Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018
Abstract
1. Introduction
2. Results
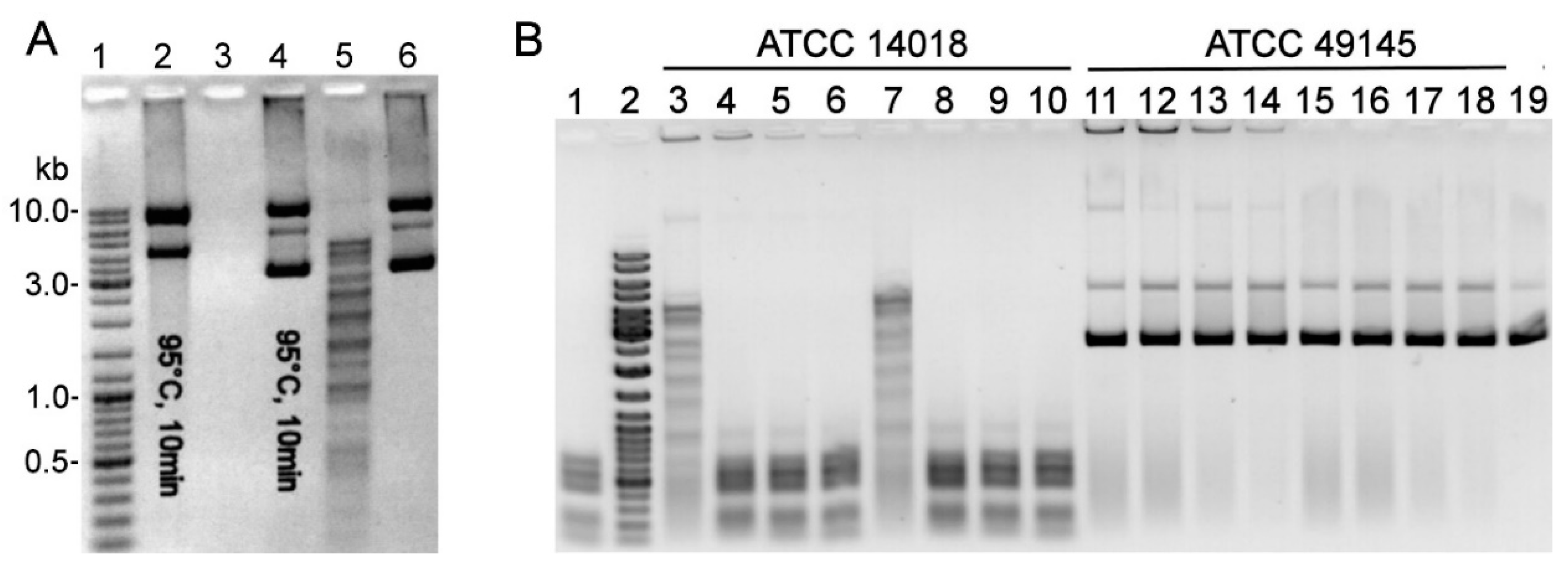
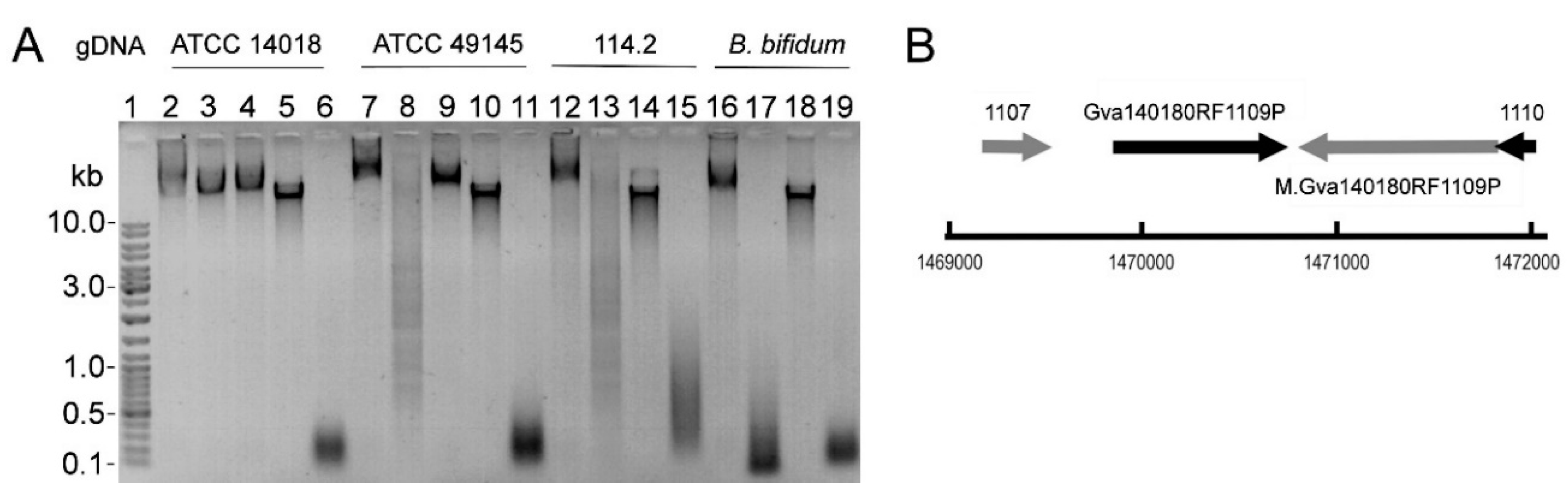
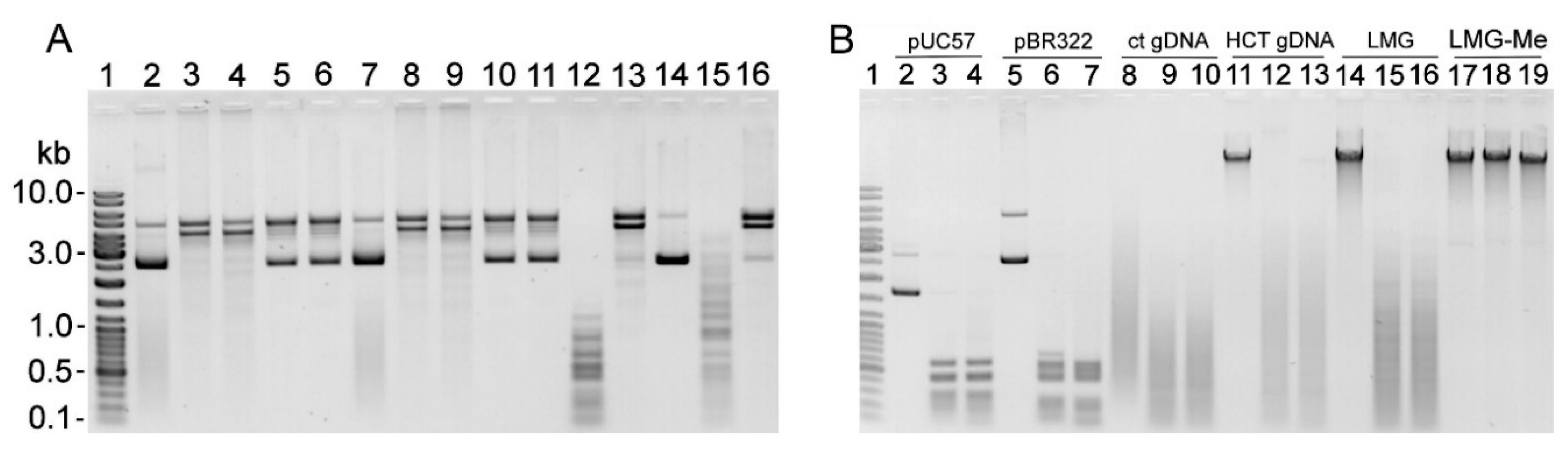
2.1. R-M Systems in Gardnerella spp. and Detection of Restriction Endonuclease Activity
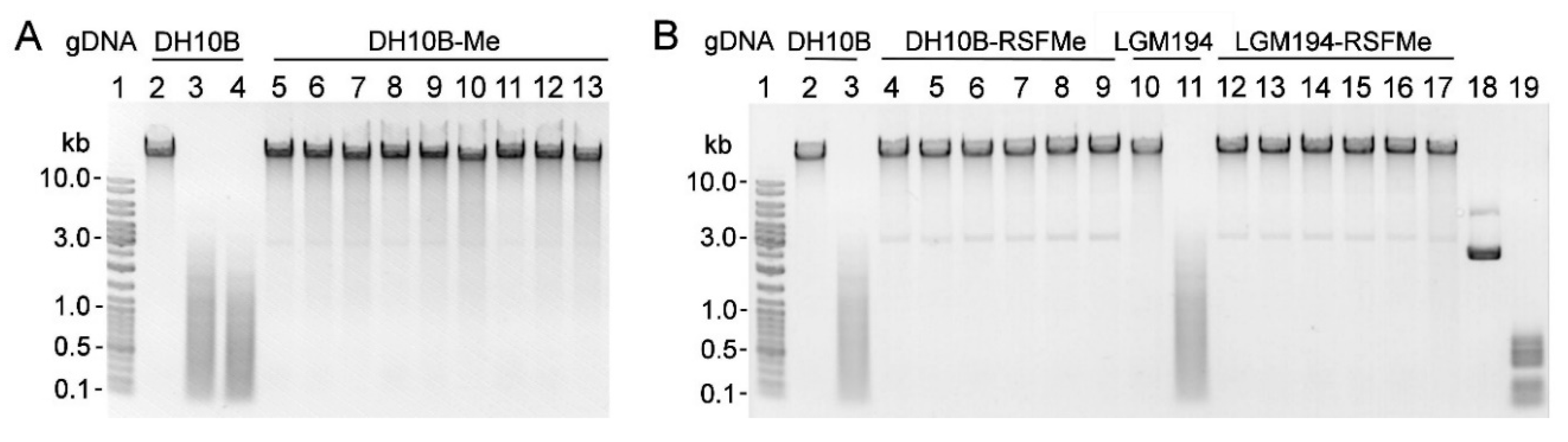
2.2. Cloning and Expression of gva14018IM and gva14018IR in E. coli
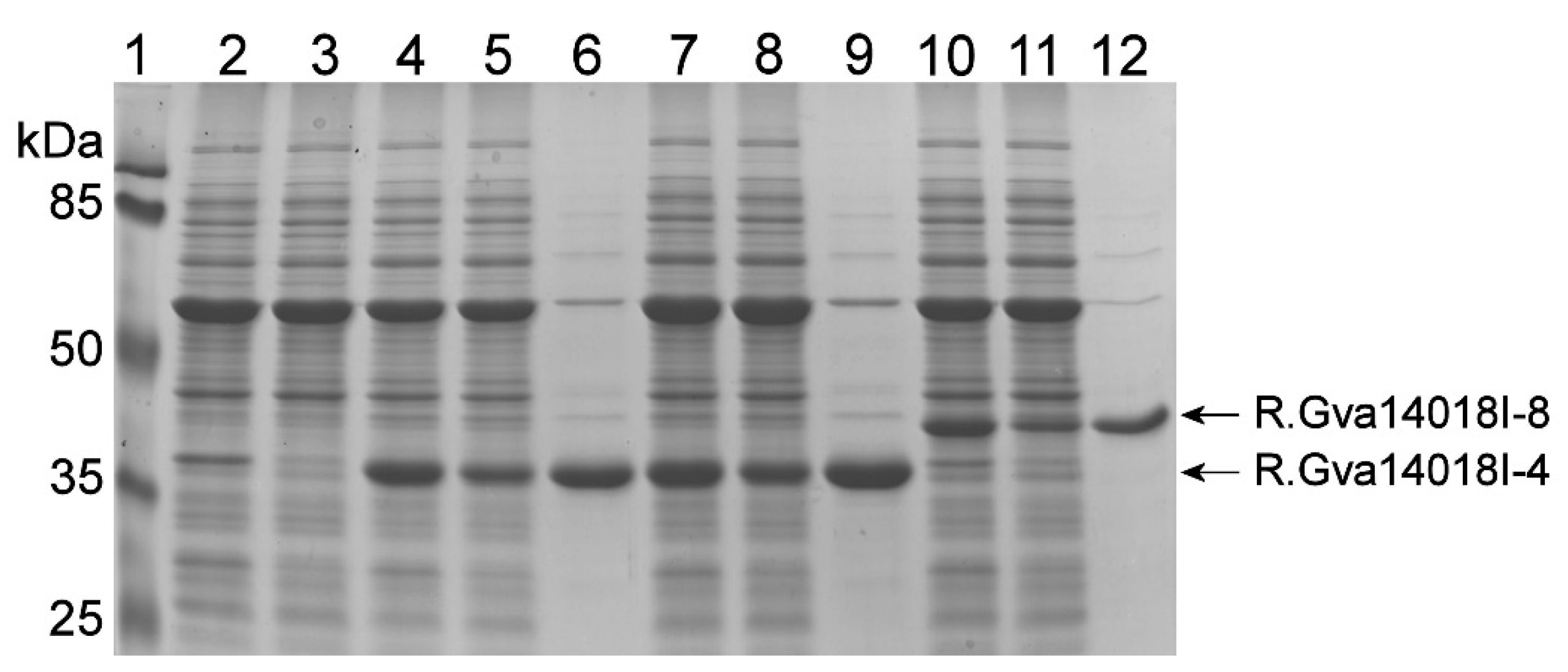
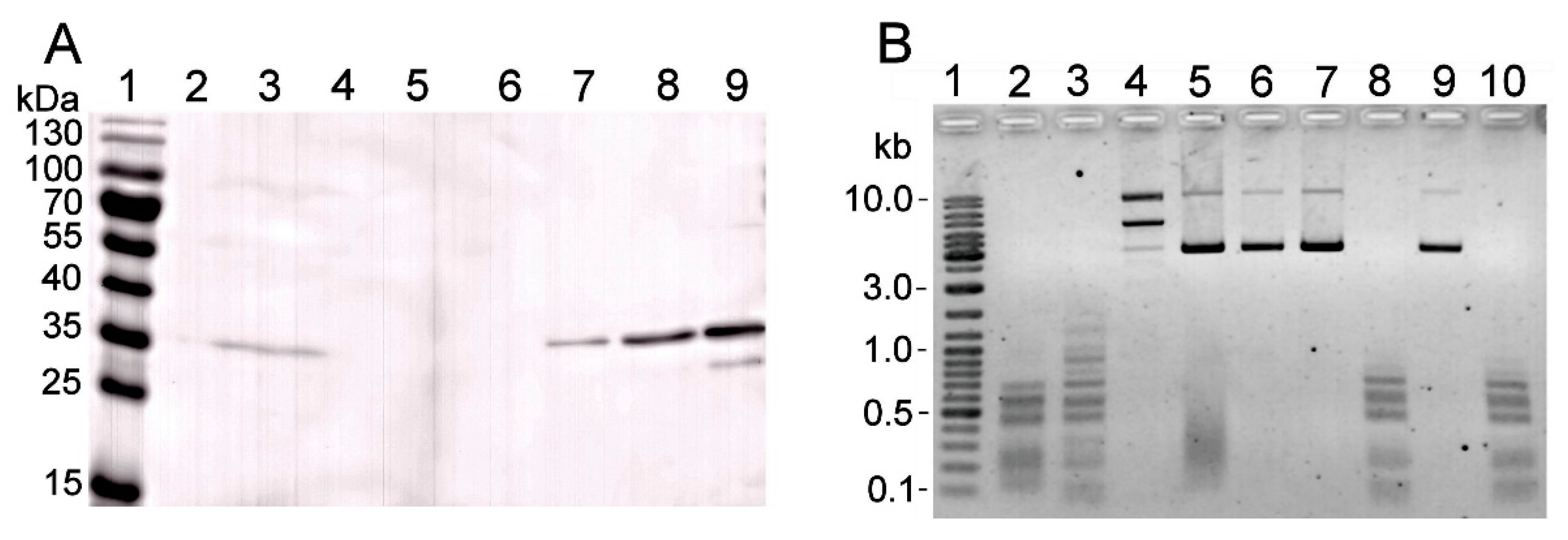
2.3. Expression of Mutated gva14018IR Genes in E. coli
2.4. Polyclonal Antibodies against Recombinant R.Gva14018I-4 also Detected R.Gva14018I
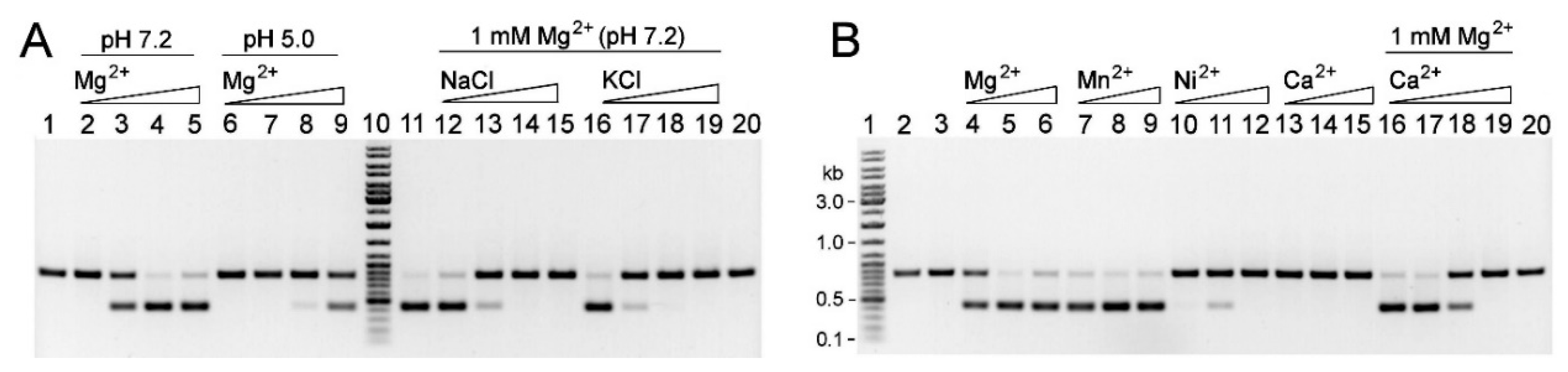
2.5. Characterization of the R.Gva14018I-4 Protein
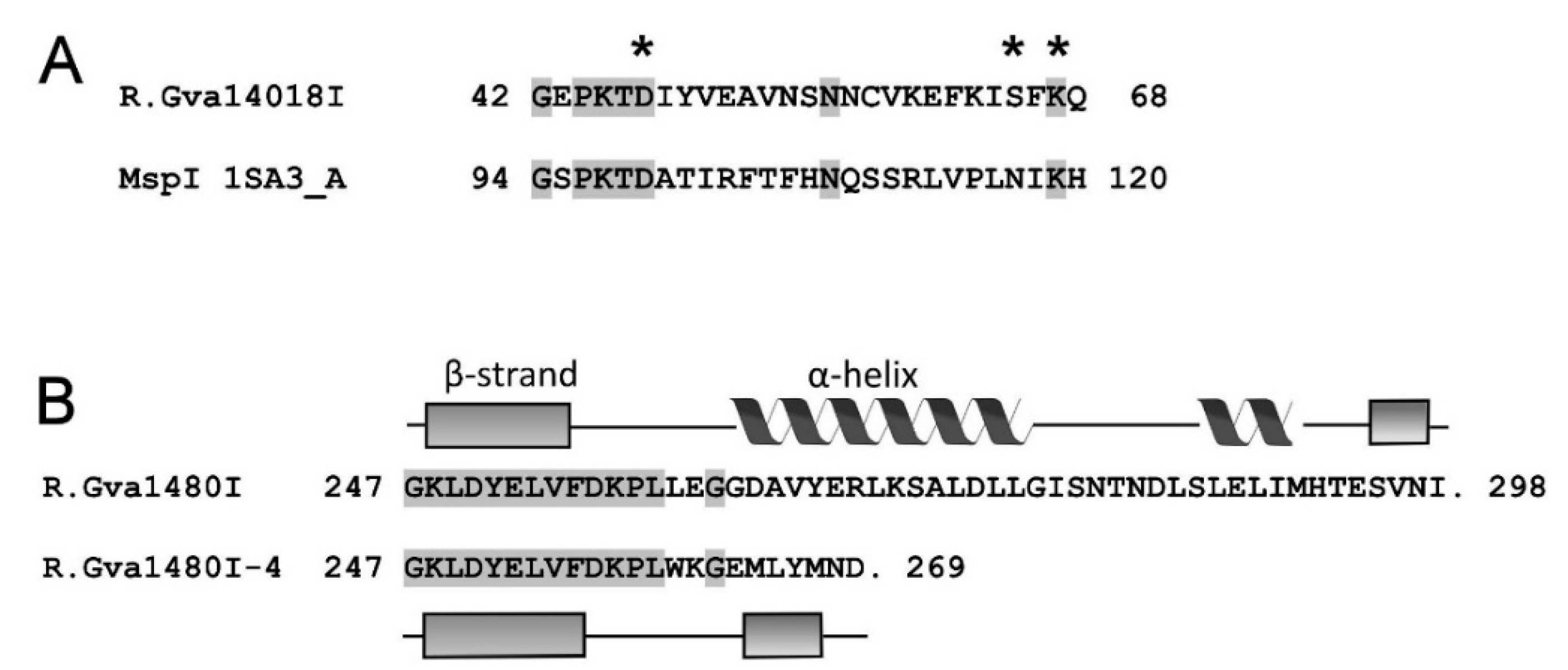
2.6. Bioinformatics Analyses of M.Gva14018I, R.Gva14018I, and R.Gva14018I-4
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultivation Conditions
4.2. Amplification of gva14018IR and gva14018IM Genes
4.3. Cloning of the gva14018IM Gene
4.4. Cloning the gva14018IR Gene
4.5. Expression of R.Gva14018I Mutants in E. coli
4.6. Small-Scale Synthesis of Recombinant Proteins
4.7. Large-scale Protein Synthesis of Gva14018RI-4 and Protein Purification
4.8. Determination of REase Cleavage Site
4.9. REase Activity Assays
4.10. MTase Activity Assay
4.11. Generation of Murine Polyclonal Antibodies Against Recombinant R.Gva14018I-4
4.12. Immunoblot Detection of R.Gva14018I
4.13. Immunoprecipitation and Mass Spectrometry Analysis of Recombinant R.Gva14018I-4
4.14. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bilardi, J.E.; Walker, S.; Temple-Smith, M.; McNair, R.; Mooney-Somers, J.; Bellhouse, C.; Fairley, C.K.; Chen, M.Y.; Bradshaw, C. The burden of bacterial vaginosis: Women’s experience of the physical, emotional, sexual and social impact of living with recurrent bacterial vaginosis. PLoS ONE 2013, 8, e74378. [Google Scholar] [CrossRef] [PubMed]
- Leitich, H.; Kiss, H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract. Res. Clin. Obstet. Gynaecol. 2007, 21, 375–390. [Google Scholar] [CrossRef]
- Dingens, A.S.; Fairfortune, T.S.; Reed, S.; Mitchell, C. Bacterial vaginosis and adverse outcomes among full-term infants: A cohort study. BMC Pregnancy Childbirth 2016, 16, 278. [Google Scholar] [CrossRef]
- Shipitsyna, E.; Khusnutdinova, T.; Budilovskaya, O.; Krysanova, A.; Shalepo, K.; Savicheva, A.; Unemo, M. Bacterial vaginosis-associated vaginal microbiota is an age-independent risk factor for Chlamydia trachomatis, Mycoplasma genitalium and Trichomonas vaginalis infections in low-risk women, St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Mendling, W.; Loening-Baucke, V.; Ladhoff, A.; Swidsinski, S.; Hale, L.P.; Lochs, H. Adherent biofilms in bacterial vaginosis. Obstet. Gynecol. 2005, 106, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Mendling, W.; Dörffel, Y.; Schilling, J.; Halwani, Z.; Jiang, X.F.; Verstraelen, H.; Swidsinski, S. Infection through structured polymicrobial Gardnerella biofilms (StPM-GB). Histol. Histopathol. 2014, 29, 567–587. [Google Scholar] [CrossRef] [PubMed]
- Gelber, S.E.; Aguilar, J.L.; Lewis, K.L.T.; Ratner, A.J. Functional and phylogenetic characterization of vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J. Bacteriol. 2008, 190, 3896–3903. [Google Scholar] [CrossRef]
- Patterson, J.L.; Stull-Lane, A.; Girerd, P.H.; Jefferson, K.K. Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 2010, 156, 392–399. [Google Scholar] [CrossRef]
- Robinson, L.S.; Schwebke, J.; Lewis, W.G.; Lewis, A.L. Identification and characterization of NanH2 and NanH3, enzymes responsible for sialidase activity in the vaginal bacterium Gardnerella vaginalis. J. Biol. Chem. 2019, 294, 5230–5245. [Google Scholar] [CrossRef]
- Zozaya-Hinchliffe, M.; Lillis, R.; Martin, D.H.; Ferris, M.J. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J. Clin. Microbiol. 2010, 48, 1812–1819. [Google Scholar] [CrossRef]
- Srinivasan, S.; Hoffman, N.G.; Morgan, M.T.; Matsen, F.A.; Fiedler, T.L.; Hall, R.W.; Ross, F.J.; McCoy, C.O.; Bumgarner, R.; Marrazzo, J.M.; et al. Bacterial communities in women with bacterial vaginosis: High resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE 2012, 7, e37818. [Google Scholar] [CrossRef] [PubMed]
- Janulaitiene, M.; Paliulyte, V.; Grinceviciene, S.; Zakareviciene, J.; Vladisauskiene, A.; Marcinkute, A.; Pleckaityte, M. Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis. BMC Infect. Dis. 2017, 17, 394. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Earl, J.; Retchless, A.; Hillier, S.L.; Rabe, L.K.; Cherpes, T.L.; Powell, E.; Janto, B.; Eutsey, R.; Hiller, N.L.; et al. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J. Bacteriol. 2012, 194, 3922–3937. [Google Scholar] [CrossRef] [PubMed]
- Hickey, R.J.; Forney, L.J. Gardnerella vaginalis does not always cause bacterial vaginosis. J. Infect. Dis. 2014, 210, 1682–1683. [Google Scholar] [CrossRef]
- Janulaitiene, M.; Gegzna, V.; Baranauskiene, L.; Bulavaitė, A.; Simanavicius, M.; Pleckaityte, M. Phenotypic characterization of Gardnerella vaginalis subgroups suggests differences in their virulence potential. PLoS ONE 2018, 13, e0200625. [Google Scholar] [CrossRef]
- Castro, J.; Jefferson, K.K.; Cerca, N. Genetic heterogeneity and taxonomic diversity among Gardnerella species. Trends Microbiol. 2020, 28, 202–211. [Google Scholar] [CrossRef]
- Schellenberg, J.J.; Paramel Jayaprakash, T.; Withana Gamage, N.; Patterson, M.H.; Vaneechoutte, M.; Hill, J.E. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS ONE 2016, 11, e0146510. [Google Scholar] [CrossRef]
- Vaneechoutte, M.; Guschin, A.; Van Simaey, L.; Gansemans, Y.; Van Nieuwerburgh, F.; Cools, P. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int. J. Syst. Evol. Microbiol. 2019, 69, 679–687. [Google Scholar] [CrossRef]
- Khan, S.; Voordouw, M.J.; Hill, J.E. Competition among Gardnerella subgroups from the human vaginal microbiome. Front. Microbiol. 2019, 9, 374. [Google Scholar] [CrossRef]
- Castro, J.; Machado, D.; Cerca, N. Unveiling the role of Gardnerella vaginalis in polymicrobial bacterial vaginosis biofilms: The impact of other vaginal pathogens living as neighbors. ISME J. 2019, 13, 1306–1317. [Google Scholar] [CrossRef]
- Castro, J.; Rosca, A.S.; Cools, P.; Vaneechoutte, M.; Cerca, N. Gardnerella vaginalis enhances Atopobium vaginae viability in an in vitro model. Front. Microbiol. 2020, 10, 83. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.G. Gene transfer in bacteria: Speciation without species? Theor. Popul. Biol. 2002, 61, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef] [PubMed]
- Terns, M.P.; Terns, R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011, 14, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. Regulation of genetic flux between bacteria by restriction–modification systems. Proc. Natl. Acad. Sci. USA 2016, 113, 5658–5663. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18. [Google Scholar] [CrossRef]
- Pleckaityte, M.; Zilnyte, M.; Zvirbliene, A. Insights into the CRISPR/Cas system of Gardnerella vaginalis. BMC Microbiol. 2012, 12, 301. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014, 42, 10618–10631. [Google Scholar] [CrossRef]
- Stein, D.C.; Gunn, J.S.; Radlinska, M.; Piekarowicz, A. Restriction and modification systems of Neisseria gonorrhoeae. Gene 1995, 157, 19–22. [Google Scholar] [CrossRef]
- Alm, R.A.; Trust, T.J. Analysis of the genetic diversity of Helicobacter pylori: The tale of two genomes. J. Mol. Med. 1999, 77, 834–846. [Google Scholar] [CrossRef]
- Ando, T.; Xu, Q.; Torres, M.; Kusugami, K.; Israel, D.A.; Blaser, M.J. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 2000, 37, 1052–1065. [Google Scholar] [CrossRef] [PubMed]
- Vasu, K.; Nagaraja, V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Belfort, M.; Bestor, T.; Bhagwat, A.S.; Bickle, T.A.; Bitinaite, J.; Blumenthal, R.M.; Degtyarev, S.K.; Dryden, D.T.F.; Dybvig, K.; et al. Survey and summary: A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003, 31, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, A.; Wilson, G.G.; Wende, W. Type II restriction endonucleases—A historical perspective and more. Nucleic Acids Res. 2014, 42, 7489–7527. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
- Tankovic, J.; Timinskas, A.; Janulaitiene, M.; Zilnyte, M.; Baudel, J.-L.; Maury, E.; Zvirbliene, A.; Pleckaityte, M. Gardnerella vaginalis bacteremia associated with severe acute encephalopathy in a young female patient. Anaerobe 2017, 47, 132–134. [Google Scholar] [CrossRef]
- Gunthert, U.; Storm, K.; Bald, R. Restriction and modification in Bacillus subtilis. Localization of the methylated nucleotide in the BsuRI recognition sequence. Eur. J. Biochem. 1978, 90, 581–583. [Google Scholar] [CrossRef]
- Raskó, T.; Dér, A.; Klement, É.; Ślaska-Kiss, K.; Pósfai, E.; Medzihradszky, K.F.; Marshak, D.R.; Roberts, R.J.; Kiss, A. BspRI restriction endonuclease: Cloning, expression in Escherichia coli and sequential cleavage mechanism. Nucleic Acids Res. 2010, 38, 7155–7166. [Google Scholar] [CrossRef][Green Version]
- Laganeckas, M.; Margelevičius, M.; Venclovas, Č. Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile–profile alignments. Nucleic Acids Res. 2011, 39, 1187–1196. [Google Scholar] [CrossRef]
- Xu, Q.S.; Roberts, R.J.; Guo, H.-C. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005, 14, 2590–2600. [Google Scholar] [CrossRef][Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V.A. Completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Buchan, D.W.A.; Minneci, F.; Nugent, T.C.O.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.-H.; Huddleston, J.R.; Farkas, J.; Westpheling, J. Identification and characterization of CbeI, a novel thermostable restriction enzyme from Caldicellulosiruptor bescii DSM 6725 and a member of a new subfamily of HaeIII-like enzymes. J. Ind. Microbiol. Biotechnol. 2011, 38, 1867–1877. [Google Scholar] [CrossRef]
- Skowron, P.M.; Vitkute, J.; Ramanauskaite, D.; Mitkaite, G.; Jezewska-Frackowiak, J.; Zebrowska, J.; Zylicz-Stachula, A.; Lubys, A. Three-stage biochemical selection: Cloning of prototype class IIS/IIC/IIG restriction endonuclease-methyltransferase TsoI from the thermophile Thermus scotoductus. BMC Mol. Biol. 2013, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Sasnauskas, G.; Tamulaitis, G.; Urbanke, C.; Razaniene, D.; Siksnys, V. Tetrameric restriction enzymes: Expansion to the GIY-YIG nuclease family. Nucleic Acids Res. 2007, 36, 938–949. [Google Scholar] [CrossRef]
- Chan, S.-H.; Opitz, L.; Higgins, L.; O’Loane, D.; Xu, S. Cofactor requirement of HpyAV restriction endonuclease. PLoS ONE 2010, 5, e9071. [Google Scholar] [CrossRef][Green Version]
- Bohr, L.L.; Mortimer, T.D.; Pepperell, C.S. Lateral gene transfer shaper diversity of Gardnerella spp. Front. Cell. Infect. Microbiol. 2020, 10, 293. [Google Scholar] [CrossRef]
- Budroni, S.; Siena, E.; Hotopp, J.C.D.; Seib, K.L.; Serruto, D.; Nofroni, C.; Comanducci, M.; Riley, D.R.; Daugherty, S.C.; Angiuoli, S.V.; et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. USA 2011, 108, 4494–4499. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Yildirim, S.; Thomas, S.M.; Durkin, A.S.; Torralba, M.; Sutton, G.; Buhay, C.J.; Ding, Y.; Dugan-Rocha, S.P.; Muzny, D.M.; et al. Comparative genomics of Gardnerella vaginalis strains reveal substantial differences in metabolic and virulence potential. PLoS ONE 2010, 5, e12411. [Google Scholar] [CrossRef]
- Herriot, R.M.; Connolly, J.H.; Gupta, S. Blood nucleases and infectious viral nucleic acids. Nature 1961, 189, 817–820. [Google Scholar] [CrossRef]
- Novy, R.; Morris, B. Use of glucose to control basal expression in the pET system. InNovations 2001, 13, 8–10. [Google Scholar]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Tolia, N.H.; Joshua-Tor, L. Strategies for protein coexpression in Escherichia coli. Nat. Methods 2006, 3, 55–64. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer Sequence 5′→3′ | T Annealing (°C) | Amplicon Size (bp) |
|---|---|---|---|
| GvMe5 | TACGTAATGAAGGTATTAAGCTTATTTAG | 56 | 1024 |
| GvMe3 | TACGTACTTATCTGAATTATGATAGAACT | ||
| GVRe5 | GCTAGCATGGCAAAAACATTTGGTT | 58 | 917 |
| GVRe3 | GTCGACATCTATGCCTATATGTTCACAG | ||
| GvRe5-NheI | GTCTAGCTAGCATGGCAAAAACATTTGGTT | 55 | 917 |
| GvRe3-XhoI | GATCTCGAGTTATATGTTCACAGATTCAGT | ||
| GV-pUC57-For | CCGCGTTGCTGGCGTTTTTC | 65 | 868 |
| GV-pUC57-Rev | GTAGTTATCTACACGACGGGGAGTC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulavaitė, A.; Dalgediene, I.; Michailoviene, V.; Pleckaityte, M. Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018. Pathogens 2020, 9, 703. https://doi.org/10.3390/pathogens9090703
Bulavaitė A, Dalgediene I, Michailoviene V, Pleckaityte M. Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018. Pathogens. 2020; 9(9):703. https://doi.org/10.3390/pathogens9090703
Chicago/Turabian StyleBulavaitė, Aistė, Indre Dalgediene, Vilma Michailoviene, and Milda Pleckaityte. 2020. "Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018" Pathogens 9, no. 9: 703. https://doi.org/10.3390/pathogens9090703
APA StyleBulavaitė, A., Dalgediene, I., Michailoviene, V., & Pleckaityte, M. (2020). Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018. Pathogens, 9(9), 703. https://doi.org/10.3390/pathogens9090703





