fertilization: Implications for theories on human evolution. Proc. Natl. Acad. Sci. USA 1996, 93, 13859–13863. [Google Scholar] [CrossRef] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Eggs. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Sperm. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Sato, M.; Sato, K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1833, 1979–1984. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Henkel, R.; Sengupta, P.; Agarwal, A. Physiological Role of ROS in Sperm Function; Springer Science and Business Media LLC: Berlin, Germany, 2020; pp. 337–345. [Google Scholar]
- Latorre-Pellicer, A.; Lechuga-Vieco, A.V.; Johnston, I.G.; Hämäläinen, R.H.; Pellico, J.; Justo-Méndez, R.; Fernández-Toro, J.M.; Clavería, C.; Guaras, A.; Sierra, R.; et al. Regulation of Mother-to-Offspring Transmission of mtDNA Heteroplasmy. Cell Metab. 2019, 30, 1120–1130.e5. [Google Scholar] [CrossRef]
- Goodenough, U.; Heitman, J. Origins of Eukaryotic Sexual Reproduction. Cold Spring Harb. Perspect. Biol. 2014, 6, a016154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ephrussi, B. Nucleo-Cytoplasmic Relations in Micro-Organisms. Their Bearing on Cell Heredity and Differentiation; Oxford University Press: Oxford, UK, 1953. [Google Scholar]
- Birky, C.W. The Inheritance of Genes in Mitochondria and Chloroplasts: Laws, Mechanisms, and Models. Annu. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Feretzaki, M.; Sun, S.; Wang, X.; Heitman, J. Sex in Fungi. Ann. Rev. Genet. 2011, 45, 405–430. [Google Scholar] [CrossRef]
- Berger, K.H.; Yaffe, M.P. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 2000, 8, 508–513. [Google Scholar] [CrossRef]
- Dujon, B. Mitochondrial genetics revisited. Yeast 2020, 37, 191–205. [Google Scholar] [CrossRef]
- Xu, J.; Ali, R.Y.; Gregory, D.A.; Amick, D.; Lambert, S.E.; Yoell, H.J.; Vilgalys, R.; Mitchell, T.G. Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr. Microbiol. 2000, 40, 269–273. [Google Scholar] [CrossRef]
- Yan, Z.; Hull, C.M.; Sun, S.; Heitman, J.; Xu, J. The mating type-specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet. 2006, 51, 187–195. [Google Scholar] [CrossRef]
- McClelland, C.M.; Chang, Y.C.; Varma, A.; Kwon-Chung, K. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 2004, 12, 208–212. [Google Scholar] [CrossRef]
- Sun, S.; Fu, C.; Ianiri, G.; Heitman, J. The Pheromone and Pheromone Receptor Mating-Type Locus Is Involved in Controlling Uniparental Mitochondrial Inheritance in Cryptococcus. Genet. 2020, 214, 703–717. [Google Scholar] [CrossRef]
- Nishimura, Y.; Shikanai, T.; Kawamoto, S.; Toh-E, A. Step-wise elimination of α-mitochondrial nucleoids and mitochondrial structure as a basis for the strict uniparental inheritance in Cryptococcus neoformans. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Hull, C.M.; Heitman, J. Genetics ofCryptococcus neoformans. Ann. Rev. Genet. 2002, 36, 557–615. [Google Scholar] [CrossRef] [PubMed]
- Lackner, L.L.; Ping, H.; Graef, M.; Murley, A.; Nunnari, J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA 2013, 110, E458–E467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chacko, L.A.; Mehta, K.; Ananthanarayanan, V. Cortical tethering of mitochondria by the anchor protein Mcp5 enables uniparental inheritance. J. Cell Biol. 2019, 218, 3560–3571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.; Sun, S.; Shahid, M.; Xu, J. Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet. Biol. 2007, 44, 315–322. [Google Scholar] [CrossRef]
- Gyawali, R.; Lin, X. Prezygotic and Postzygotic Control of Uniparental Mitochondrial DNA Inheritance in Cryptococcus neoformans. mBio 2013, 4, e00112-13. [Google Scholar] [CrossRef] [Green Version]
- Kruzel, E.K.; Giles, S.S.; Hull, C.M. Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity. Genetics 2012, 191, 435–449. [Google Scholar] [CrossRef] [Green Version]
- Galgoczy, D.J.; Cassidy-Stone, A.; Llinás, M.; O’Rourke, S.M.; Herskowitz, I.; DeRisi, J.L.; Johnson, A.D. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2004, 101, 18069–18074. [Google Scholar] [CrossRef] [Green Version]
- Mead, M.E.; Stanton, B.C.; Kruzel, E.K.; Hull, C.M. Targets of the Sex Inducer homeodomain proteins are required for fungal development and virulence in Cryptococcus neoformans. Mol. Microbiol. 2015, 95, 804–818. [Google Scholar] [CrossRef] [Green Version]
- Hull, C.M.; Boily, M.-J.; Heitman, J. Sex-Specific Homeodomain Proteins Sxi1α and Sxi2a Coordinately Regulate Sexual Development in Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 526–535. [Google Scholar] [CrossRef] [Green Version]
- Hull, C.M.; Davidson, R.C.; Heitman, J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1alpha. Genes Dev. 2002, 16, 3046–3060. [Google Scholar] [CrossRef] [Green Version]
- Mead, M.E.; Hull, C.M. Transcriptional control of sexual development in Cryptococcus neoformans. J. Microbiol. 2016, 54, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner-Vogel, G.; Lämmer, F.; Kämper, J.; Basse, C.W. Uniparental mitochondrial DNA inheritance is not affected in Ustilago maydis Δatg11 mutants blocked in mitophagy. BMC Microbiol. 2015, 15, 23. [Google Scholar] [CrossRef] [Green Version]
- Fedler, M.; Luh, K.-S.; Stelter, K.; Nieto-Jacobo, F.; Basse, C.W. The a2 Mating-Type Locus Genes lga2 and rga2 Direct Uniparental Mitochondrial DNA (mtDNA) Inheritance and Constrain mtDNA Recombination During Sexual Development of Ustilago maydis. Genetics 2008, 181, 847–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberto, T.N.; Lima, R.F.; Pascon, R.C.; Idnurm, A.; Vallim, M.A. Biological functions of the autophagy-related proteins Atg4 and Atg8 in Cryptococcus neoformans. PLoS ONE 2020, 15, e0230981. [Google Scholar] [CrossRef] [Green Version]
- Umen, J.G.; Goodenough, U.W. Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev. 2001, 15, 2585–2597. [Google Scholar] [CrossRef] [PubMed]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitin tag for sperm mitochondria. Nature 1999, 402, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 2000, 63, 582–590. [Google Scholar] [CrossRef]
| Cross Description | Cell Type Tested | mtDNA Inheritance %, a/α | Reference |
|---|---|---|---|
| Aα × Da | Basidiospores | Uniparental from MATa 100/0, n = 446 | [20] |
| Aα × Da | Diploid zygote | Uniparental from MATa 82/4, n = 50 | [29] |
| Aα MAT2OE × Da | Diploid zygote | Biparental (mostly from MATα) 35/60, n = 87 | [29] |
| Dα × Aa sxi2aΔ | Diploid zygote | Biparental (mostly from MATα) 24.5/69.4, n = 49 | [29] |
| Dα sxi1αΔ × Aa | Diploid zygote | Biparental (mostly from MATα) 25/75, n = 48 | [29] |
| Dα sxi1αΔ × Aa MAT2OE | Diploid zygote | Uniparental from MATa 73.8/23.8, n = 42 | [29] |
| Da (mtA) atg8Δ × Dα (mtD) atg8Δ | Diploid zygote | Uniparental from MATa 92.3/7.7, n = 52 | [24] |
| Da (mtA) nuc1Δ × Dα (mtD) nuc1Δ | Diploid zygote | Uniparental from MATa 95.7/4.3, n = 47 | [24] |
| Da (mtA) × Dα (mtD) UV radiation | Diploid zygote | Biparental 54/40, n = 163 | [28] |
| Da (mtA) × Dα (mtD) High temperature (33 °C) | Diploid zygote | Biparental 53/40, n = 184 | [28] |
| Aα crg1Δ × Aa crg1Δ | Basidiospores | Biparental (mostly from MATa) 80/20, n = 40 | [23] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matha, A.R.; Lin, X. Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Pathogens 2020, 9, 743. https://doi.org/10.3390/pathogens9090743
Matha AR, Lin X. Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Pathogens. 2020; 9(9):743. https://doi.org/10.3390/pathogens9090743
Chicago/Turabian StyleMatha, Amber R., and Xiaorong Lin. 2020. "Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans" Pathogens 9, no. 9: 743. https://doi.org/10.3390/pathogens9090743
APA StyleMatha, A. R., & Lin, X. (2020). Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Pathogens, 9(9), 743. https://doi.org/10.3390/pathogens9090743





