Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand
Abstract
1. Introduction
2. Results
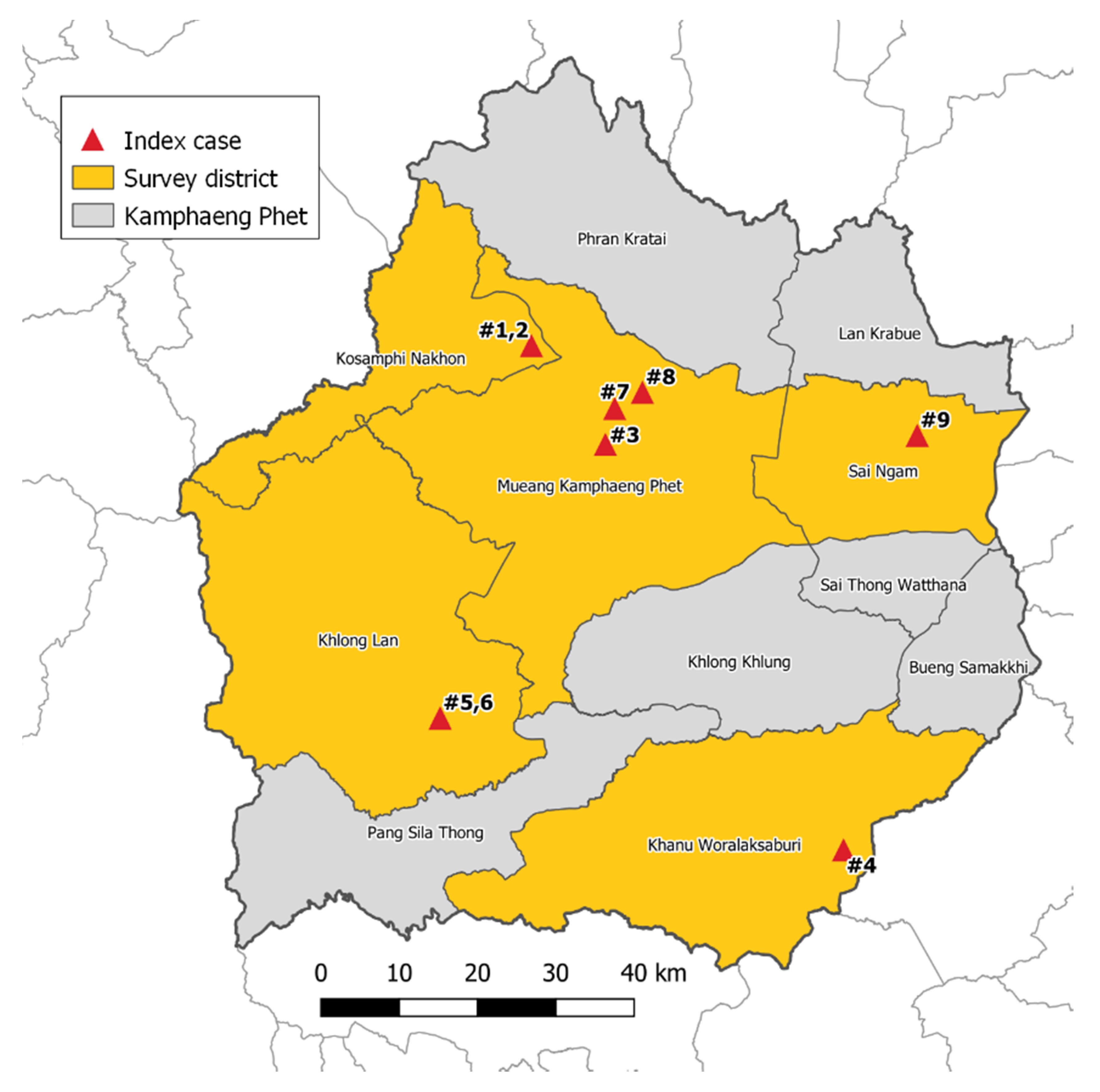
2.1. Mosquito Collection
2.2. Virus Detection
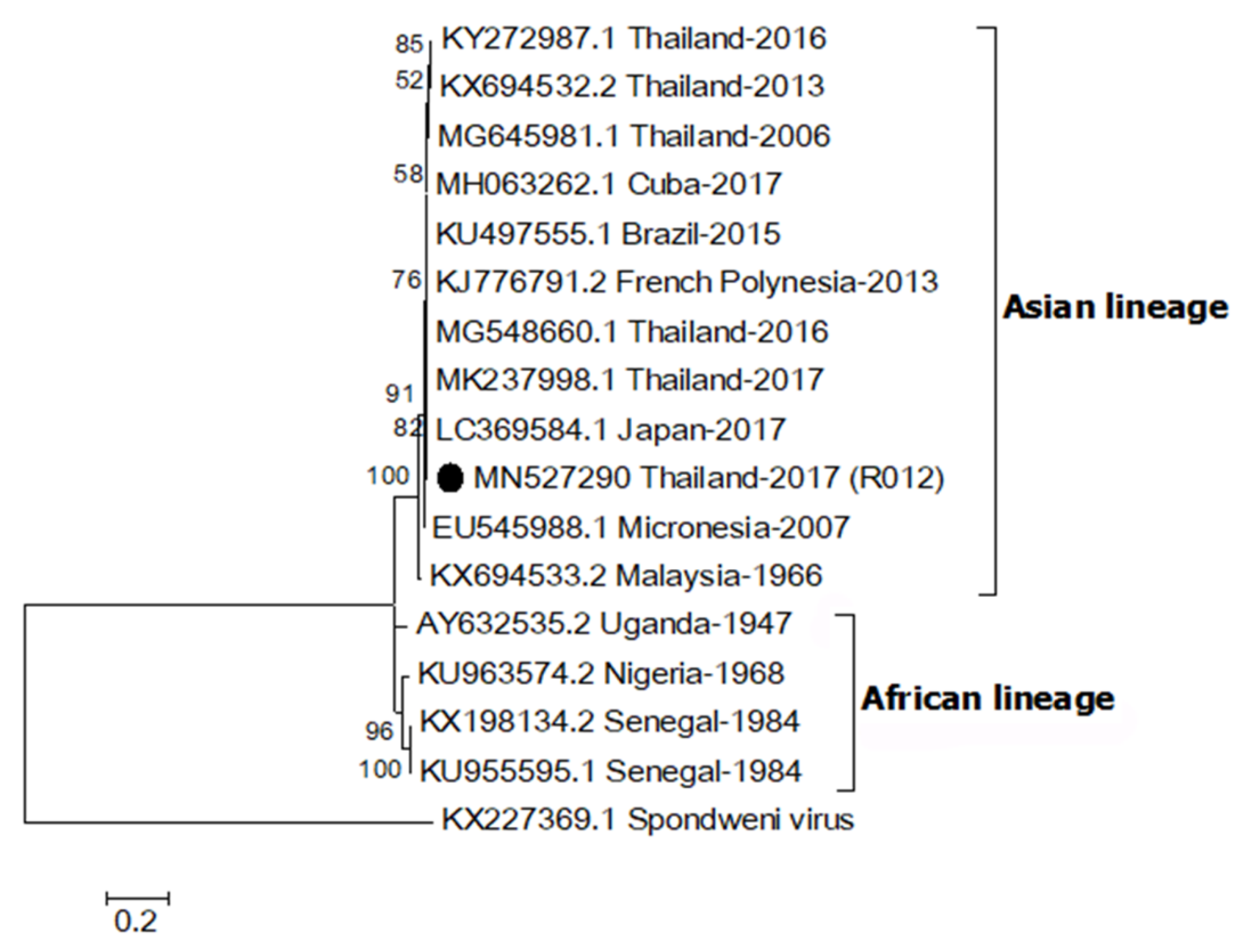
2.3. Phylogenetic Relationship of ZIKV
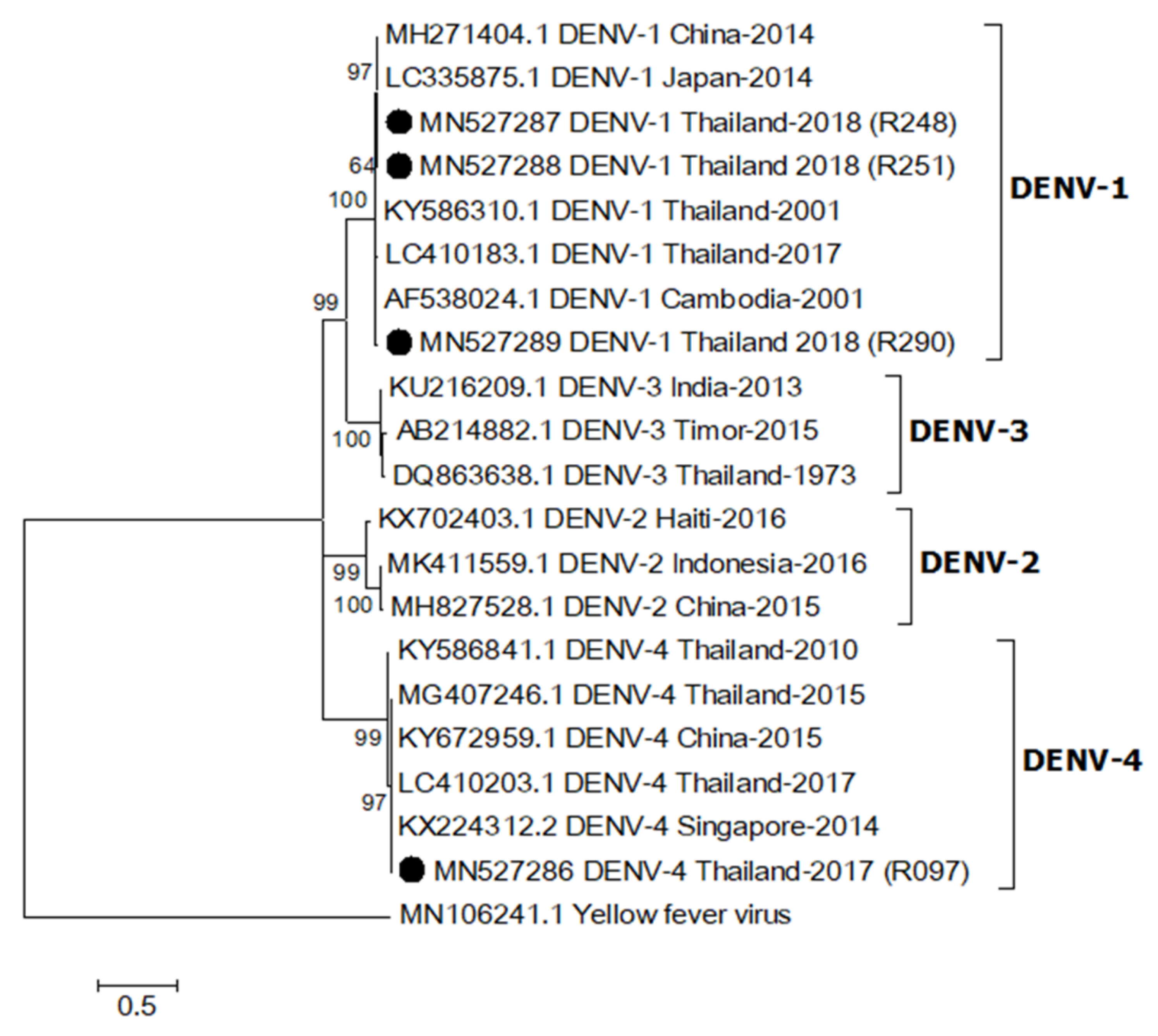
2.4. Phylogenetic Relationship of DENV
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Site
5.2. Mosquito Collection
5.3. Detection of ZIKV and DENV by Real-Time RT-PCR
5.4. DNA Sequencing and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Dengue and severe dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 3 April 2020).
- Buathong, R.; Hermann, L.; Thaisomboonsuk, B.; Rutvisuttinunt, W.; Klungthong, C.; Chinnawirotpisan, P.; Manasatienkij, W.; Nisalak, A.; Fernandez, S.; Yoon, I.K.; et al. Detection of Zika virus infection in Thailand, 2012–2014. Am. J. Trop. Med. Hyg. 2015, 93, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.P.; Chen, L.H.; Hamer, D.H. Evolving epidemiology of Japanese Encephalitis: Implications for vaccination. Curr. Infect. Dis. Rep. 2018, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Halverson, S.; Ezinwa, N. Mosquito-borne diseases. Prim. Care. 2018, 45, 393–407. [Google Scholar] [CrossRef]
- Boyer, S.; Calvez, E.; Chouin-Carneiro, T.; Diallo, D.; Failloux, A.B. An overview of mosquito vectors of Zika virus. Microbes. Infect. 2018, 20, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet. Infect. Dis. 2016, 16, e119–e126. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastere, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet. 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Carteaux, G.; Maquart, M.; Bedet, A.; Contou, D.; Brugières, P.; Fourati, S.; Cleret de Langavant, L.; de Broucker, T.; Brun-Buisson, C.; Leparc-Goffart, I.; et al. Zika virus associated with meningoencephalitis. N. Engl. J. Med. 2016, 374, 1595–1596. [Google Scholar] [CrossRef]
- Kleber de Oliveira, W.; Cortez-Escalante, J.; De Oliveira, W.T.; do Carmo, G.M.; Henriques, C.M.; Coelho, G.E.; Araújo de França, G.V. Increase in reported prevalence of microcephaly in infants born to women living in areas with confirmed Zika virus transmission during the first trimester of pregnancy—Brazil, 2015. MMWR. Morb. Mortal. Wkly. Rep. 2016, 65, 242–247. [Google Scholar] [CrossRef]
- Holmes, E.C.; Twiddy, S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef]
- Hotta, S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J. Infect. Dis. 1952, 90, 1–9. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Bhatt, S.; Katzelnick, L.; Howes, R.E.; Battle, K.E.; et al. Global spread of dengue virus types: Mapping the 70 years history. Trends. Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef]
- World Health Organization. Global strategy for dengue prevention and control 2012–2020. Available online: http://apps.who.int/iris/bitstream/handle/10665/75303/9789241504034_eng.pdf;jsessionid=8BD7C5C13E1CC23A760ADD649728C11C?sequence=1 (accessed on 3 April 2020).
- Phadungsombat, J.; Lin, M.Y.; Srimark, N.; Yamanaka, A.; Nakayama, E.E.; Moolasart, V.; Suttha, P.; Shioda, T.; Uttayamakul, S. Emergence of genotype cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. PLoS. ONE. 2018, 13, e0207220. [Google Scholar] [CrossRef]
- Sittivicharpinyo, T.; Wonnapinij, P.; Surat, W. Phylogenetic analyses of DENV-3 isolated from field-caught mosquitoes in Thailand. Virus. Res. 2018, 244, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Hayes, E.B. Zika virus outside Africa. Emerg. Infect. Dis. 2009, 15, 1347–1350. [Google Scholar] [CrossRef]
- Moore, D.L.; Causey, O.R.; Carey, D.E.; Reddy, S.; Cooke, A.R.; Akinkugbe, F.M.; David-West, T.S.; Kemp, G.E. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann. Trop. Med. Parasitol. 1975, 69, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.; Ong, S.; Leang, R.; Huy, R.; Ly, S.; Mounier, U.; Ou, T.; In, S.; Peng, B.; Ken, S.; et al. Low circulation of Zika virus, Cambodia, 2007–2016. Emerg. Infect. Dis. 2017, 23, 296–299. [Google Scholar] [CrossRef]
- Perkasa, A.; Yudhaputri, F.; Haryanto, S.; Hayati, R.F.; Ma’roef, C.N.; Antonjaya, U.; Yohan, B.; Myint, K.S.; Ledermann, J.P.; Rosenberg, R.; et al. Isolation of Zika virus from febrile patient, Indonesia. Emerg. Infect. Dis. 2016, 22, 924–925. [Google Scholar] [CrossRef]
- Tappe, D.; Nachtigall, S.; Kapaun, A.; Schnitzler, P.; Günther, S.; Schmidt-Chanasit, J. Acute Zika virus infection after travel to Malaysian Borneo, September 2014. Emerg. Infect. Dis. 2015, 21, 911–913. [Google Scholar] [CrossRef]
- Alera, M.T.; Hermann, L.; Tac-An, I.A.; Klungthong, C.; Rutvisuttinunt, W.; Manasatienkij, W.; Villa, D.; Thaisomboonsuk, B.; Velasco, J.M.; Chinnawirotpisan, P.; et al. Zika virus infection, Philippines, 2012. Emerg. Infect. Dis. 2015, 21, 722–724. [Google Scholar] [CrossRef]
- Singapore Zika Study Group. Outbreak of Zika virus infection in Singapore: An epidemiological, entomological, virological, and clinical analysis. Lancet. Infect. Dis. 2017, 17, 813–821. [Google Scholar] [CrossRef]
- Moi, M.L.; Nguyen, T.T.T.; Nguyen, C.T.; Vu, T.B.H.; Tun, M.M.N.; Pham, T.D.; Pham, N.T.; Tran, T.; Morita, K.; Le, T.Q.M.; et al. Zika virus infection and microcephaly in Vietnam. Lancet. Infect. Dis. 2017, 17, 805–806. [Google Scholar] [CrossRef]
- Fonseca, K.; Meatherall, B.; Zarra, D.; Drebot, M.; MacDonald, J.; Pabbaraju, K.; Wong, S.; Webster, P.; Lindsay, R.; Tellier, R. First case of Zika virus infection in a returning Canadian traveler. Am. J. Trop. Med. Hyg. 2014, 91, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Tappe, D.; Rissland, J.; Gabriel, M.; Emmerich, P.; Gunther, S.; Held, G.; Smola, S.; Schmidt-Chanasit, J. First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Eur. Surveill. 2014, 19, 20685. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, K.; Kutsuna, S.; Takasaki, T.; Moi, M.L.; Ikeda, M.; Kotaki, A.; Yamamoto, K.; Fujiya, Y.; Mawatari, M.; Takeshita, N.; et al. Zika fever imported from Thailand to Japan, and diagnosed by PCR in the urines. J. Travel. Med. 2016, 23, tav011. [Google Scholar] [CrossRef]
- Khongwichit, S.; Wikan, N.; Auewarakul, P.; Smith, D.R. Zika virus in Thailand. Microbes. Infect. 2018, 20, 670–675. [Google Scholar] [CrossRef]
- Phumee, A.; Buathong, R.; Boonserm, R.; Intayot, P.; Aungsananta, N.; Jittmittraphap, A.; Joyjinda, Y.; Wacharapluesadee, S.; Siriyasatien, P. Molecular epidemiology and genetic diversity of Zika virus from field-caught mosquitoes in various regions of Thailand. Pathogens 2019, 8, 30. [Google Scholar] [CrossRef]
- Wongsurawat, T.; Athipanyasilp, N.; Jenjaroenpun, P.; Jun, S.R.; Kaewnapan, B.; Wassenaar, T.M.; Leelahakorn, N.; Angkasekwinai, N.; Kantakamalakul, W.; Ussery, D.W.; et al. Case of microcephaly after congenital infection with Asian lineage Zika virus, Thailand. Emerg. Infect. Dis. 2018, 24, 1758. [Google Scholar] [CrossRef]
- Faye, O.; Freire, C.C.; Iamarino, A.; Faye, O.; de Oliveira, J.V.; Diallo, M.; Zanotto, P.M.; Sall, A.A. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS. Negl. Trop. Dis. 2014, 8, e2636. [Google Scholar] [CrossRef]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS. Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef]
- Ferreira-de-Lima, V.H.; Lima-Camara, T.N. Natural vertical transmission of dengue virus in Aedes aegypti and Aedes albopictus: A systematic review. Parasit. Vectors. 2018, 11, 77. [Google Scholar] [CrossRef]
- Rattanarithikul, R.; Harbach, R.E.; Harrison, B.A.; Panthusiri, P.; Coleman, R.E.; Richardson, J.H. Illustrated keys to the mosquitoes of Thailand. VI. Tribe Aedini. Southeast. Asian. J. Trop. Med. Public. Health. 2010, 41, 1–225. [Google Scholar]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
| Species | Female | Male | Total |
|---|---|---|---|
| Ae. aegypti | 130 | 57 | 187 |
| Ae. albopictus | 11 | 1 | 12 |
| An. peditaeniatus | 1 | - | 1 |
| An. tessellatus | 1 | - | 1 |
| An. sp. | 1 | - | 1 |
| Ar. subalbatus | 4 | - | 4 |
| Cx. brevipalpis | 3 | - | 3 |
| Cx. quinquefasciatus | 62 | 123 | 185 |
| Cx. vishnui | 88 | 5 | 93 |
| Cx. sp | 1 | - | 1 |
| Primer | Sequence (5′→3′) | Position (nt) a | Virus | Region |
|---|---|---|---|---|
| Real-Time RT-PCR | ||||
| ZIKV835 | TTGGTCATGATACTGCTGATTGC | 835–857 | ZIKV | Matrix |
| ZIKV911c | CCTTCCACAAAGTCCCTATTGC | 911–890 | Envelope | |
| ZIKV860-FAM b | CGGCATACAGCATCAGGTGCATAGGAG | 860–886 | Envelope | |
| ZIKV1086 | CCGCTGCCCAACACAAG | 1086–1102 | ZIKV | Envelope |
| ZIKV1162c | CCACTAACGTTCTTTTGCAGACAT | 1162–1139 | Envelope | |
| ZIKV1107-FAM b | AGCCTACCTTGACAAGCAGTCAGACACTCAA | 1107–1137 | Envelope | |
| RT-PCR for Sequencing | ||||
| ZIKENVF | GCTGGDGCRGACACHGGRACT | 1643–1663 | ZIKV | Envelope |
| ZIKENVR | RTCYACYGCCATYTGGRCTG | 1989–2008 | Envelope | |
| D1 | TCAATATGCTGAAACGCGCGAGAAACCG | 132–159 | DENV | Capsid/prM |
| D2 | TTGCACCAACAGTCAATGTCTTCAGGTTC | 614–642 | Capsid/prM | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosoltanapiwat, N.; Tongshoob, J.; Singkhaimuk, P.; Nitatsukprasert, C.; Davidson, S.A.; Ponlawat, A. Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand. Pathogens 2020, 9, 442. https://doi.org/10.3390/pathogens9060442
Kosoltanapiwat N, Tongshoob J, Singkhaimuk P, Nitatsukprasert C, Davidson SA, Ponlawat A. Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand. Pathogens. 2020; 9(6):442. https://doi.org/10.3390/pathogens9060442
Chicago/Turabian StyleKosoltanapiwat, Nathamon, Jarinee Tongshoob, Preeraya Singkhaimuk, Chanyapat Nitatsukprasert, Silas A. Davidson, and Alongkot Ponlawat. 2020. "Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand" Pathogens 9, no. 6: 442. https://doi.org/10.3390/pathogens9060442
APA StyleKosoltanapiwat, N., Tongshoob, J., Singkhaimuk, P., Nitatsukprasert, C., Davidson, S. A., & Ponlawat, A. (2020). Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand. Pathogens, 9(6), 442. https://doi.org/10.3390/pathogens9060442





