Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy
Abstract
1. Introduction
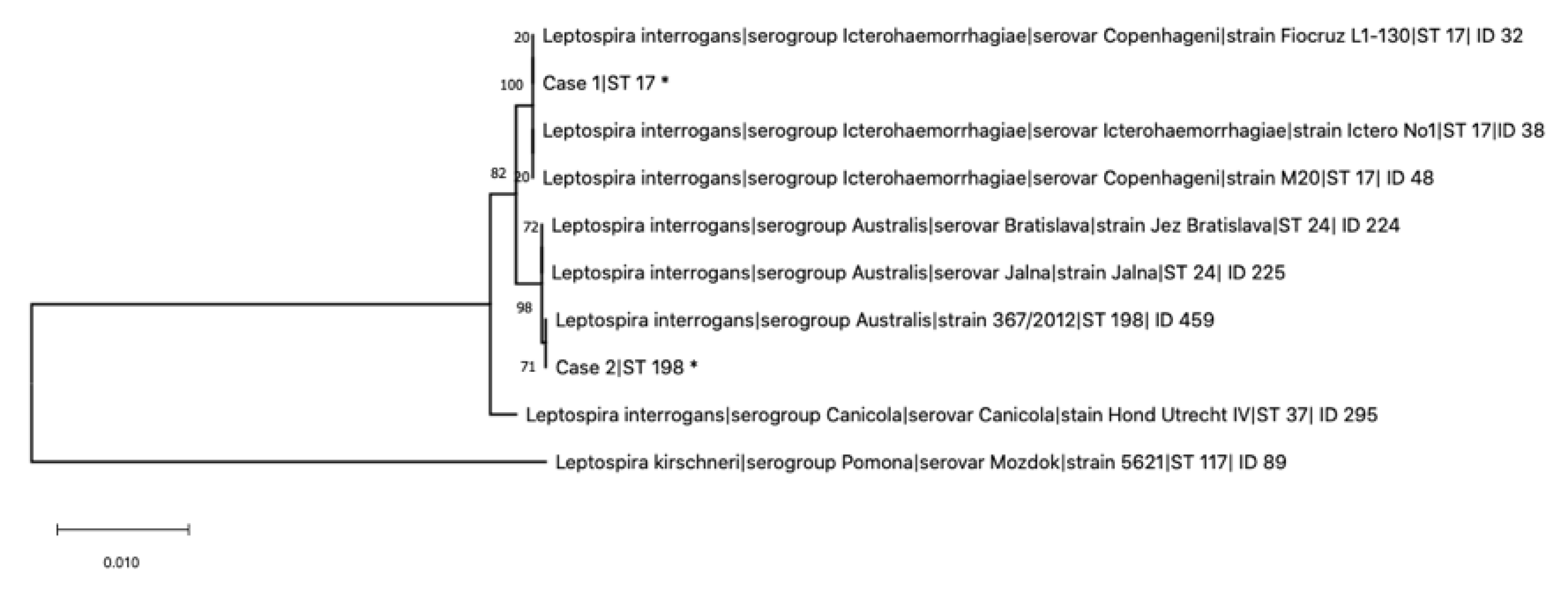
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection, Inclusion Criteria, and Diagnosis of Leptospirosis
4.2. Multi-Locus Sequence Typing
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Goldstein, R.E.; Lin, R.C.; Langston, C.E.; Scrivani, P.V.; Erb, H.N.; Barr, S.C. Influence of infecting serogroup on clinical features of leptospirosis in dogs. J. Vet. Intern. Med. 2006, 20, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Kohn, B.; Steinicke, K.; Arndt, G.; Gruber, A.D.; Guerra, B.; Jansen, A.; Kaser-Hotz, B.; Klopfleisch, R.; Lotz, F.; Luge, E.; et al. Pulmonary abnormalities in dogs with leptospirosis. J. Vet. Intern. Med. 2010, 24, 1277–1282. [Google Scholar] [CrossRef] [PubMed]
- Delaude, A.; Rodriguez-Campos, S.; Dreyfus, A.; Counotte, M.J.; Francey, T.; Schweighauser, A.; Lettry, S.; Schuller, S. Canine leptospirosis in Switzerland-A prospective cross-sectional study examining seroprevalence, risk factors and urinary shedding of pathogenic leptospires. Prev. Vet. Med. 2017, 141, 48–60. [Google Scholar] [CrossRef]
- López, M.C.; Vila, A.; Rodón, J.; Roura, X. Leptospira seroprevalence in owned dogs from Spain. Heliyon 2019, 5, 02373. [Google Scholar] [CrossRef]
- Tagliabue, S.; Figarolli, B.M.; D’Incau, M.; Foschi, G.; Gennero, M.S.; Giordani, R.; Natale, A.; Papa, P.; Ponti, N.; Scaltrito, D.; et al. Serological surveillance of Leptospirosis in Italy: Two-year national data (2010–2011). Vet. Ital. 2016, 52, 129–138. [Google Scholar]
- Bertelloni, F.; Cilia, G.; Turchi, B.; Pinzauti, P.; Cerri, D.; Fratini, F. Epidemiology of leptospirosis in North-Central Italy: Fifteen years of serological data (2002–2016). Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 14–22. [Google Scholar] [CrossRef]
- Schuller, S.; Francey, T.; Hartmann, K.; Hugonnard, M.; Kohn, B.; Nally, J.E.; Sykes, J. European consensus statement on leptospirosis in dogs and cats. J. Small Anim. Pract. 2015, 56, 159–179. [Google Scholar] [CrossRef]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef]
- Miller, M.D.; Annis, K.M.; Lappin, M.R.; Lunn, K.F. Variability in results of the microscopic agglutination test in dogs with clinical leptospirosis and dogs vaccinated against leptospirosis. J. Vet. Intern. Med. 2006, 25, 426–432. [Google Scholar] [CrossRef]
- Troìa, R.; Balboni, A.; Zamagni, S.; Frigo, S.; Magna, L.; Perissinotto, L.; Battilani, M.; Dondi, F. Prospective evaluation of rapid point-of-care tests for the diagnosis of acute leptospirosis in dogs. Vet. J. 2018, 237, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.A.; Tozzi, B.F.; Penteado, M.S.; Guilloux, A.G.A.; Moreno, L.Z.; Heinemann, M.B.; Moreno, A.M.; Lilenbaum, W.; Hagiwara, M.K. Diagnosis of acute canine leptospirosis using multiple laboratory tests and characterization of the isolated strains. BMC Vet. Res. 2018, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Muto, M.M.; Akachi, S.; Okano, S.; Yamamoto, S.; Horikawa, K.; Harada, S.; Funatsumaru, S.; Ohnishi, M. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J. Med. Microbiol. 2013, 62, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Muto, M.M.; Izumiya, H.; Suzuki, M.; Ohnishi, M. Multiple-locus variable-number tandem repeat analysis and clinical characterization of Leptospira interrogans canine isolates. J. Med. Microbiol. 2015, 64, 288–294. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Larson, C.R.; Dennis, M.; Nair, R.V.; Llanes, A.; Peda, A.; Welcome, S.; Rajeev, S. Isolation and characterization of Leptospira interrogans serovar Copenhageni from a dog from Saint Kitts. JMM Case Rep. 2017, 4, e005120. [Google Scholar] [CrossRef]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop Dis. 2013, 7, e1954. [Google Scholar] [CrossRef]
- Day, M.J.; Horzinek, M.C.; Schultz, R.D.; Squires, R.A. Vaccination Guidelines Group (VGG) of the World Small Animal Veterinary Association (WSAVA). WSAVA Guidelines for the vaccination of dogs and cats. J. Small Anim. Pract. 2016, 57, E1–E45. [Google Scholar] [CrossRef]
- Ferreira, A.S.; Ahmed, A.; Rocha, T.; Vieira, M.L.; Paiva-Cardoso, M.D.N.; Mesquita, J.R.; van der Linden, H.; Goris, M.; Thompson, G.; Hartskeerl, R.A.; et al. Genetic diversity of pathogenic leptospires from wild, domestic and captive host species in Portugal. Transbound. Emerg. Dis. 2020, 67, 852–864. [Google Scholar] [CrossRef]
- Arent, Z.; Gilmore, C.; Brem, S.; Ellis, W.A. Molecular studies on European equine isolates of Leptospira interrogans serovars Bratislava and Muenchen. Infect. Genet. Evol. 2015, 34, 26–31. [Google Scholar] [CrossRef]
- Renaud, C.; Andrews, S.; Djelouadji, Z.; Lecheval, S.; Corrao-Revol, N.; Buff, S.; Demont, P.; Kodjo, A. Prevalence of the Leptospira serovars bratislava, grippotyphosa, mozdok and pomona in French dogs. Vet. J. 2013, 196, 126–127. [Google Scholar] [CrossRef]
- Mastrorilli, C.; Dondi, F.; Agnoli, C.; Turba, M.E.; Vezzali, E.; Gentilini, F. Clinicopathologic features and outcome predictors of Leptospira interrogans Australis serogroup infection in dogs: A retrospective study of 20 cases (2001–2004). J. Vet. Intern. Med. 2007, 21, 3–10. [Google Scholar] [CrossRef]
- Boniotti, M.B.; Gelmini, L.; Carra, E.; Figarolli, B.M.; D’incau, M.; Tagliabue, S. Leptospira Interrogans Serogroup Australis In Hedgehog In Northern Italy. In Proceedings of the International Leptospirsis Society, Fukuoka, Japan, 7–11 October 2013. [Google Scholar]
- Nielsen, J.N.; Armstrong, C.H.; Nielsen, N.C. Relationship among selected Leptospira interrogans serogroups as determined by nucleic acid hybridization. J. Clin. Microbiol. 1989, 27, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Menezes, A.; Woods, K.; Chanthongthip, A.; Dittrich, S.; Opoku-Boateng, A.; Kimuli, M.; Chalker, V. An Extended Multilocus Sequence Typing (MLST) Scheme for Rapid Direct Typing of Leptospira from Clinical Samples. PLoS Negl. Trop. Dis. 2016, 10, e0004996. [Google Scholar] [CrossRef] [PubMed]
- Leptospira spp. MLST Database. Available online: https://pubmlst.org/leptospira/ (accessed on 9 April 2020).
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balboni, A.; Zamagni, S.; Bertasio, C.; Boniotti, M.B.; Troìa, R.; Battilani, M.; Dondi, F. Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy. Pathogens 2020, 9, 351. https://doi.org/10.3390/pathogens9050351
Balboni A, Zamagni S, Bertasio C, Boniotti MB, Troìa R, Battilani M, Dondi F. Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy. Pathogens. 2020; 9(5):351. https://doi.org/10.3390/pathogens9050351
Chicago/Turabian StyleBalboni, Andrea, Silvia Zamagni, Cristina Bertasio, Maria Beatrice Boniotti, Roberta Troìa, Mara Battilani, and Francesco Dondi. 2020. "Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy" Pathogens 9, no. 5: 351. https://doi.org/10.3390/pathogens9050351
APA StyleBalboni, A., Zamagni, S., Bertasio, C., Boniotti, M. B., Troìa, R., Battilani, M., & Dondi, F. (2020). Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy. Pathogens, 9(5), 351. https://doi.org/10.3390/pathogens9050351




