Serial Passage of Cryptococcus neoformans in Galleria mellonella Results in Increased Capsule and Intracellular Replication in Hemocytes, but Not Increased Resistance to Hydrogen Peroxide
Abstract
:1. Introduction
2. Results
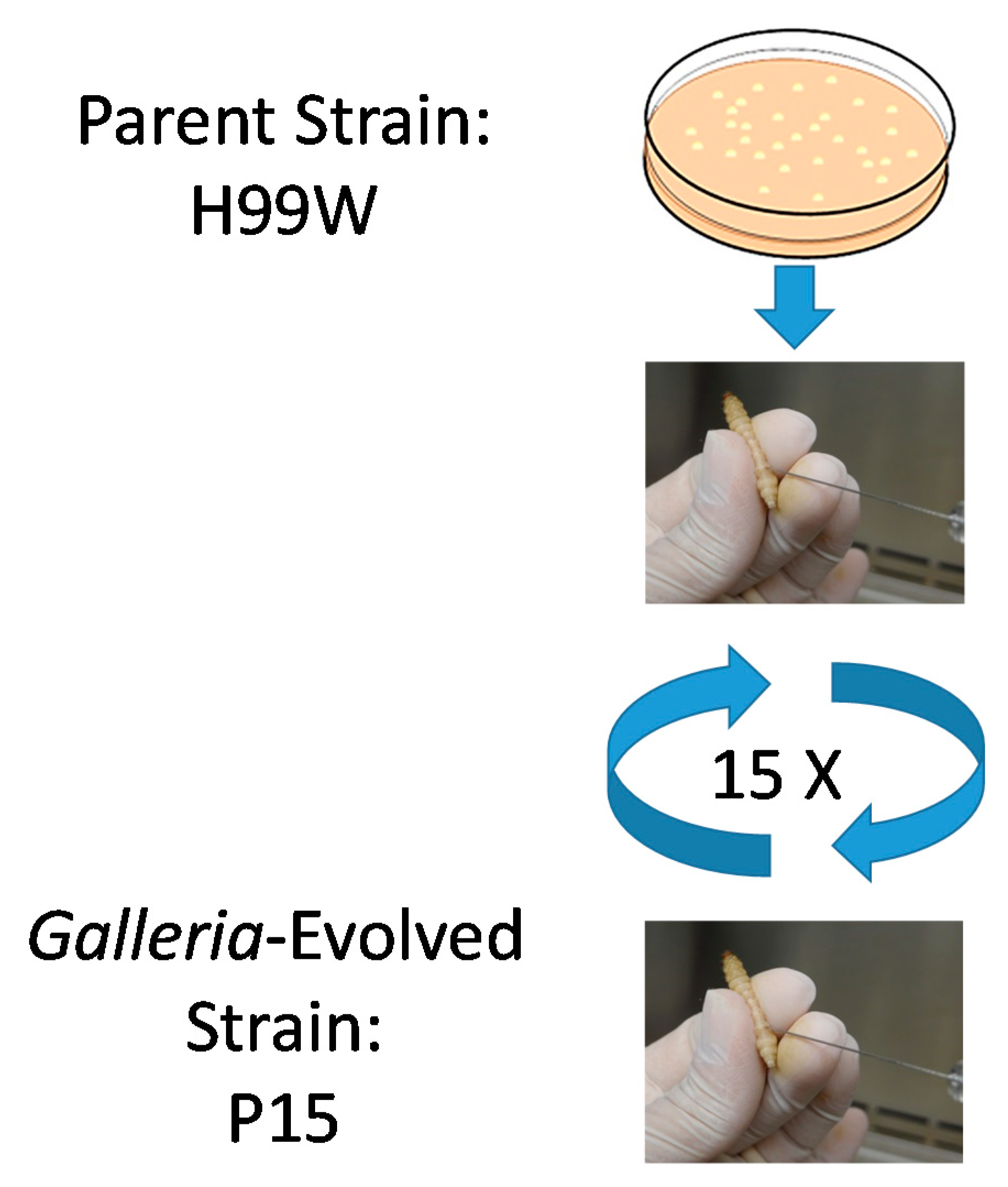
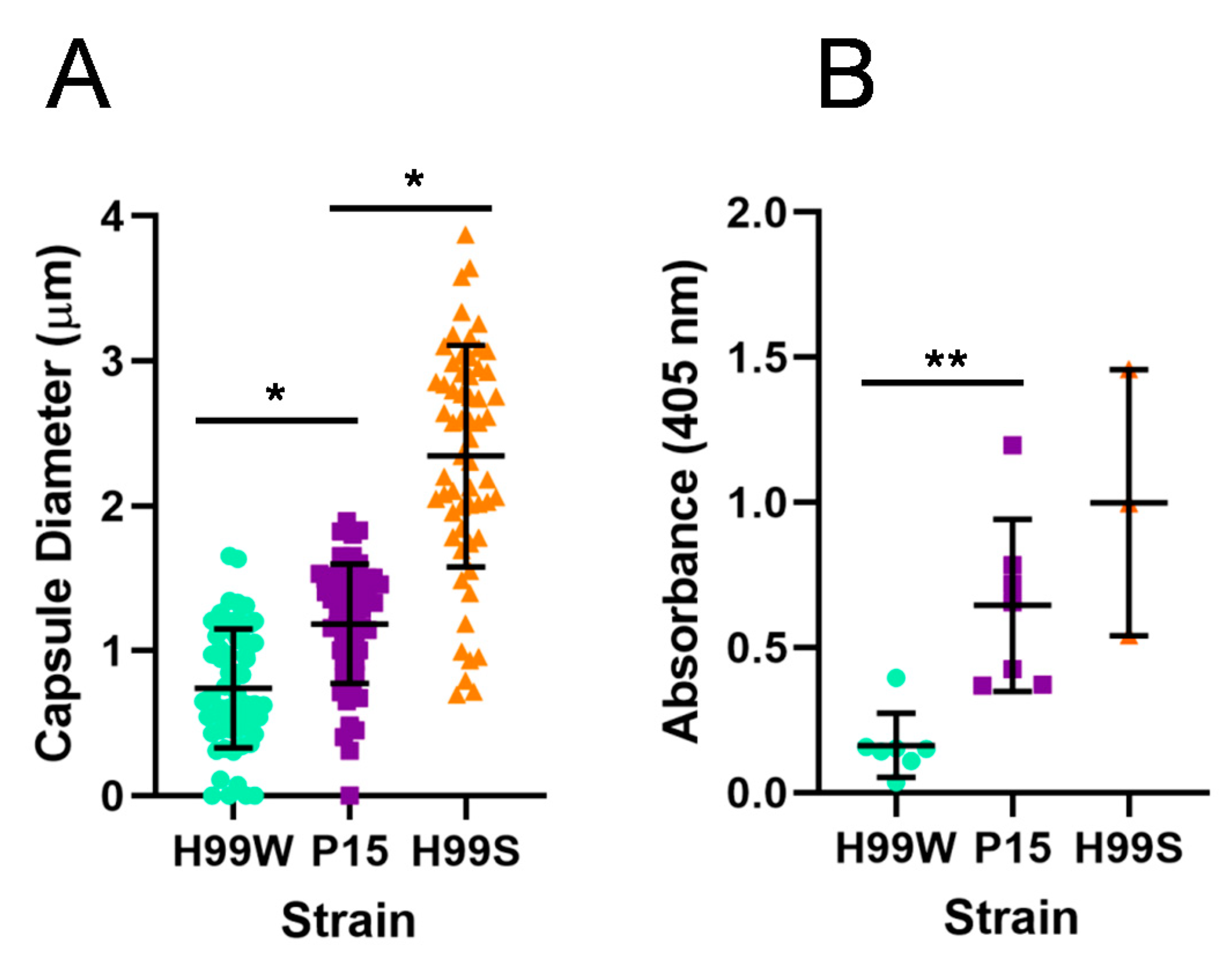
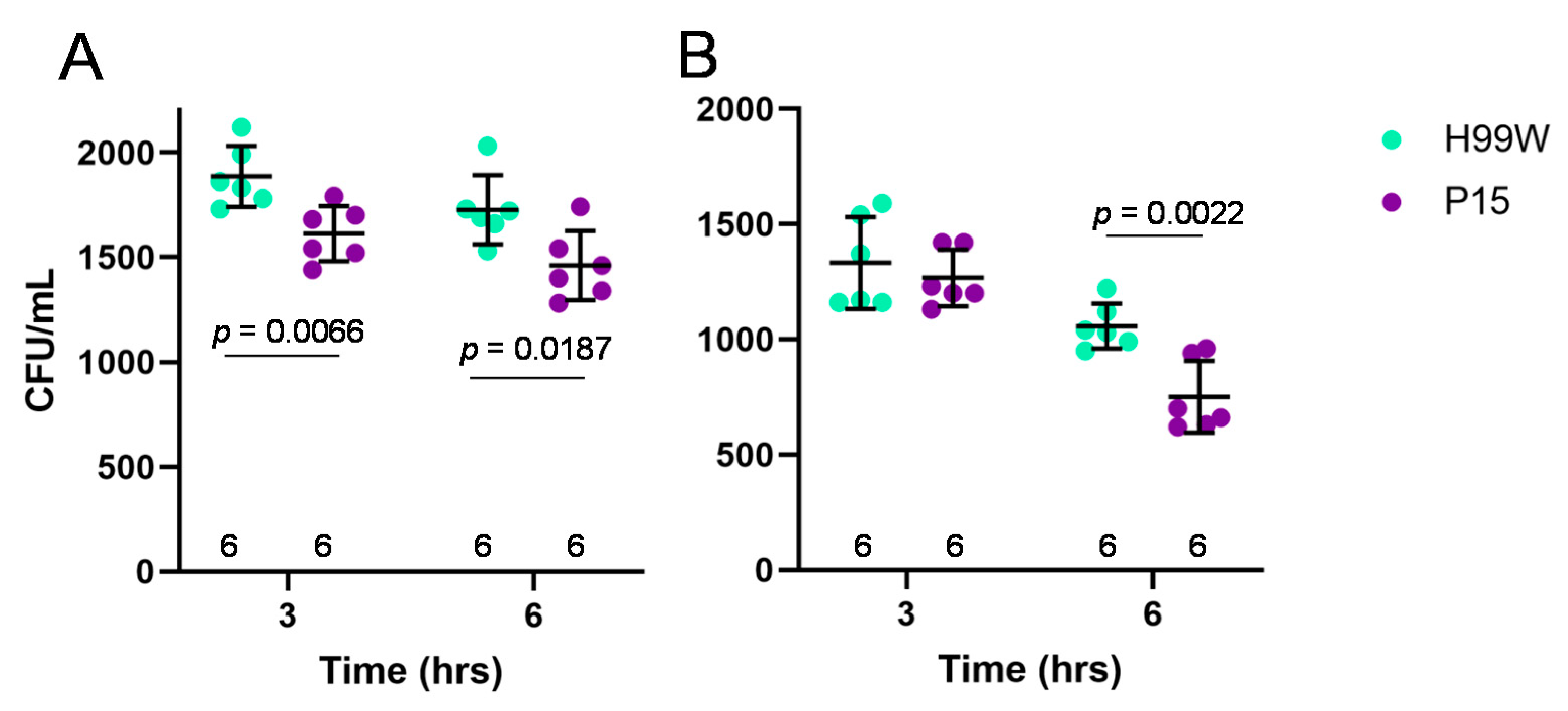
2.1. Evidence for Adaptation
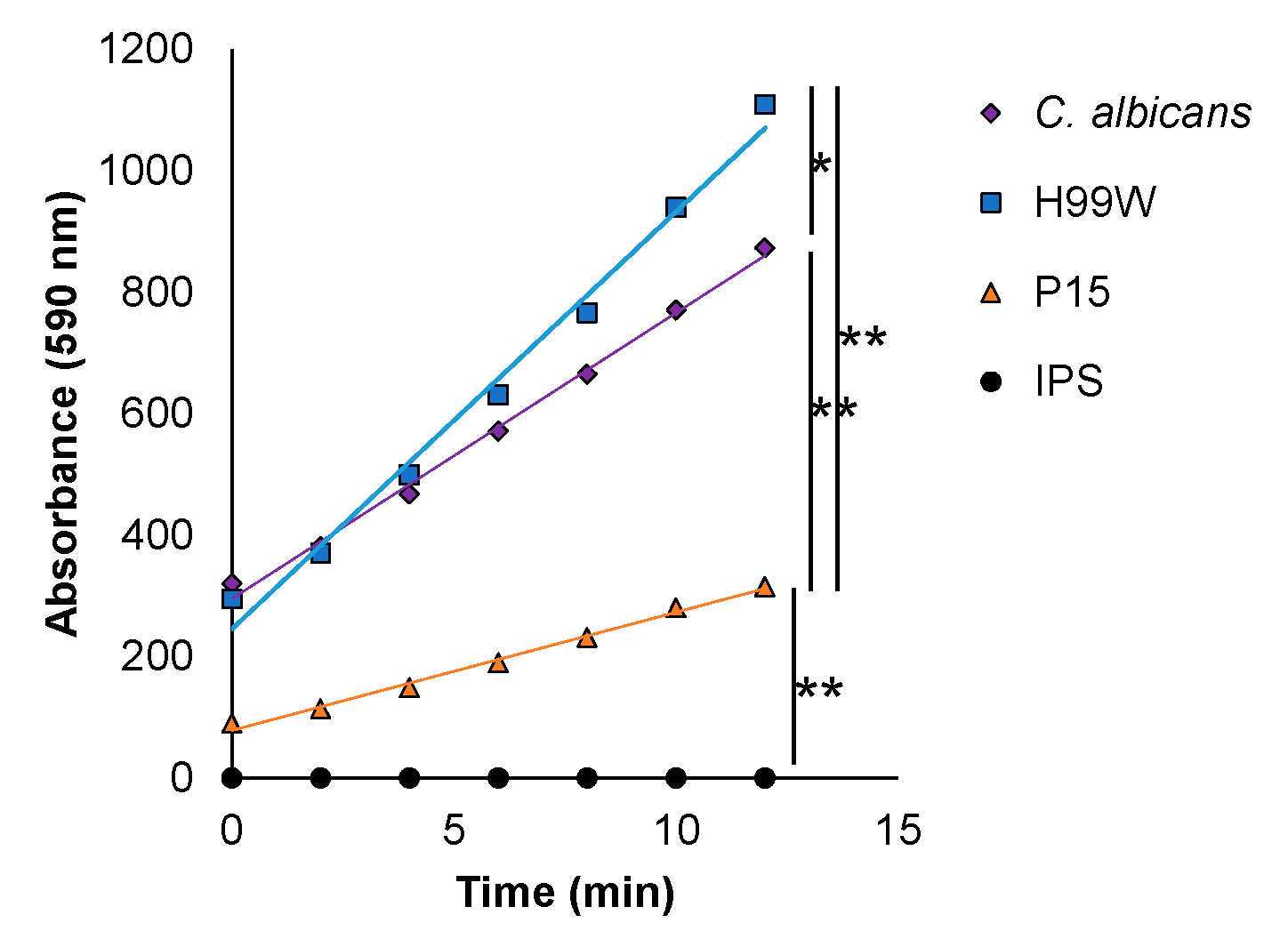
2.2. Identifying a Putative Adaptation Mechanism
2.3. Validating the Putative Adaptation Mechanism
2.4. Experimental Evidence for Mechanism
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Passages
4.3. In Vitro Growth Curves
4.4. In Vivo Galleria Fungal Burden
4.5. Capsule Size
4.6. GXM Release
4.7. Mouse Macrophage Death and C. neoformans Intracellular Replication
4.8. Isolation and Culture of Human Monocytes
4.9. Human Macrophage Death and C. neoformans Intracellular Replication
4.10. Resistance to Hydrogen Peroxide
4.11. Resistance to Nitrosative Stress
4.12. Microarray Analysis
4.13. STRING Analysis
4.14. qRT-PCR
4.15. Hemocyte Hydrogen Peroxide Production
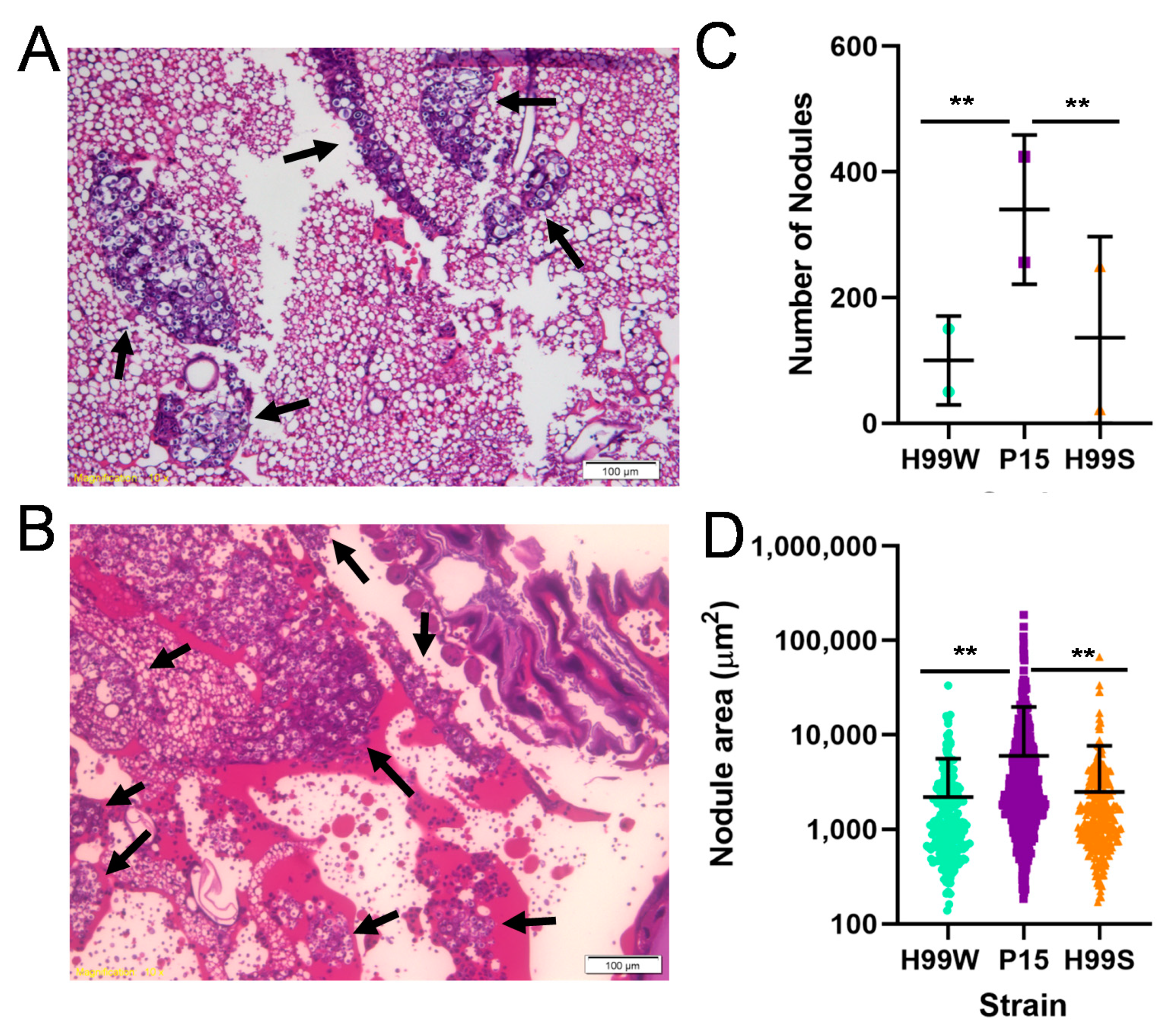
4.16. Histology
4.17. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J., 3rd; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Mylonakis, E.; Moreno, R.; El Khoury, J.B.; Idnurm, A.; Heitman, J.; Calderwood, S.B.; Ausubel, F.M.; Diener, A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 2005, 73, 3842–3850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slepneva, I.A.; Glupov, V.V.; Sergeeva, S.V.; Khramtsov, V.V. EPR detection of reactive oxygen species in hemolymph of Galleria mellonella and Dendrolimus superans sibiricus (Lepidoptera) larvae. Biochem. Biophys. Res. Commun. 1999, 264, 212–215. [Google Scholar] [CrossRef] [PubMed]
- McClelland, E.E.; Casadevall, A.; Eisenman, H.C. Pathogenesis of Cryptococcus neoformans. In New Insights in Medical Mycology; Kavanagh, K., Ed.; Springer: Dordrecht, The Netherlands, 2007; Volume 1, pp. 131–157. [Google Scholar]
- Grijpstra, J.; Tefsen, B.; van Die, I.; de Cock, H. The Cryptococcus neoformans cap10 and cap59 mutant strains, affected in glucuronoxylomannan synthesis, differentially activate human dendritic cells. FEMS Immunol. Med. Microbiol. 2009, 57, 142–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.C.; Kwon-Chung, K.J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994, 14, 4912–4919. [Google Scholar] [CrossRef]
- Upadhya, R.; Campbell, L.T.; Donlin, M.J.; Aurora, R.; Lodge, J.K. Global transcriptome profile of Cryptococcus neoformans during exposure to hydrogen peroxide induced oxidative stress. PLoS ONE 2013, 8, e55110. [Google Scholar] [CrossRef] [Green Version]
- Missall, T.A.; Pusateri, M.E.; Lodge, J.K. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 2004, 51, 1447–1458. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995, 64, 97–112. [Google Scholar] [CrossRef]
- Minagawa, N.; Koga, S.; Nakano, M.; Sakajo, S.; Yoshimoto, A. Possible involvement of superoxide anion in the induction of cyanide-resistant respiration in Hansenula anomala. FEBS Lett. 1992, 302, 217–219. [Google Scholar] [CrossRef] [Green Version]
- Fenton, H.J.H. Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 1894, 65, 899–910. [Google Scholar] [CrossRef] [Green Version]
- Jolly, W.L. The inorganic chemistry of nitrogen. In The Inorganic Chemistry of Nitrogen; W.A. Benjamin, Inc.: New York, NY, USA, 1964; pp. 68–79. [Google Scholar] [CrossRef]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef] [PubMed]
- Denham, S.T.; Verma, S.; Reynolds, R.C.; Worne, C.L.; Daugherty, J.M.; Lane, T.E.; Brown, J.C.S. Regulated release of cryptococcal polysaccharide drives virulence and suppresses immune cell infiltration into the central nervous system. Infect. Immun. 2018, 86, e00662-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neely, M.N.; Pfeifer, J.D.; Caparon, M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 2002, 70, 3904–3914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, A.; Saraceni, P.R.; Merino, S.; Figueras, A.; Tomas, J.M.; Novoa, B. The animal model determines the results of Aeromonas virulence factors. Front. Microbiol. 2016, 7, 1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mylonakis, E.; Aballay, A. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect. Immun. 2005, 73, 3833–3841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.W.; Rahme, L.G.; Sternberg, J.A.; Tompkins, R.G.; Ausubel, F.M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 1999, 96, 2408–2413. [Google Scholar] [CrossRef] [Green Version]
- Sebald, J.; Morettini, S.; Podhraski, V.; Lass-Florl, C.; Lusser, A. CHD1 contributes to intestinal resistance against infection by P. aeruginosa in Drosophila melanogaster. PLoS ONE 2012, 7, e43144. [Google Scholar] [CrossRef]
- Lionakis, M.S.; Kontoyiannis, D.P. The growing promise of Toll-deficient Drosophila melanogaster as a model for studying Aspergillus pathogenesis and treatment. Virulence 2010, 1, 488–499. [Google Scholar] [CrossRef] [Green Version]
- Swanson, M.S.; Hammer, B.K. Legionella pneumophila pathogesesis: A fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 2000, 54, 567–613. [Google Scholar] [CrossRef]
- Steenbergen, J.N.; Shuman, H.A.; Casadevall, A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 2001, 98, 15245–15250. [Google Scholar] [CrossRef] [Green Version]
- Bergin, D.; Brennan, M.; Kavanagh, K. Fluctuations in haemocyte density and microbial load may be used as indicators of fungal pathogenicity in larvae of Galleria mellonella. Microbes Infect. 2003, 5, 1389–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, D. Experimental evolution of parasites. Science 1998, 282, 1432–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersohn, A.; Brigulla, M.; Haas, S.; Hoheisel, J.D.; Volker, U.; Hecker, M. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 2001, 183, 5617–5631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergin, D.; Reeves, E.P.; Renwick, J.; Wientjes, F.B.; Kavanagh, K. Superoxide production in Galleria mellonella hemocytes: Identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 2005, 73, 4161–4170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Little, J.R. Deactivation of macrophage oxidative burst in vitro by different strains of Histoplasma capsulatum. Mycopathologia 1995, 132, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 11, 275–288. [Google Scholar] [CrossRef]
- Zaragoza, O.; Chrisman, C.J.; Castelli, M.V.; Frases, S.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L.; Casadevall, A. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell. Microbiol. 2008, 10, 2043–2057. [Google Scholar] [CrossRef] [Green Version]
- Fries, B.C.; Taborda, C.P.; Serfass, E.; Casadevall, A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 2001, 108, 1639–1648. [Google Scholar] [CrossRef]
- Fernandes, K.E.; Brockway, A.; Haverkamp, M.; Cuomo, C.A.; van Ogtrop, F.; Perfect, J.R.; Carter, D.A. Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from Botswanan HIV/AIDS patients. mBio 2018, 9, e02016-18. [Google Scholar] [CrossRef] [Green Version]
- Charlier, C.; Chretien, F.; Baudrimont, M.; Mordelet, E.; Lortholary, O.; Dromer, F. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 2005, 166, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.H.; McClelland, E.E.; McFeeters, H.; McFeeters, R.L. Novel antifungal activity for the lectin scytovirin: Inhibition of Cryptococcus neoformans and Cryptococcus gattii. Front. Microbiol. 2017, 8, 755. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Mukherjee, J.; Scharff, M.D. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 1992, 154, 27–35. [Google Scholar] [CrossRef]
- McClelland, E.E.; Hobbs, L.M.; Rivera, J.; Casadevall, A.; Potts, W.K.; Smith, J.M.; Ory, J.J. The role of host gender in the pathogenesis of Cryptococcus neoformans infections. PLoS ONE 2013, 8, e63632. [Google Scholar] [CrossRef] [Green Version]
- Alspaugh, J.A.; Granger, D.L. Inhibition of Cryptococcus neoformans replication by nitrogen oxides supports the role of these molecules as effectors of macrophage-mediated cytostasis. Infect. Immun. 1991, 59, 2291–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- McClelland, E.E.; Casadevall, A. Strain-related differences in antibody-mediated changes in gene expression are associated with differences in capsule and location of binding. Fungal Genet. Biol. 2012, 49, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Vasicek, O.; Papezikova, I.; Hyrsl, P. Fluorimetric determination of hydrogen peroxide production by the haemocytes of the wax moth Galleria mellonella (Lepidoptera: Pyralidae). Eur. J. Entomol. 2011, 108, 481–485. [Google Scholar] [CrossRef] [Green Version]




| Strain | Experiment 1 | Experiment 2 | Experiment 3 |
|---|---|---|---|
| H99S | 3.49 | 7.26 | 10.53 |
| P15 | 2.14 | 2.03 | 2.50 |
| H99W | 3.16 | 3.30 | 0.95 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.F.; Tansie, S.M.; Shahan, J.R.; Seipelt-Thiemann, R.L.; McClelland, E.E. Serial Passage of Cryptococcus neoformans in Galleria mellonella Results in Increased Capsule and Intracellular Replication in Hemocytes, but Not Increased Resistance to Hydrogen Peroxide. Pathogens 2020, 9, 732. https://doi.org/10.3390/pathogens9090732
Ali MF, Tansie SM, Shahan JR, Seipelt-Thiemann RL, McClelland EE. Serial Passage of Cryptococcus neoformans in Galleria mellonella Results in Increased Capsule and Intracellular Replication in Hemocytes, but Not Increased Resistance to Hydrogen Peroxide. Pathogens. 2020; 9(9):732. https://doi.org/10.3390/pathogens9090732
Chicago/Turabian StyleAli, Muhammad Fariz, Stephen M. Tansie, John R. Shahan, Rebecca L. Seipelt-Thiemann, and Erin E. McClelland. 2020. "Serial Passage of Cryptococcus neoformans in Galleria mellonella Results in Increased Capsule and Intracellular Replication in Hemocytes, but Not Increased Resistance to Hydrogen Peroxide" Pathogens 9, no. 9: 732. https://doi.org/10.3390/pathogens9090732
APA StyleAli, M. F., Tansie, S. M., Shahan, J. R., Seipelt-Thiemann, R. L., & McClelland, E. E. (2020). Serial Passage of Cryptococcus neoformans in Galleria mellonella Results in Increased Capsule and Intracellular Replication in Hemocytes, but Not Increased Resistance to Hydrogen Peroxide. Pathogens, 9(9), 732. https://doi.org/10.3390/pathogens9090732





