Functional and Mass Spectrometric Evaluation of an Anti-Tick Antigen Based on the P0 Peptide Conjugated to Bm86 Protein
Abstract
1. Introduction
2. Results
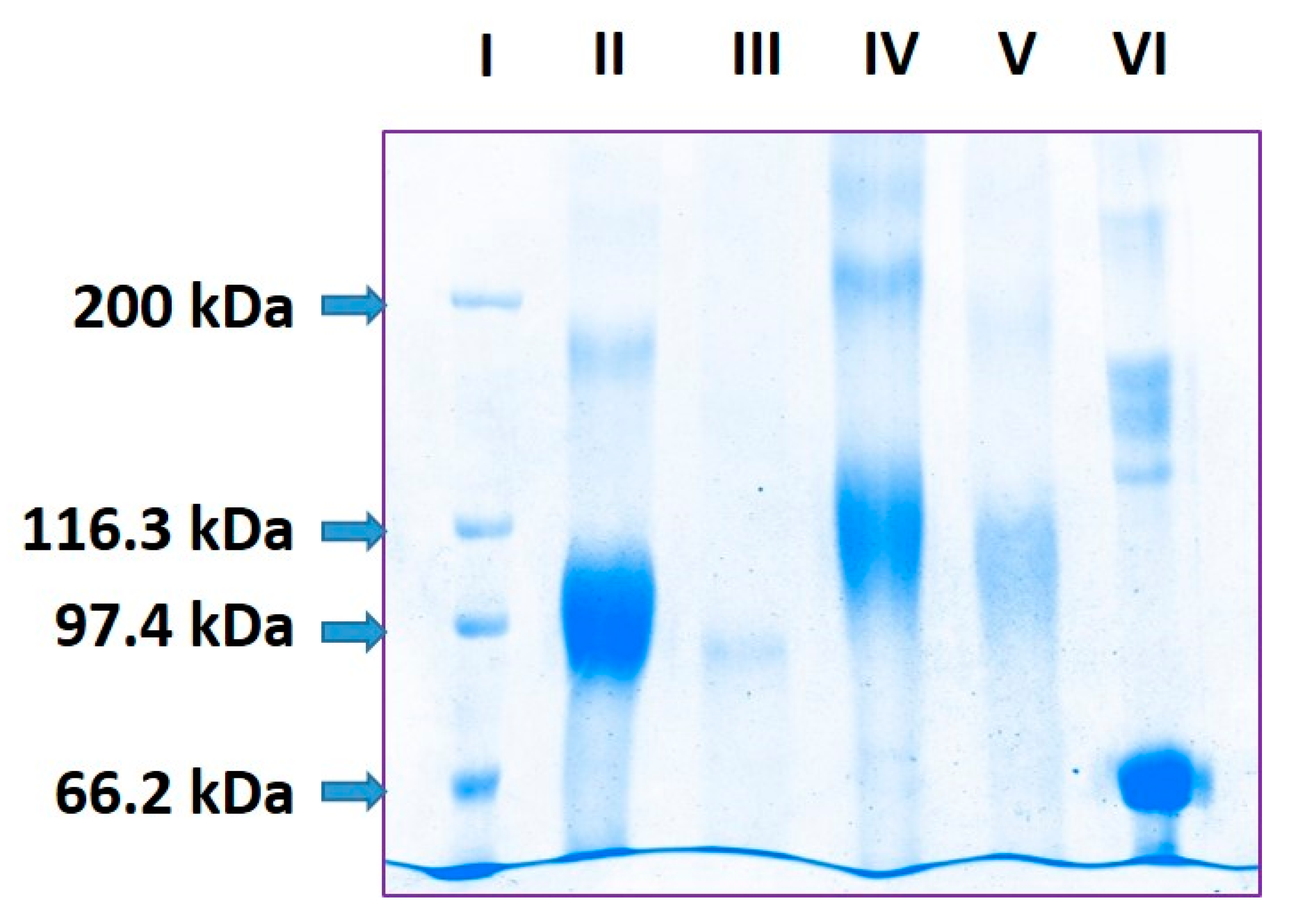
2.1. Estimation of pP0 Molecules Linked to Bm86 Carrier Protein
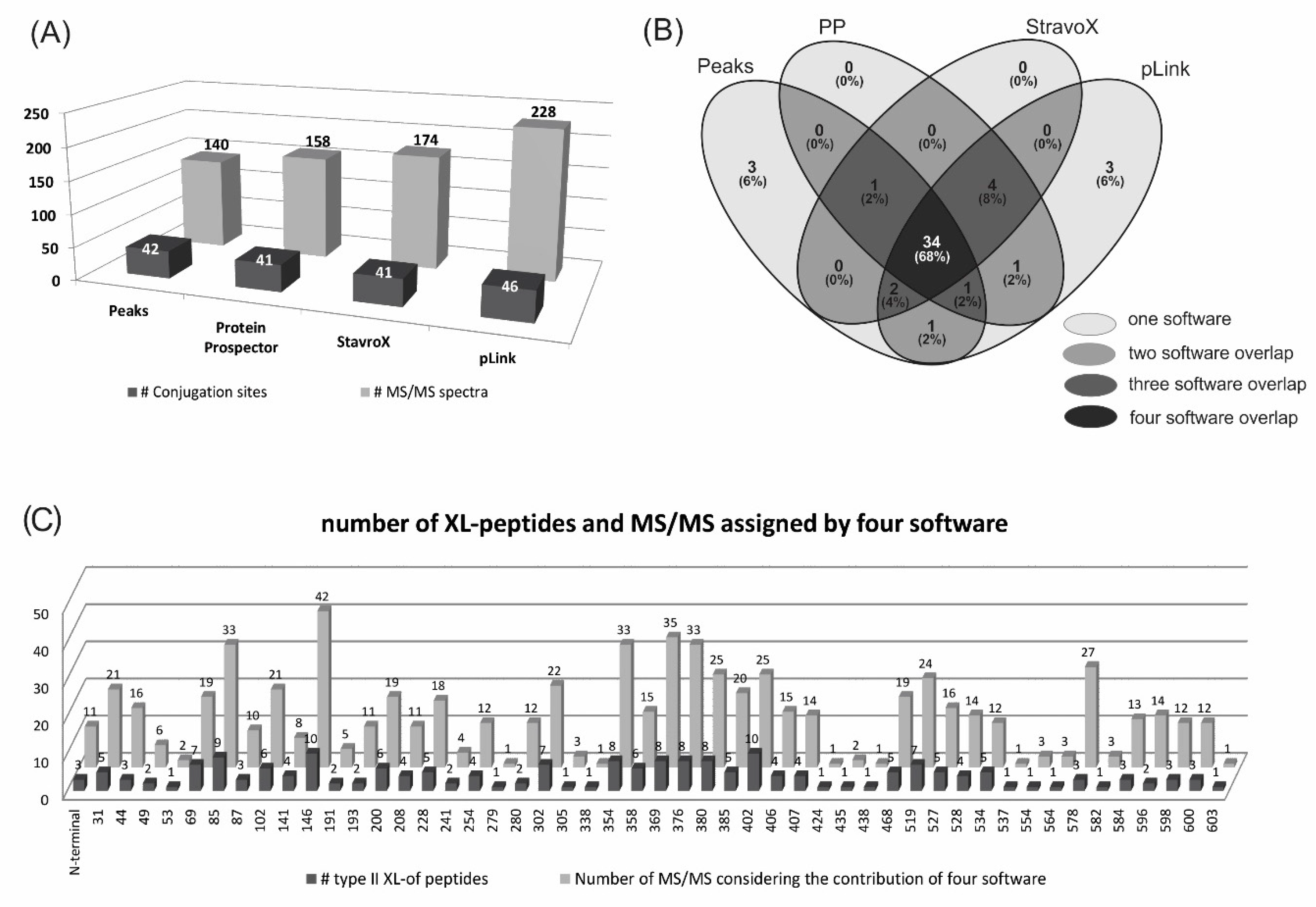
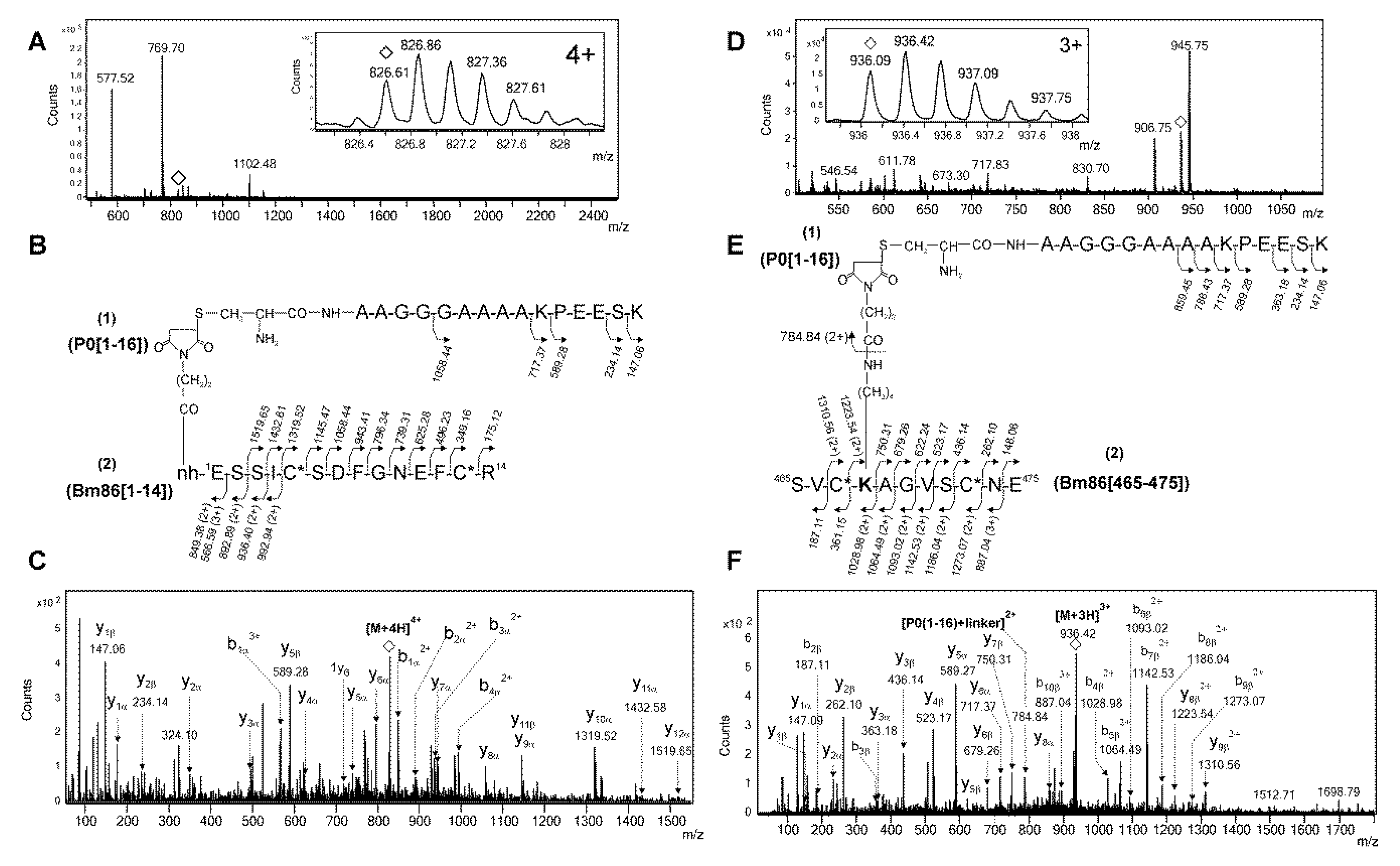
2.2. Identification of Conjugation Sites in pP0–Bm86 Conjugate
2.3. Post-Translational Modifications of the Bm86 Protein
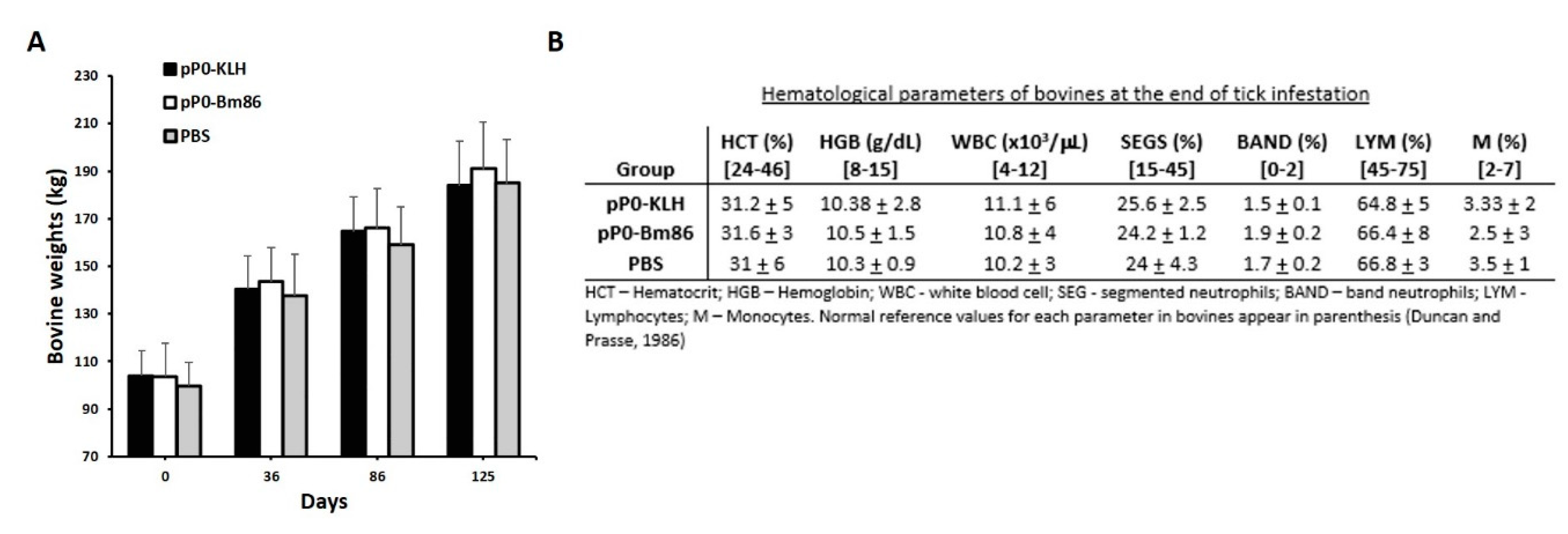
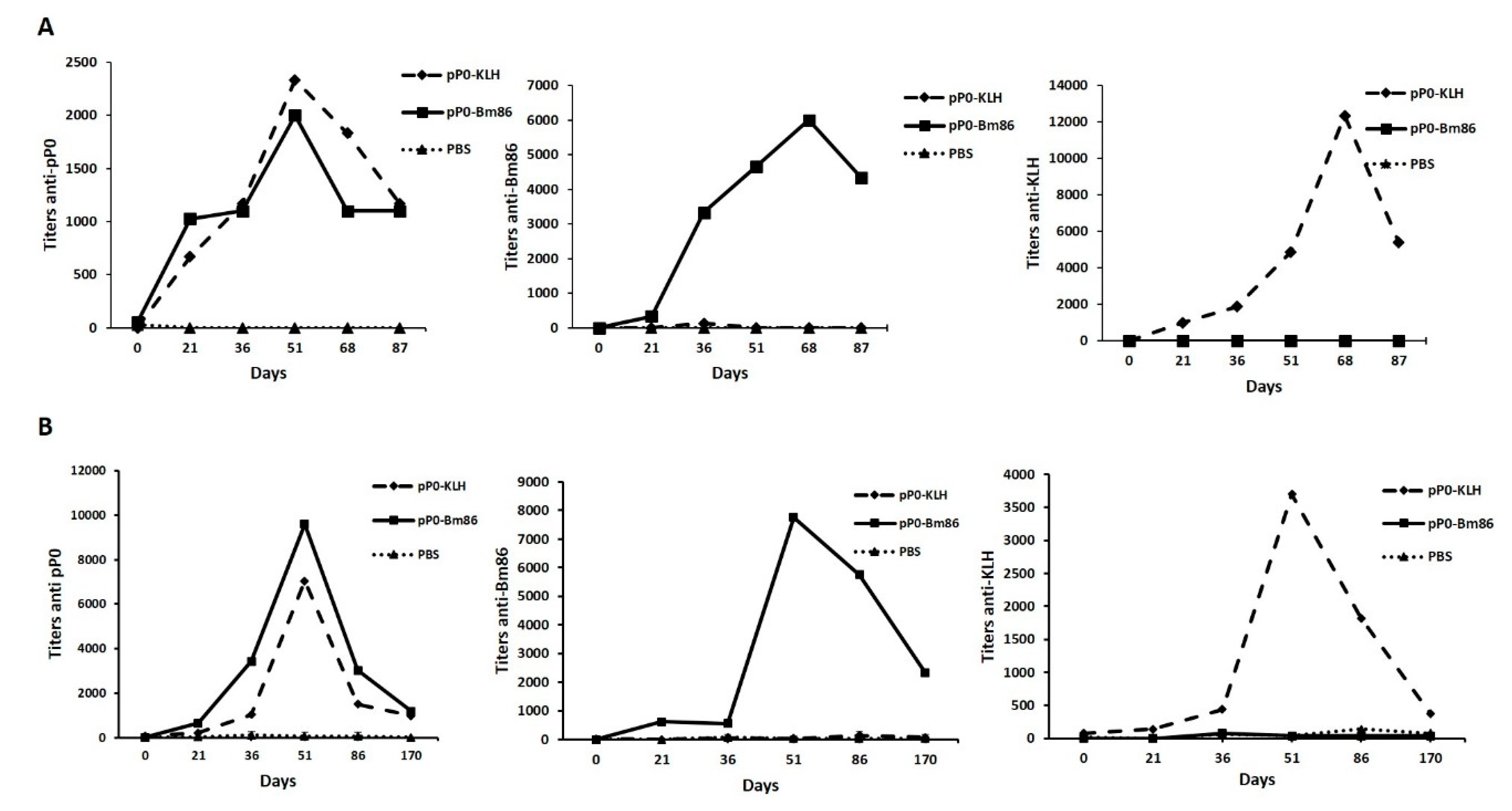
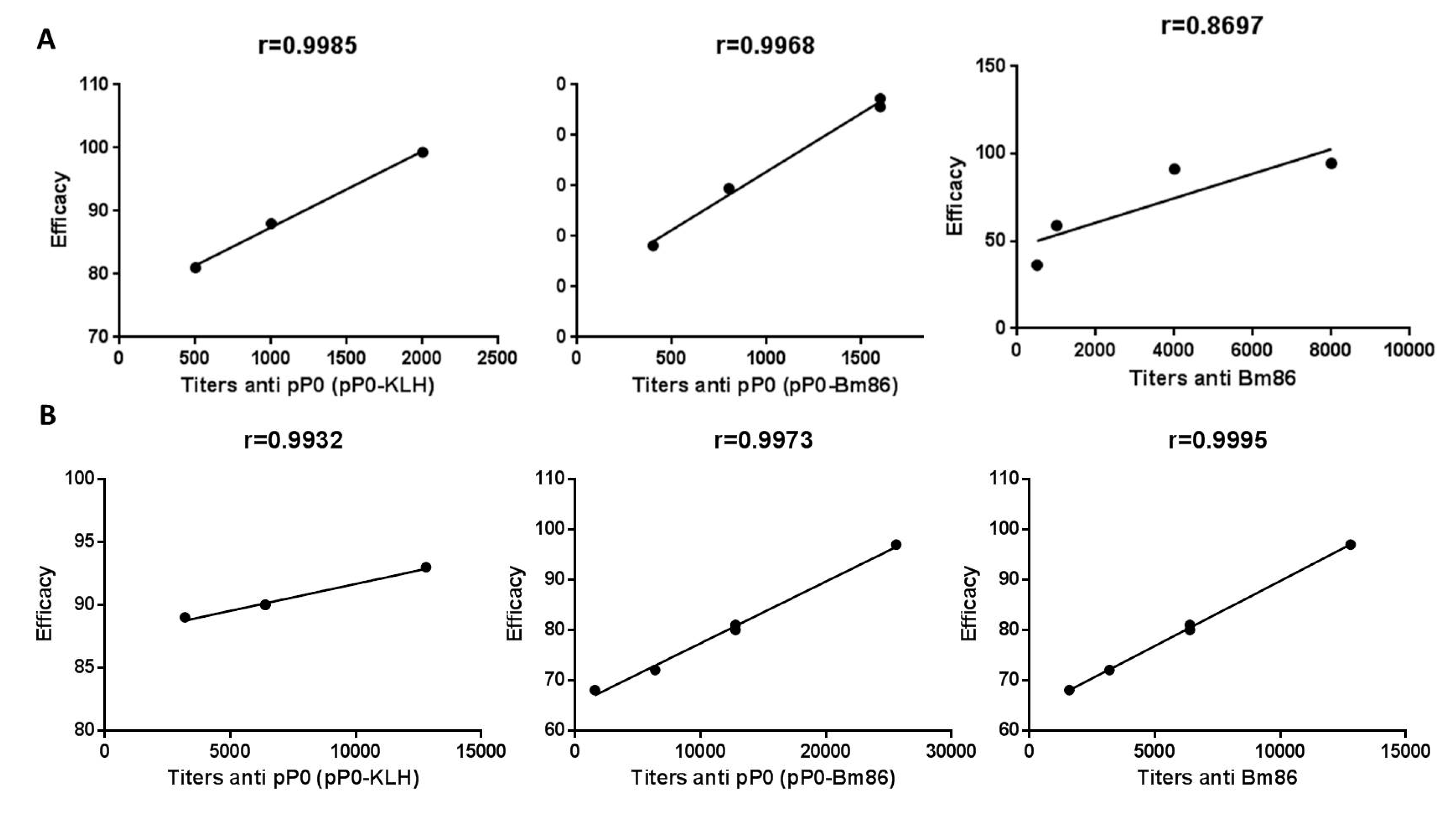
2.4. Efficacy of Immunization with pP0–Bm86 Conjugate against R. sanguineus and R. microplus Ticks
3. Discussion
4. Materials and Methods
4.1. Chemical Synthesis and Conjugation of pP0 Peptide to Bm86 and KLH as Carrier Proteins
4.2. pP0-Bm86 Deglycosylation with PNGase-F
4.3. SDS-PAGE Analysis and pP0 Payload Estimation of the pP0–Bm86 Conjugate
4.4. Reduction and S-Alkylation of pP0-Bm86
4.5. Proteolytic Digestions of pP0–Bm86 Conjugate
4.6. Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS) Analysis
4.7. Identification of Conjugation Sites on pP0–Bm86 Conjugate
4.8. Bioinformatic Analysis
4.9. Immunization and Challenge Experiment in Dogs
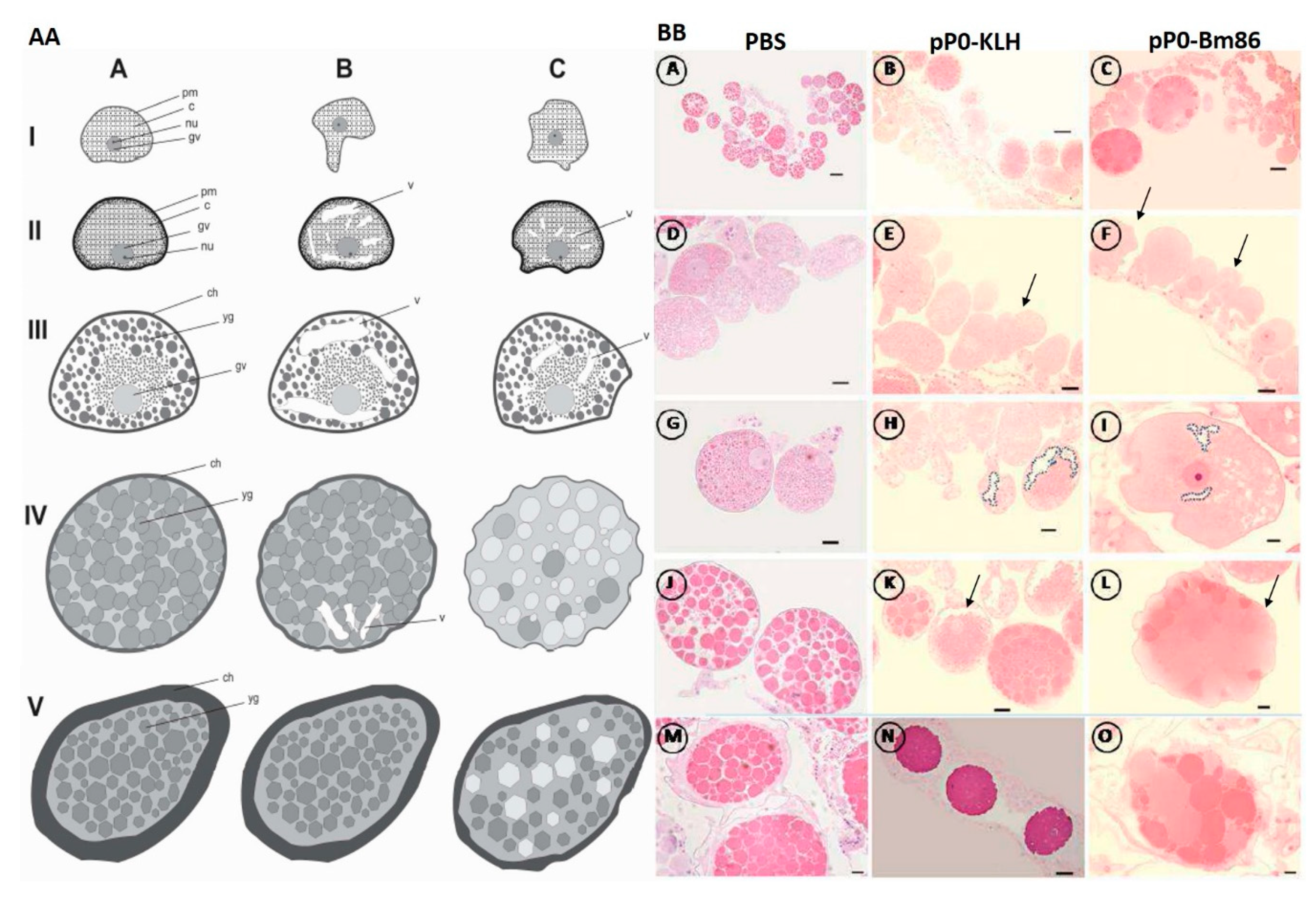
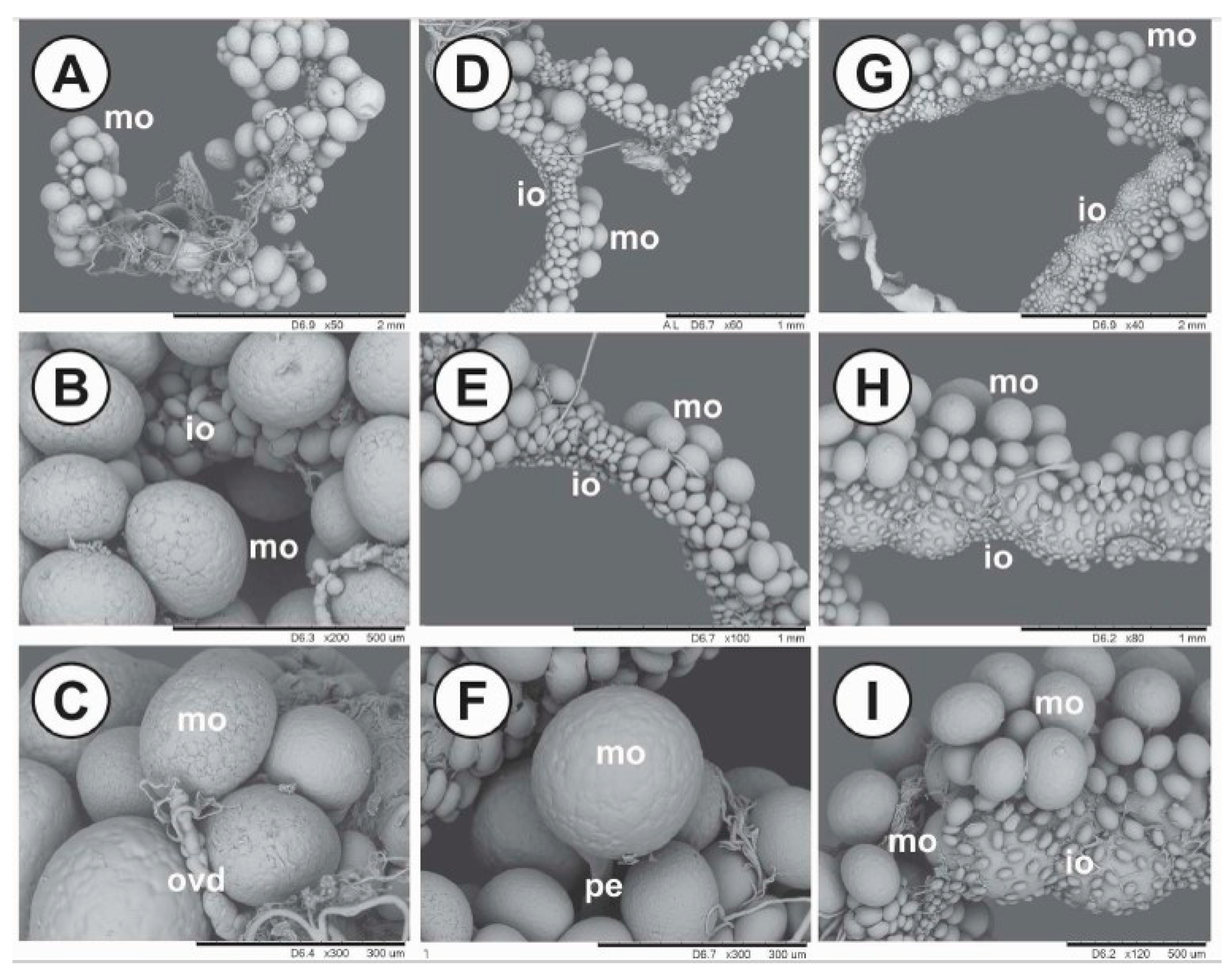
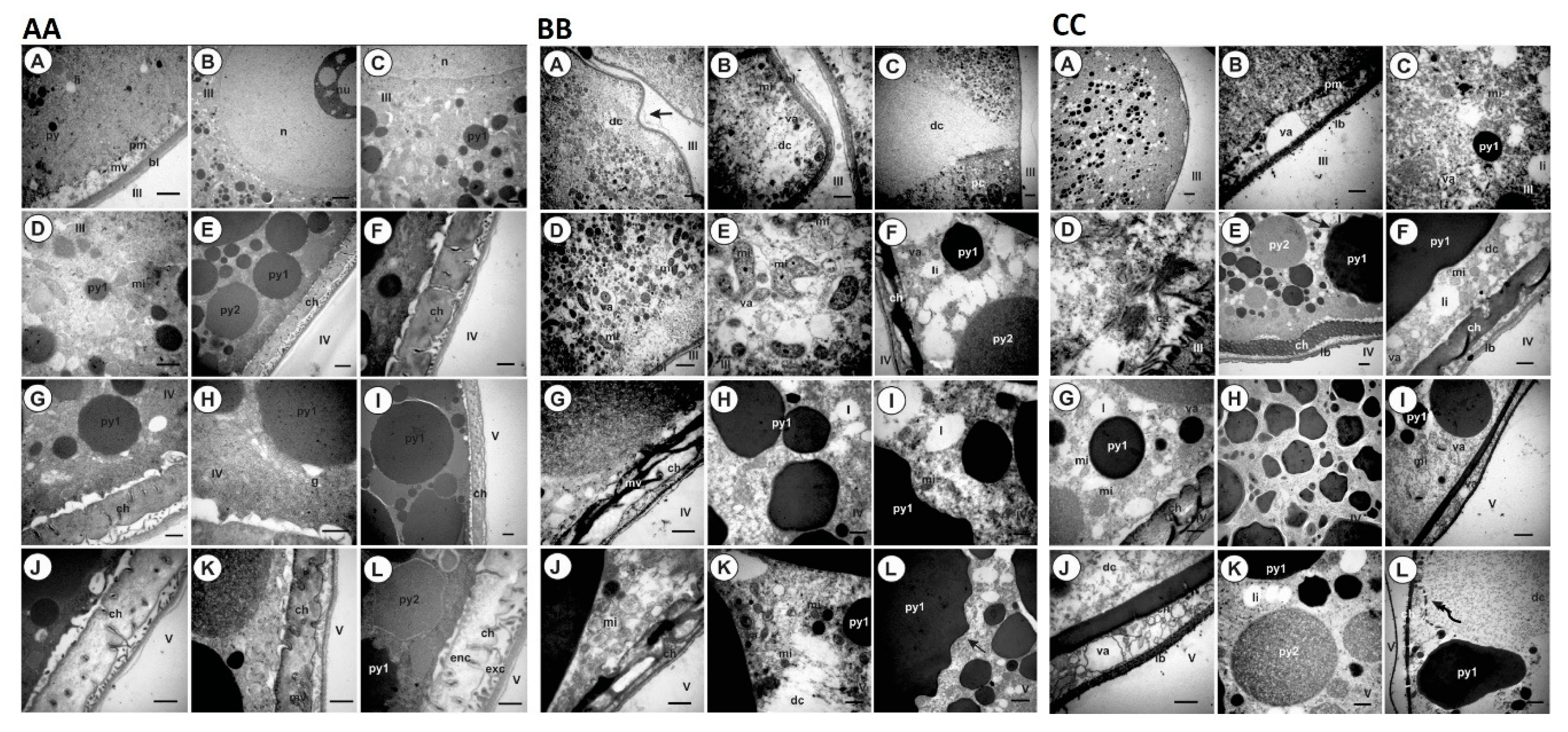
4.10. Histology and Ultrastructure of Ovaries from Ticks Fed on Immunized Dogs
4.11. Immunization and Challenge Experiment in Cattle
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- de la Fuente, J.; Antunes, S.; Bonnet, S.; Cabezas-Cruz, A.; Domingos, A.G.; Estrada-Pena, A.; Johnson, N.; Kocan, K.M.; Mansfield, K.L.; Nijhof, A.M.; et al. Tick-Pathogen Interactions and Vector Competence: Identification of Molecular Drivers for Tick-Borne Diseases. PLoS ONE 2017, 7, 114. (In English) [Google Scholar] [CrossRef] [PubMed]
- Schetters, T.P. Vaccination Against Ticks. Ectoparasites: Drug Discovery Against Moving Targets; Wiley-VCH: Weinheim, Germany, 2018; p. 25. [Google Scholar]
- Willadsen, P.; Riding, G.A.; McKenna, R.V.; Kemp, D.H.; Tellam, R.L.; Nielsen, J.N.; Lahnstein, J.; Cobon, G.S.; Gough, J.M. Immunologic control of a parasitic arthropod. Identification of a protective antigen from Boophilus microplus. J. Immunol. 1989, 143, 1346–1351. [Google Scholar] [PubMed]
- Braun-Galleani, S.; Dias, J.A.; Aisling, Y.; Coughlan, A.P.; Byrne, K.P.; Wolfe, K.H. Genomic diversity and meiotic recombination among isolates of the biotech yeast Komagataella phaffii (Pichia pastoris). Microb. Cell Factories 2019, 18, 211. [Google Scholar] [CrossRef] [PubMed]
- Canales, M.; Enriquez, A.; Ramos, E.; Cabrera, D.; Dandie, H.; Soto, A.; Falcon, V.; Rodriguez, M.; de la Fuente, J. Large-scale production in Pichia pastoris of the recombinant vaccine Gavac against cattle tick. Vaccine 1997, 15, 414–422. [Google Scholar] [CrossRef]
- de la Fuente, J.; Rodriguez, M.; Montero, C.; Redondo, M.; Garcia-Garcia, J.C.; Mendez, L.; Serrano, E.; Valdes, M.; Enriquez, A.; Canales, M.; et al. Vaccination against ticks (Boophilus spp.): The experience with the Bm86-based vaccine Gavac. Genet. Anal. 1999, 15, 143–148. [Google Scholar] [CrossRef]
- de la Fuente, J.; Rodriguez, M.; Redondo, M.; Montero, C.; Garcia-Garcia, J.C.; Mendez, L.; Serrano, E.; Valdes, M.; Enriquez, A.; Canales, M.; et al. Field studies and cost-effectiveness analysis of vaccination with Gavac against the cattle tick Boophilus microplus. Vaccine 1998, 16, 366–373. [Google Scholar] [CrossRef]
- Suarez, M.; Rubi, J.; Pérez, D.; Cordova, V.; Salazar, Y.; Vielma, A.; Barrios, F.; Gil, C.A.; Segura, N.; Carrillo, Y.; et al. High impact and effectiveness of Gavac™ vaccine in the national program for control of bovine ticks Rhipicephalus microplus in Venezuela. Livest. Sci. 2016, 187, 48–52. [Google Scholar] [CrossRef]
- Rodríguez-Valle, M.; Mendez, L.; Valdez, M.; Redondo, M.; Espinosa, C.M.; Vargas, M.; Cruz, R.L.; Barrios, H.P.; Seoane, G.; Ramirez, E.S. Integrated control of Boophilus microplus ticks in Cuba based on vaccination with the anti-tick vaccine Gavac. Exp. Appl. Acarol. 2004, 34, 375–382. [Google Scholar] [CrossRef]
- Rodríguez-Mallon, A.; Encinosa, P.E.; Méndez-Pérez, L.; Bello, Y.; Fernández, R.R.; Garay, H.; Cabrales, A.; Méndez, L.; Borroto, C.; Estrada, M. High efficacy of a 20 amino acid peptide of the acidic ribosomal protein P0 against the cattle tick, Rhipicephalus Microplus. Ticks Tick-Borne Dis. 2015, 6, 530–537. [Google Scholar] [CrossRef]
- Rodríguez-Mallon, A.; Fernández, E.; Encinosa, P.E.; Bello, Y.; Méndez-Pérez, L.; Cepero, L.; Pérez, D.; González, M.; Garay, H.; Reyes, O. A novel tick antigen shows high vaccine efficacy against the dog tick, Rhipicephalus Sanguineus. Vaccine 2012, 30, 1782–1789. [Google Scholar] [CrossRef]
- Rodríguez-Mallon, A.; Encinosa Guzmán, P.E.; Bello Soto, Y.; Perdomo, K.R.; Espinosa, C.M.; Vargas, M.; García, M.P.E. A chemical conjugate of the tick P0 peptide is efficacious against Amblyomma Mixtum. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.T. Electrophoresis: Theory, Techniques and Biochemical and Clinical Applications, 2nd ed.; Oxford University Press: Oxford, UK, 1986. [Google Scholar]
- Shi, Y.; Mowery, R.A.; Ashley, J.; Hentz, M.; Ramirez, A.J.; Bilgicer, B.; Slunt-Brown, H.; Borchelt, D.R.; Shaw, B.F. Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci. 2012, 21, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, J.C.; Montero, C.; Rodriguez, M.; Soto, A.; Redondo, M.; Valdes, M.; Mendez, L.; de la Fuente, J. Effect of particulation on the immunogenic and protective properties of the recombinant Bm86 antigen expressed in Pichia pastoris. Vaccine 1998, 16, 374–380. [Google Scholar] [CrossRef]
- Shi, Q.; Jackowski, G. Gel Electrophoresis of Proteins: A Practical Approach, 3rd ed.; Hames, B.D., Ed.; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Dennler, P.; Fischer, E.; Schibli, R. Antibodies Conjugates: From heterogeneous populations to defined reagents. Antibodies 2015, 4, 197–224. [Google Scholar] [CrossRef]
- Schilling, B.; Row, R.H.; Gibson, B.W.; Guo, X.; Young, M.M. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003, 14, 834–850. [Google Scholar] [CrossRef]
- Giese, S.H.; Fischer, L.; Rappsilber, J. A study into the collision-induced dissociation (CID) behavior of cross-linked peptides. Mol. Cell. Proteom. 2016, 15, 1094–1104. [Google Scholar] [CrossRef]
- Kolbowski, L.; Mendes, M.L.; Rappsilber, J. Optimizing the parameters governing the fragmentation of cross-linked peptides in a tribrid mass spectrometer. Anal. Chem. 2017, 89, 5311–5318. [Google Scholar] [CrossRef]
- Gomes, A.M.V.; Carmo, T.S.; Carvalho, L.S.; Bahia, F.M.; Parachin, N.S. Comparison of yeasts as hosts for recombinant protein production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef]
- Trimble, R.B.; Lubowski, C.; Hauer, C.R.; Stack, R.; McNaughton, L.; Gemmill, T.R.; Kumar, S.A. Characterization of N-and O-linked glycosylation of recombinant human bile salt–stimulated lipase secreted by Pichia pastoris. Glycobiology 2004, 14, 265–274. [Google Scholar] [CrossRef]
- Willer, T.; Valero, M.C.; Tanner, W.; Cruces, J.; Strahl, S. O-mannosyl glycans: From yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 2003, 13, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Edrissi, B.; Taghizadeh, K.; Dedon, P.C. Quantitative analysis of histone modifications: Formaldehyde is a source of pathological N6-formyllysine that is refractory to histone deacetylases. PLoS Genet. 2013, 9, e1003328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Freeman, L.E.B.; Nakamura, J.; Hecht, S.S.; Vandenberg, J.J.; Smith, M.T.; Sonawane, B.R. Formaldehyde and leukemia: Epidemiology, potential mechanisms, and implications for risk assessment. Environ. Mol. Mutagenesis 2010, 51, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Edrissi, B.; Taghizadeh, K.; Moeller, B.C.; Kracko, D.; Doyle-Eisele, M.; Swenberg, J.A.; Dedon, P.C. Dosimetry of N 6-Formyllysine Adducts Following [13C2H2]-Formaldehyde Exposures in Rats. Chem. Res. Toxicol. 2013, 26, 1421–1423. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; van Veelen, P.A.; Mahler, M.; Janssen, G.M.C.; Drijfhout, J.W.; Huizinga, T.W.J.; Toes, R.E.M.; Trouw, L.A. Carbamylation and antibodies against carbamylated proteins in autoimmunity and other pathologies. Autoimmun. Rev. 2014, 13, 225–230. [Google Scholar] [CrossRef]
- Verheul, M.K.; van Erp, S.J.; van der Woude, H.D.; Levarht, E.W.N.; Mallat, M.J.K.; Verspaget, H.W.; Stolk, J.; Toes, R.E.M.; van der Meulen-de Jong, A.E.P.; Hiemstra, S. Anti-carbamylated protein antibodies: A specific hall mark for rheumatoid arthritis. Comparison to conditions known for enhanced carbamylation; renal failure, smoking and chronicinflammation. Ann. Rheum. Dis. 2016, 75, 1575–1576. [Google Scholar] [CrossRef]
- Rodriguez, M.; Rubiera, R.; Penichet, M.; Montesinos, R.; Cremata, J.; Falcon, V.; Sanchez, G.; Bringas, R.; Cordoves, C.; Valdes, M.; et al. High level expression of the B. microplus Bm86 antigen in the yeast Pichia pastoris forming highly immunogenic particles for cattle. J. Biotechnol. 1994, 33, 135–146. [Google Scholar] [CrossRef]
- Roland, L.; Drillich, M.; Iwersen, M. Hematology as a diagnostic tool in bovine medicine. J. Vet. Diagn. Investig. 2014, 26, 592–598. [Google Scholar] [CrossRef]
- Vargas, M.; Montero, C.; Sanchez, D.; Perez, D.; Valdes, M.; Alfonso, A.; Joglar, M.; Machado, H.; Rodriguez, E.; Mendez, L. Two initial vaccinations with the Bm86-based Gavacplus vaccine against Rhipicephalus (Boophilus) microplus induce similar reproductive suppression to three initial vaccinations under production conditions. BMC Vet. Res. 2010, 6, 43. [Google Scholar] [CrossRef]
- de Oliveira, P.R.; Bechara, G.H.; Denardi, S.E.; Nunes, E.T.; Mathias, M.I.C. Morphological characterization of the ovary and oocytes vitellogenesis of the tick Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae). Exp. Parasitol. 2005, 110, 146–156. [Google Scholar] [CrossRef]
- de Oliveira, P.R.; Bechara, G.H.; Camargo-Mathias, M.I. Evaluation of cytotoxic effects of fipronil on ovaries of semi-engorged Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae) tick female. Food Chem. Toxicol. 2008, 46, 2459–2465. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Almazan, C.; Canales, M.; de la Lastra, J.M.P.; Kocan, K.M.; Willadsen, P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 2007, 8, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Sprong, H.; Trentelman, J.; Seemann, I.; Grubhoffer, L.; Rego, R.O.; Hajdusek, O.; Kopacek, P.; Sima, R.; Nijhof, A.M.; Anguita, J. ANTIDotE: Anti-tick vaccines to prevent tick-borne diseases in Europe. Parasit. Vectors 2014, 7, 77. (In English) [Google Scholar] [CrossRef] [PubMed]
- Schetters, T.; Bishop, R.; Crampton, M.; Kopáček, P.; Lew-Tabor, A.; Maritz-Olivier, C.; Miller, R.; Mosqueda, J.; Patarroyo, J.; Rodriguez-Valle, M. Cattle tick vaccine researchers join forces in CATVAC. Parasit. Vectors 2016, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, J.C.; Soto, A.; Nigro, F.; Mazza, M.; Joglar, M.; Hechevarria, M.; Lamberti, J.; de la Fuente, J. Adjuvant and immunostimulating properties of the recombinant Bm86 protein expressed in Pichia pastoris. Vaccine 1998, 16, 1053–1055. [Google Scholar] [CrossRef]
- Rodriguez-Gabriel, M.A.; Remacha, M.; Ballesta, J.P. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J. Biol. Chem. 2000, 275, 2130–2136. [Google Scholar] [CrossRef]
- Wang, J.S.; Yang, X.; Li, R.; Zhou, P.; Zhang, M.L.; Han, H. Analysis of nuclear localization signal (NLS) in ribosomal protein L6/ Taxreb107. Prog. Biochem. Biophys. 2002, 29, 144–148. [Google Scholar]
- Nijhof, A.M.; Taoufik, A.; de la Fuente, J.; Kocan, K.M.; de Vries, E.; Jongejan, F. Gene silencing of the tick protective antigens, Bm86, Bm91 and subolesin, in the one-host tick Boophilus microplus by RNA interference. Int. J. Parasitol. 2007, 37, 653–662. [Google Scholar] [CrossRef]
- Nijhof, A.M.; Balk, J.A.; Postigo, M.; Rhebergen, A.M.; Taoufik, A.; Jongejan, F. Bm86 homologues and novel ATAQ proteins with multiple epidermal growth factor (EGF)-like domains from hard and soft ticks. Int. J. Parasitol. 2010, 40, 1587–1597. [Google Scholar] [CrossRef][Green Version]
- Liao, M.; Zhou, J.; Hatta, T.; Umemiya, R.; Miyoshi, T.; Tsuji, N.; Xuan, X.; Fujisaki, K. Molecular characterization of Rhipicephalus (Boophilus) microplus Bm86 homologue from Haemaphysalis longicornis ticks. Vet. Parasitol. 2007, 146, 148–157. [Google Scholar] [CrossRef]
- Singh, S.; Sehgal, A.; Waghmare, S.; Chakraborty, T.; Goswami, A.; Sharma, S. Surface expression of the conserved ribosomal protein P0 on parasite and other cells. Mol. Biochem. Parasitol. 2002, 119, 121–124. [Google Scholar] [CrossRef]
- Radulović, Ž.M.; Kim, T.K.; Porter, L.M.; Sze, S.H.; Lewis, L.; Mulenga, A. A 24–48 h fed Amblyomma americanum tick saliva immuno-proteome. BMC Genom. 2014, 15, 518. [Google Scholar] [CrossRef] [PubMed]
- Tirloni, L.; Reck, J.; Terra, R.M.S.; Martins, J.R.; Mulenga, A.; Sherman, N.E.; Fox, J.W.; Yates, J.R.; Termignoni, C.; Pinto, A.F.M.; et al. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: A comparison between partially and fully engorged females. PLoS ONE 2014, 9, e94831. [Google Scholar] [CrossRef] [PubMed]
- Parizi, L.F.; Reck, J.J.; Oldiges, D.P.; Guizzo, M.G.; Seixas, A.; Logullo, C.; de Oliveira, P.L.; Termignoni, C.; Martins, J.R.; Ida, S.V.J. Multi-antigenic vaccine against the cattle tick Rhipicephalus (Boophilus) microplus: A field evaluation. Vaccine 2012, 30, 6912–6917. (In Eng) [Google Scholar] [CrossRef] [PubMed]
- Almazán, C.; Lagunes, R.; Villar, M.; Canales, M.; Rosario-Cruz, R.; Jongejan, F.; de la Fuente, J. Identification and characterization of Rhipicephalus (Boophilus) microplus candidate protective antigens for the control of cattle tick infestations. Parasitol. Res. 2010, 106, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Almazan, C.; Moreno-Cantu, O.; Moreno-Cid, J.A.; Galindo, R.C.; Canales, M.; Villar, M.; de la Fuente, J. Control of tick infestations in cattle vaccinated with bacterial membranes containing surface-exposed tick protective antigens. Vaccine 2012, 30, 265–272. (In English) [Google Scholar] [CrossRef]
- Willadsen, P.; Smith, D.; Cobon, G.; McKenna, R.V. Comparative vaccination of cattle against Boophilus microplus with recombinant antigen Bm86 alone or in combination with recombinant Bm91. Parasite Immunol. 1996, 18, 241–246. [Google Scholar] [CrossRef]
- Tabor, A.E.; Ali, A.; Rehman, G.; Garcia, G.R.; Zangirolamo, A.F.; Malardo, T.; Jonsson, N.N. Cattle Tick Rhipicephalus microplus-Host Interface: A Review of Resistant and Susceptible Host Responses. Front. Cell. Infect. Microbiol. 2017, 7. (In English) [Google Scholar] [CrossRef]
- de la Fuente, J.; Kocan, K.M.; Contreras, M. Prevention and control strategies for ticks and pathogen transmission. Rev. Sci. Tech. 2015, 34, 249–264. (In English) [Google Scholar] [CrossRef]
- Field, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 2009, 35, 161–214. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein oncentration by using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Bigge, J.C.; Patel, T.P.; Bruce, J.A.; Goulding, P.N.; Charles, S.M.; Parekh, R.B. Nonselective and efficient fluorescent labeling of glycans using 2-aminobenzamide and anthranilic acid. Anal. Biochem. 1995, 230, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, G.; Salazar, V.; Montesino, R.; Támbara, Y.; Struwe, W.B.; Leon, E.; Harvey, D.J.; Antoine, L.; Rincón, M.; Domon, B. Structural characterization and biological implications of sulfated N-glycans in a serine protease from the neotropical moth Hylesia metabus (Cramer, 1775) (Lepidoptera: Saturniidae). Glycobiology 2016, 26, 230–250. [Google Scholar] [PubMed]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. Peaks: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wu, Y.J.; Zhu, M.; Fan, S.B.; Lin, J.; Zhang, K.; Li, S.; Chi, H.; Li, Y.X.; Chen, H.F.; et al. Identification of cross-linked peptides from complex samples. Nat. Methods 2012, 9, 904–906. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Fan, S.B.; Yang, B.; Li, Y.X.; Meng, J.M.; Wu, L.; Li, P.; Zhang, K.; Zhang, M.J.; Fu, Y.; et al. Mapping native disulfide bonds at a proteome scale. Nat. Methods 2015, 12, 329–331. [Google Scholar] [CrossRef]
- Götze, M.; Pettelkau, J.; Schaks, S.; Bosse, K.; Ihling, C.H.; Krauth, F.; Fritzsche, R.; Kühn, U.; Sinz, A. StavroX—A software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 2012, 23, 76–87. [Google Scholar] [CrossRef]
- Chu, F.; Baker, P.R.; Burlingame, A.L.; Chalkley, R.J. Finding Chimeras: A Bioinformatics Strategy for Identification of Cross-linked Peptides. Mol. Cell. Proteom. 2010, 9, 25–31. [Google Scholar] [CrossRef]
- Trnka, M.J.; Baker, P.R.; Robinson, P.J.J.; Burlingame, A.L.; Chalkley, R.J. Matching Cross-linked Peptide Spectra: Only as Good as the Worse Identification. Mol. Cell. Proteom. 2013, 13, 420–434. [Google Scholar] [CrossRef]
- Guide for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Press: Colombia, WA, USA, 2011.
- Bechara, G.H.; Szabó, M.P.J.; Ferreira, B.R. Rhipicephalus sanguineus tick in Brazil: Feeding and reproductive aspects under laboratorial conditions. Braz. J. Vet. Parasitol. 1995, 4, 61–66. [Google Scholar]
- Perez-Perez, D.; Bechara, G.H.; Machado, R.Z.; Andrade, G.M.; del Vecchio, R.E.; Pedroso, M.S.; Hernandez, M.V.; Farnos, O. Efficacy of the Bm86 antigen against immature instars and adults of the dog tick Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae). Vet. Parasitol. 2010, 167, 321–326. [Google Scholar] [CrossRef] [PubMed]
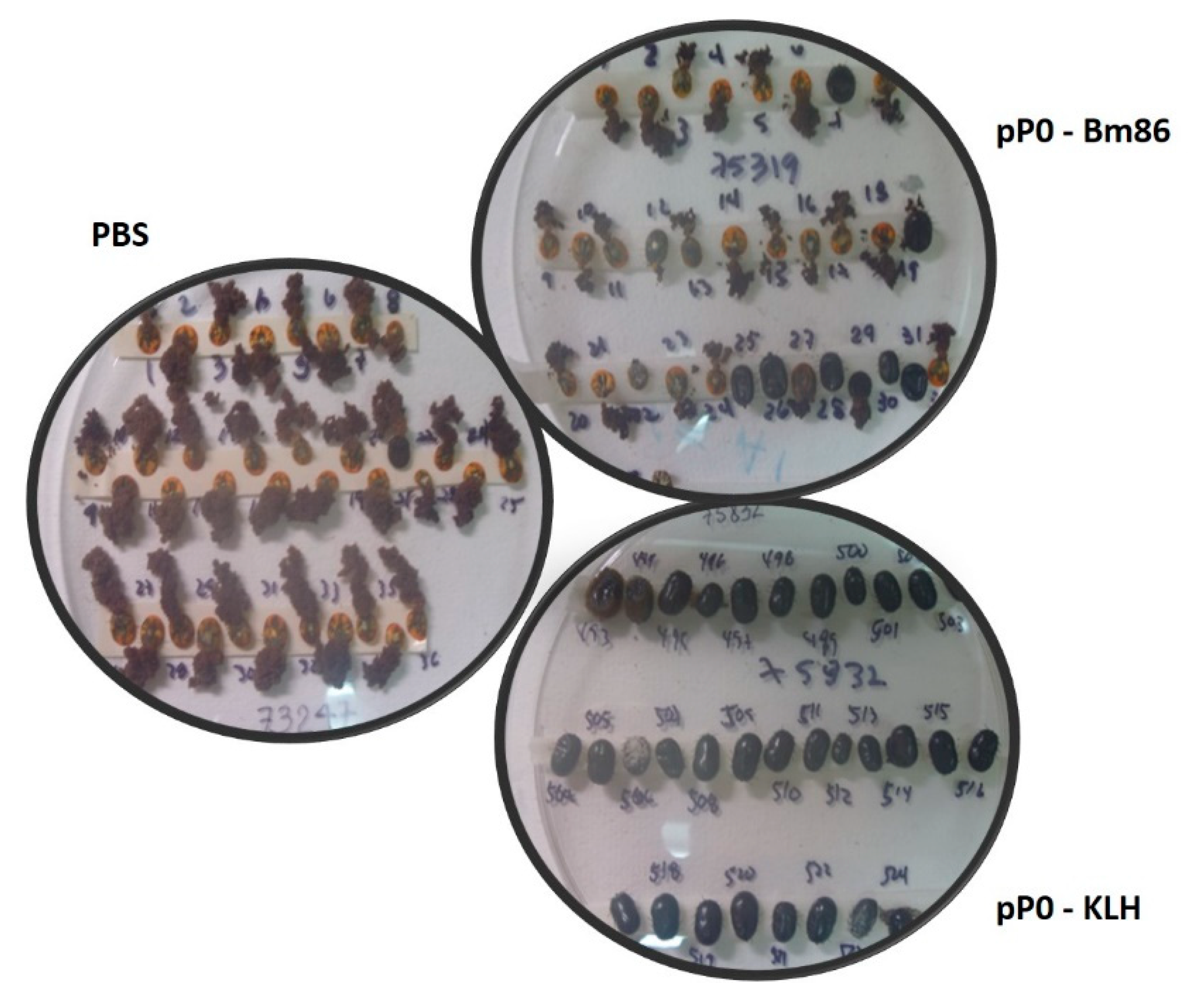
| Reduction Percent with Respect to Control of | ||||
| Groups | Larval Yield | Larval Viability (%) | Nymph Yield | Nymphs’ Viability (%) |
| pP0-KLH (n = 4) | 15% (167 ± 75) a | 0% (95 ± 2) a | 63% (40 ± 15) b | 47% (50 ± 4) b |
| pP0-Bm86 (n = 4) | 30% (138 ± 61) a | 0% (92 ± 5) a | 38% (67 ± 21) b | 33% (64 ± 9) b |
| PBS (n = 4, Control) | (196 ± 72) a | (92 ± 3) a | (108 ± 15) a | (95 ± 6) a |
| Reduction Percent with Respect to Control of | ||||
| Groups | Female Yield | Egg Mass Weight (mg) | Hatchery (%) | E (%) |
| pP0-KLH (n = 4) | 54% (11 ± 8) b | 22% (51.01 ± 8.4) b | 28% (68 ± 7) b | 95% |
| pP0-Bm86 (n = 4) | 38% (15 ± 3) b | 5% (62.06 ± 25.0) a,b | 9% (86 ± 3) b | 86% |
| PBS (n = 4, Control) | (24 ± 6) a | (64.93 ± 18.06) a | (94 ± 5) a | |
| Reduction Percent with Respect to Control of | ||||
|---|---|---|---|---|
| Groups | Female Yield | Egg Mass Weight (mg) | Hatchery (%) | E (%) |
| pP0–KLH (n = 5) | 84% (102 ± 24) b | 7% (72.89 ± 51.94) a,b | 28% (71 ± 10) b | 89% |
| pP0–Bm86 (n = 5) | 72% (177 ± 56) b | 22% (69.36 ± 44.81) b | 34% (65 ± 18) b | 84% |
| PBS (n = 5, Control) | (639 ± 222) a | (78.29 ± 44.84) a | (98 ± 5) a | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Mallón, A.; Javier González, L.; Encinosa Guzmán, P.E.; Bechara, G.H.; Sanches, G.S.; Pousa, S.; Cabrera, G.; Cabrales, A.; Garay, H.; Mejías, R.; et al. Functional and Mass Spectrometric Evaluation of an Anti-Tick Antigen Based on the P0 Peptide Conjugated to Bm86 Protein. Pathogens 2020, 9, 513. https://doi.org/10.3390/pathogens9060513
Rodríguez Mallón A, Javier González L, Encinosa Guzmán PE, Bechara GH, Sanches GS, Pousa S, Cabrera G, Cabrales A, Garay H, Mejías R, et al. Functional and Mass Spectrometric Evaluation of an Anti-Tick Antigen Based on the P0 Peptide Conjugated to Bm86 Protein. Pathogens. 2020; 9(6):513. https://doi.org/10.3390/pathogens9060513
Chicago/Turabian StyleRodríguez Mallón, Alina, Luis Javier González, Pedro Enrique Encinosa Guzmán, Gervasio Henrique Bechara, Gustavo Seron Sanches, Satomy Pousa, Gleysin Cabrera, Ania Cabrales, Hilda Garay, Raúl Mejías, and et al. 2020. "Functional and Mass Spectrometric Evaluation of an Anti-Tick Antigen Based on the P0 Peptide Conjugated to Bm86 Protein" Pathogens 9, no. 6: 513. https://doi.org/10.3390/pathogens9060513
APA StyleRodríguez Mallón, A., Javier González, L., Encinosa Guzmán, P. E., Bechara, G. H., Sanches, G. S., Pousa, S., Cabrera, G., Cabrales, A., Garay, H., Mejías, R., López Álvarez, J. R., Bello Soto, Y., Almeida, F., Guirola, O., Rodríguez Fernández, R., Fuentes Castillo, A., Méndez, L., Jiménez, S., Licea-Navarro, A., ... Estrada, M. P. (2020). Functional and Mass Spectrometric Evaluation of an Anti-Tick Antigen Based on the P0 Peptide Conjugated to Bm86 Protein. Pathogens, 9(6), 513. https://doi.org/10.3390/pathogens9060513









