Human Metapneumovirus Induces Mucin 19 Which Contributes to Viral Pathogenesis
Abstract
1. Introduction
2. Results
2.1. Predominant Expression of MUC19 in Primary Human Cells
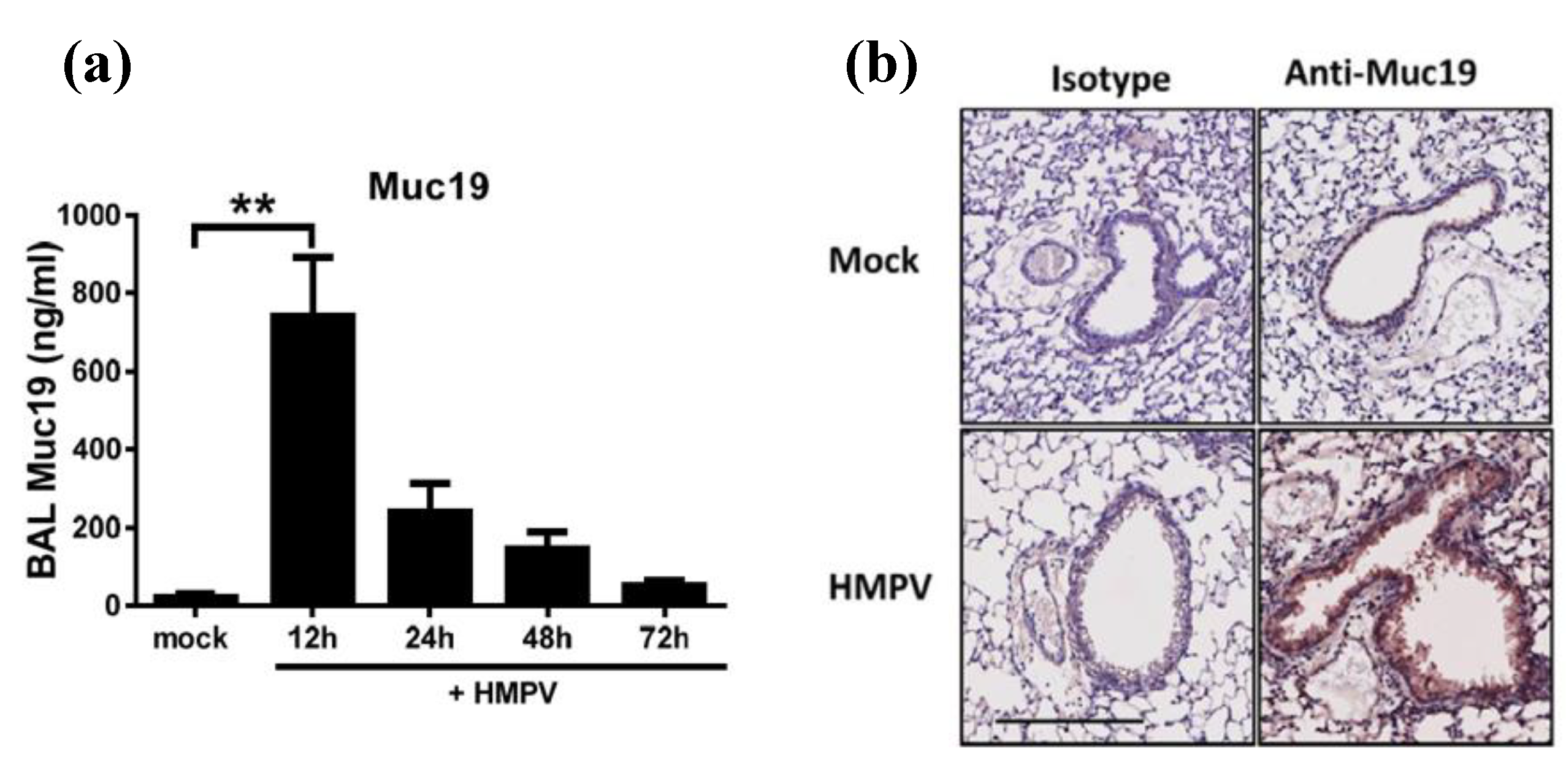
2.2. Predominant Expression of Muc19 in the Lungs of Infected Mice
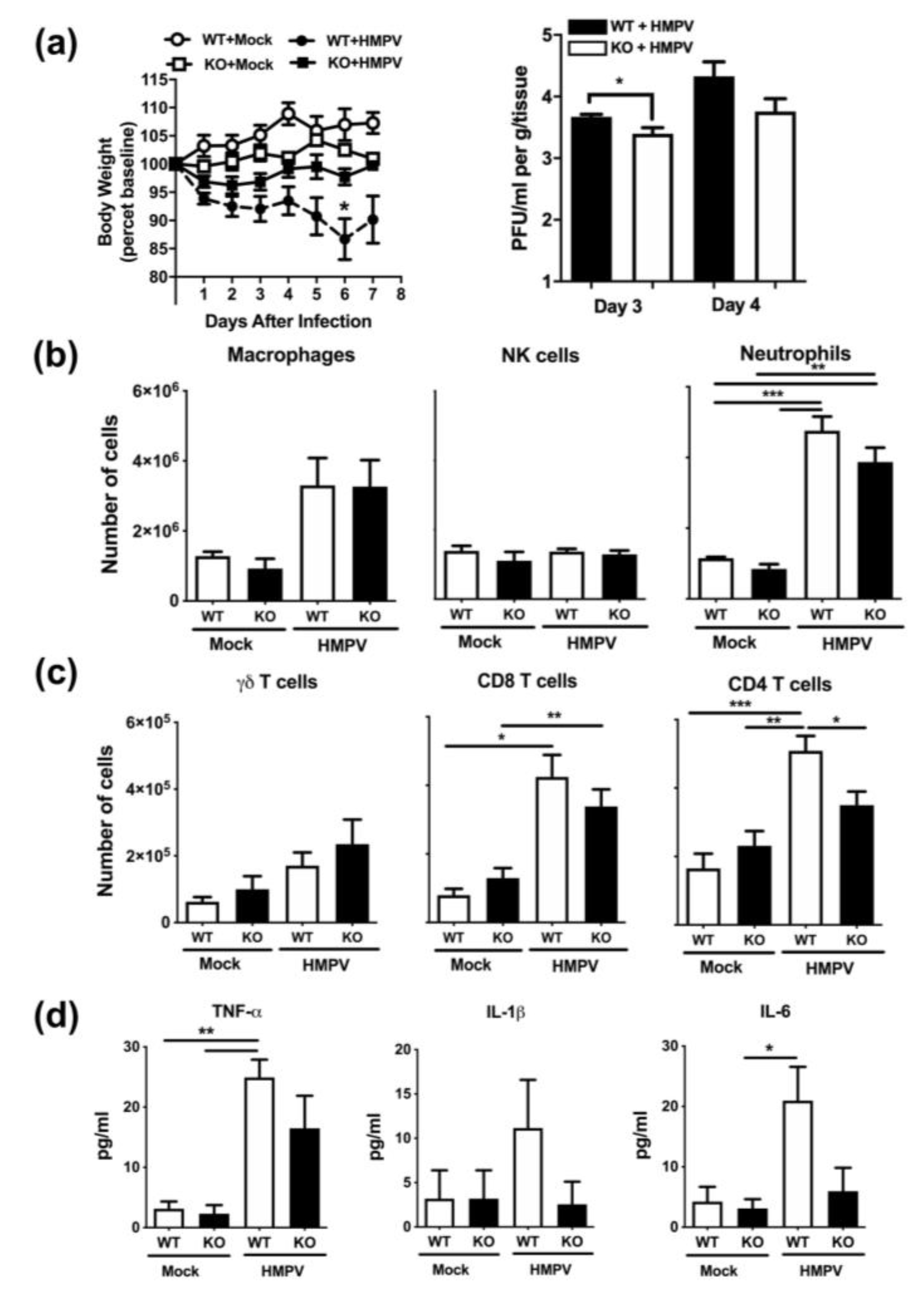
2.3. Muc19 Contributes to Viral Replication and to an Optimal CD4+ T-cell Response in the Lung of HMPV-Infected Mice
3. Discussion
4. Materials and Methods
4.1. Virus Stocks
4.2. Cell Lines and Infection in Vitro
4.3. Mice and Infection Protocol
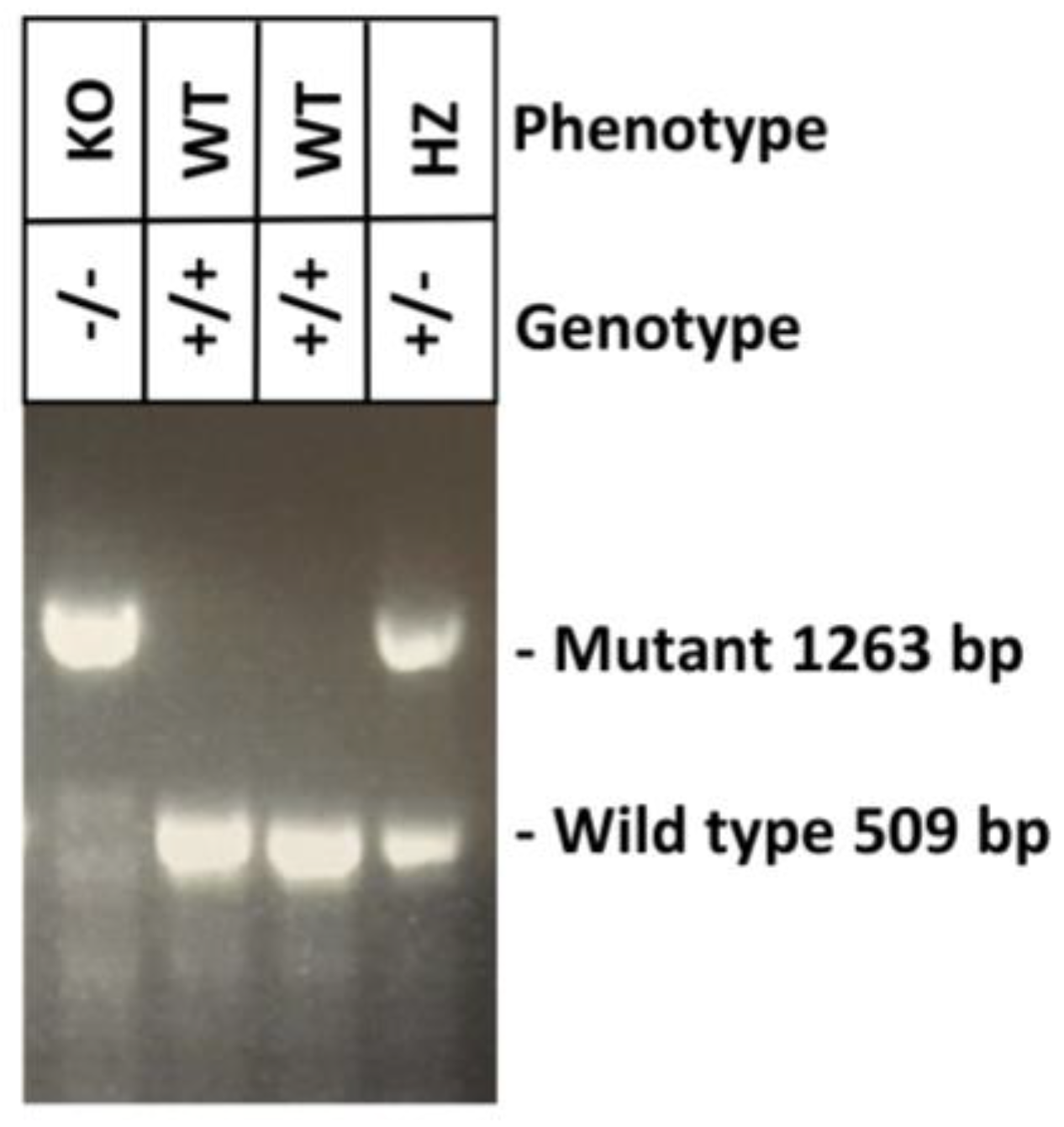
4.4. Genotyping of NFS/N.Cg-Muc19tm2.1Culp/Mmucd Mice
4.5. Sample Collection
4.6. Quantitative Real-Time RT-PCR (qRT-PCR)
4.7. ELISA
4.8. Flow Cytometry
4.9. Immunohistochemistry
4.10. Cytokine Release
4.11. Plaque Assay
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Afonso, C.L.; Amarasinghe, G.K.; Bányai, K.; Bao, Y.; Basler, C.F.; Bavari, S.; Bejerman, N.; Blasdell, K.R.; Briand, F.-X.; Brisese, T.; et al. Taxonomy of the order Mononegavirales: Update 2016. Arch. Virol. 2016, 161, 2351. [Google Scholar] [CrossRef]
- Van den Hoogen, B.G.; De Jong, J.C.; Groen, J.; Kuiken, T.; De Groot, R.; Fouchier, R.A.; Osterhaus, A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001, 7, 719–724. [Google Scholar] [CrossRef]
- Collins, P.L.; Crowe, J. Respiratory syncytial virus and metapneumovirus. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2007; Volume 2, pp. 1601–1646. [Google Scholar]
- Schildgen, V.; Van den Hoogen, B.; Fouchier, R.; Tripp, R.A.; Alvarez, R.; Manoha, C.; Schildgen, O. Human Metapneumovirus: Lessons Learned over the First Decade. Clin. Microbiol. Rev. 2011, 24, 734–754. [Google Scholar] [CrossRef]
- Panda, S.; Mohakud, N.K.; Pena, L.; Kumar, S. Human metapneumovirus: Review of an important respiratory pathogen. Int. J. Infect. Dis. 2014, 25, 45–52. [Google Scholar] [CrossRef]
- Rose, M.C. Mucins: Structure, function, and role in pulmonary diseases. Am. J. Physiol. 1992, 263, L413–L429. [Google Scholar] [CrossRef]
- Vargas, S.O.; Kozakewich, H.P.; Perez-Atayde, A.R.; McAdam, A.J. Pathology of human metapneumovirus infection: Insights into the pathogenesis of a newly identified respiratory virus. Pediatr. Dev. Pathol. 2004, 7, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Hamelin, M.E.; Prince, G.A.; Gomez, A.M.; Kinkead, R.; Boivin, G. Human metapneumovirus infection induces long-term pulmonary inflammation associated with airway onbstruction and hyperresponsiveness in mice. J. Infect. Dis. 2006, 193, 1634–1642. [Google Scholar] [CrossRef]
- Chakraborty, K.; Zhou, Z.; Wakamatsu, N.; Guerrero-Plata, A. Interleukin-12p40 Modulates Human Metapneumovirus-Induced Pulmonary Disease in an Acute Mouse Model of Infection. PLoS ONE 2012, 7, e37173. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.M.; Koo, J.S. Airways mucus: The good, the bad and the sticky. Pharmacol. Ther. 2009, 121, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Lillehoj, E.P.; Kato, K.; Lu, W.; Kim, K.C. Cellular and Molecular Biology of Airway Mucins. Int. Rev. Cell Mol. Biol. 2013, 303, 139–202. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Immunology 2008, 1, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Culp, D.J.; Robinson, B.; Cash, M.N.; Bhattacharyya, I.; Steward, C.; Cuadra-Saenz, G. Salivary mucin 19 glycoproteins innate functions in Streptococcus mutans-induced caries in mice and evidence for expression in human saliva. J. Biol. Chem. 2014, 290, 2993–3008. [Google Scholar] [CrossRef] [PubMed]
- Kolli, D.; Bataki, E.L.; Spetch, L.; Guerrero-Plata, A.; Jewell, A.M.; Piedra, P.A.; Casola, A. T Lymphocytes Contribute to Antiviral Immunity and Pathogenesis in Experimental Human Metapneumovirus Infection. J. Virol. 2008, 82, 8560–8569. [Google Scholar] [CrossRef] [PubMed]
- Jain, B.; Singh, A.K.; Dangi, T.; Agarwal, A.; Verma, A.K.; Dwivedi, M.; Jain, A. High prevalence of human metapneumovirus subtype B in cases presenting as severe acute respiratory illness: An experience at tertiary care hospital. Clin. Respir. J. 2014, 8, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Jayaweera, J.A.; Noordeen, F.; Kothalaweala, S.; Pitchai, F.N.; Rayes, M.L. A case series on common cold to severe bronchiolitis and pneumonia in children following human metapneumovirus infection in Sri Lanka. BMC Res. Notes 2018, 11. [Google Scholar] [CrossRef][Green Version]
- Rose, M.C.; Voynow, J.A. Respiratory Tract Mucin Genes and Mucin Glycoproteins in Health and Disease. Physiol. Rev. 2006, 86, 245–278. [Google Scholar] [CrossRef]
- Baños-Lara, M.D.; Piao, B.; Guerrero-Plata, A. Differential Mucin Expression by Respiratory Syncytial Virus and Human Metapneumovirus Infection in Human Epithelial Cells. Mediat. Inflamm. 2015, 1–7. [Google Scholar] [CrossRef]
- Ekstrand-Hammarström, B.; Akfur, C.M.; Andersson, P.O.; Lejon, C.; Osterlund, L.; Bucht, A. Human primary bronchial epithelial cells respond differently to titanium dioxide nanoparticles than the lung epithelial cell line A549 and BEAS-2B. Nanotoxicology 2012, 6, 623–634. [Google Scholar] [CrossRef]
- Yu, D.; Chen, Y.; Han, J.; Zhang, H.; Chen, X.; Zou, W.; Liu, Z. MUC19 expression in human ocular surface and lacrimal gland and its alteration in Sjögren syndrome patients. Exp. Eye Res. 2008, 86, 403–411. [Google Scholar] [CrossRef]
- Kerschner, J.E.; Khampang, P.; Erbe, C.B.; Kolker, A.; Cioffi, J.A. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj. J. 2009, 26, 1275–1284. [Google Scholar] [CrossRef]
- Kouznetsova, I.; Chwieralski, C.E.; Bälder, R.; Hinz, M.; Braun, A.; Krug, N.; Hoffmann, W. Induced trefoil factor family 1 expression by trans-differentiating Clara cells in a murine asthma model. Am. J. Respir. Cell Mol. Biol. 2007, 36, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Fallon, M.A.; Latchney, L.R.; Hand, A.R.; Johar, A.; Denny, P.A.; Georgel, P.T.; Denny, P.C.; Culp, D.J. The sld mutation is specific for sublingual salivary mucous cells and disrupts apomicin gene expression. Physiol. Genom. 2003, 14, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Horie, M.; Daidoji, T.; Honda, T.; Yasugi, M.; Kuno, A.; Komori, T.; Okuzaki, D.; Narimatsu, H.; Nakaya, T.; et al. Influenza A Virus-Induced Expression of a GalNac Transferase, GALNT3, via MicroRNAs Is Required for Enhanced Viral Replication. J Virol. 2016, 90, 1788–1801. [Google Scholar] [CrossRef]
- Yolken, R.H.; Peterson, J.A.; Vonderfecht, S.L.; Fouts, E.T.; Midthun, K.; Newburg, D.S. Human milk inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Invest. 1992, 90, 1984–1991. [Google Scholar] [CrossRef]
- Habte, H.H.; de Beer, C.; Lotz, Z.E.; Tyler, M.G.; Kahn, D.; Mall, A.S. Inhibition of human immunodeficiency virus type 1 activity by purified human breast milk mucin (MUC1) in an inhibition assay. Neonatology 2008, 93, 162–170. [Google Scholar] [CrossRef]
- Habte, H.H.; Kotwal, G.J.; Lotz, Z.E.; Tyler, M.G.; Abrahams, M.; Rodriques, J.; Kahn, D.; Mall, A.S. Antiviral activity of purified human breast milk mucin. Neonatology 2007, 92, 96–104. [Google Scholar] [CrossRef]
- Habte, H.H.; Mall, A.S.; de Beer, C.; Lotz, Z.E.; Kahn, D. The role of crude human saliva and purified salivary MUC5B and MUC7 mucins in the inhibition of Human Immunodeficiency Virus type 1 in an inhibition assay. Virol. J. 2006, 3, 99. [Google Scholar] [CrossRef]
- Bergey, E.J.; Cho, M.I.; Blumberg, B.M.; Hammarskjold, M.L.; Rekosh, D.; Epstein, L.G.; Levine, M.J. Interaction of HIV-1 and human salivary mucins. J. Acquir. Immune. Defic. Syndr. 1994, 7, 995–1002. [Google Scholar]
- Kennedy, S.; Davis, C.; Abrams, W.R.; Billings, P.C.; Nagashunmugam, T.; Friedman, H.; Malamud, D. Submandibular salivary proteases: Lack of a role in anti-HIV activity. J. Dent. Res. 1998, 77, 1515–1519. [Google Scholar] [CrossRef]
- Habte, H.H.; de Beer, C.; Lotz, Z.E.; Tyler, M.G.; Schoeman, L.; Kahn, D.; Mall, A.S. The inhibition of the Human Immunodeficiency Virus type 1 activity by crude and purified human pregnancy plug mucus and mucins in an inhibition assay. Virol. J. 2008, 5, 59. [Google Scholar] [CrossRef]
- Maeda, T.; Hiraki, M.; Jin, C.; Rajabi, H.; Tagde, A.; Alam, M.; Kufe, D. MUC1-C Induces PD-L1 and Immune Evasion in Triple-Negative Breast Cancer. Cancer Res. 2017, 8, 205–215. [Google Scholar] [CrossRef]
- Nishida, A.; Lau, C.W.; Zhang, M.; Andoh, A.; Shi, H.N.; Mizoguchi, E.; Mizoguchi, A. The Membrane-Bound Mucin Muc1 Regulates T Helper 17-Cell Responses and Colitis in Mice. Gastroenterology 2012, 142, 865–874. [Google Scholar] [CrossRef]
- Voynow, J.A.; Gendler, S.J.; Rose, M.C. Regulation of Mucin Genes in Chronic Inflammatory Airway Diseases. Am. J. Resp. Cell Mol. Biol. 2006, 34, 661–665. [Google Scholar] [CrossRef]
- Agrawal, B.; Longenecker, B.M. MUC1 mucin-mediated regulation of human T cells. Int. Immunol. 2005, 17, 391–399. [Google Scholar] [CrossRef]
- Agrawal, B.; Gupta, N.; Konowalchuk, J.D. MUC1 Mucin: A putative Regulatory (Checkpoint) Molecule of T Cells. Front. Immunol. 2018, 9, 2391. [Google Scholar] [CrossRef] [PubMed]
- Konowalchuk, J.D.; Agrawal, B. MUC1 mucin is expressed on human T- regulatory cells: Function in both co-stimulation and co-inhibition. Cell. Immun. 2012, 272, 193–199. [Google Scholar] [CrossRef]
- Wenzel, U.A.; Jonstrand, C.; Hansson, G.C.; Wick, M.J. CD103+CD11b+ Dendritic Cells Induce Th17 T Cells in Muc2-Deficient Mice with Extensively Spread Colitis. PLoS ONE 2015, 10, e0130750. [Google Scholar] [CrossRef]
- Guerrero-Plata, A.; Baron, S.; Poast, J.S.; Adegboyega, P.A.; Casola, A.; Garofalo, R.P. Activity and Regulation of Alpha Interferon in Respiratory Syncytial Virus and Human Metapneumovirus Experimental Infections. J. Virol. 2005, 79, 10190–10199. [Google Scholar] [CrossRef]
- Cheemarla, N.R.; Baños-Lara, M.D.; Naidu, S.; Guerrero-Plata, A. Neutrophils regulate the lung inflammatory response via γδ T cell infiltration in an experimental mouse model of human metapneumovirus infection. J. Leukoc. Biol. 2017, 101, 1383–1392. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McBride, K.; Banos-Lara, M.d.R.; Cheemarla, N.R.; Guerrero-Plata, A. Human Metapneumovirus Induces Mucin 19 Which Contributes to Viral Pathogenesis. Pathogens 2020, 9, 726. https://doi.org/10.3390/pathogens9090726
McBride K, Banos-Lara MdR, Cheemarla NR, Guerrero-Plata A. Human Metapneumovirus Induces Mucin 19 Which Contributes to Viral Pathogenesis. Pathogens. 2020; 9(9):726. https://doi.org/10.3390/pathogens9090726
Chicago/Turabian StyleMcBride, Kaitlin, Ma. del Rocio Banos-Lara, Nagarjuna R. Cheemarla, and Antonieta Guerrero-Plata. 2020. "Human Metapneumovirus Induces Mucin 19 Which Contributes to Viral Pathogenesis" Pathogens 9, no. 9: 726. https://doi.org/10.3390/pathogens9090726
APA StyleMcBride, K., Banos-Lara, M. d. R., Cheemarla, N. R., & Guerrero-Plata, A. (2020). Human Metapneumovirus Induces Mucin 19 Which Contributes to Viral Pathogenesis. Pathogens, 9(9), 726. https://doi.org/10.3390/pathogens9090726






