Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy
Abstract
1. Introduction
2. Results
2.1. Microscopic Agglutination Test (MAT)
2.2. Molecular Analysis
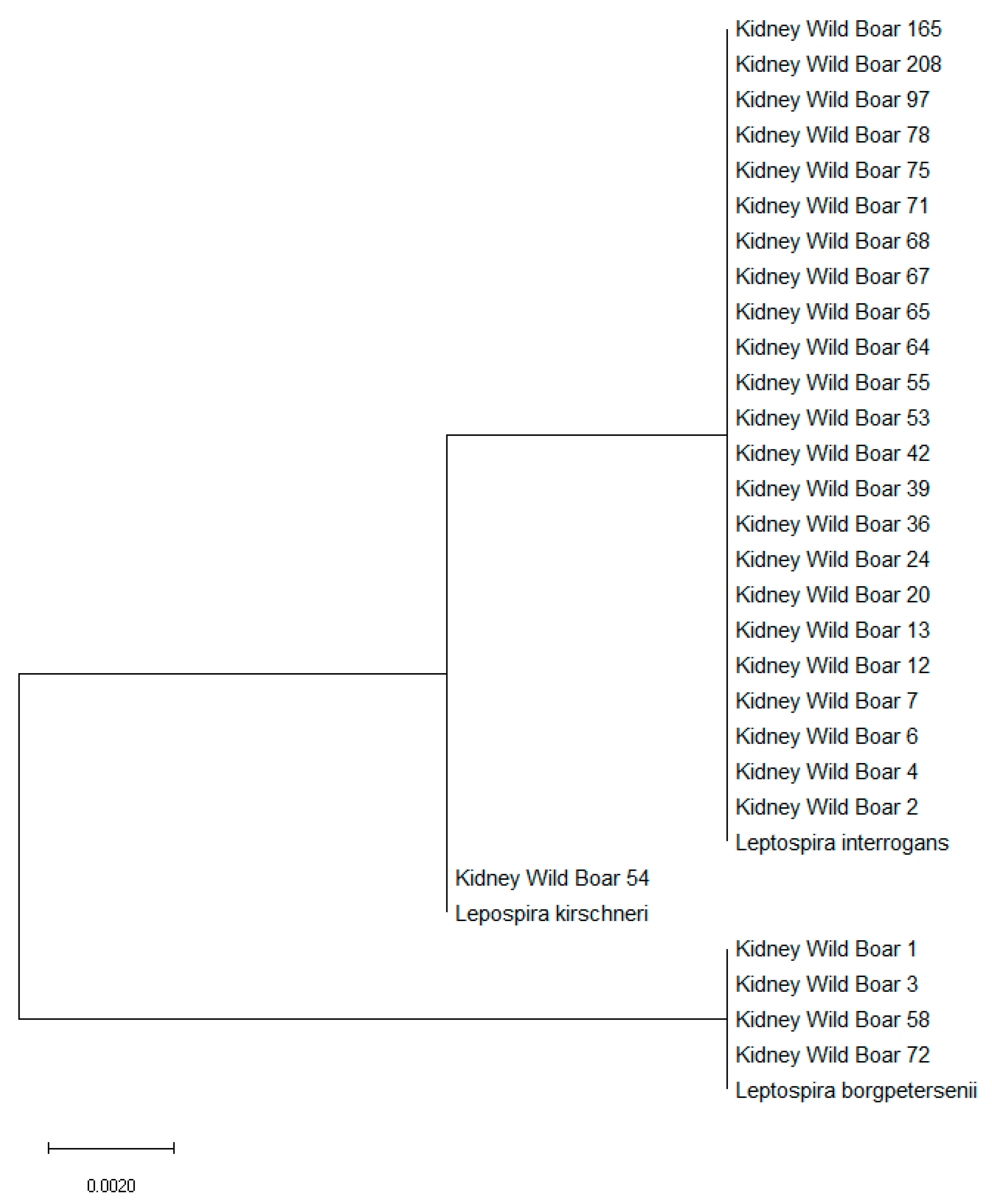
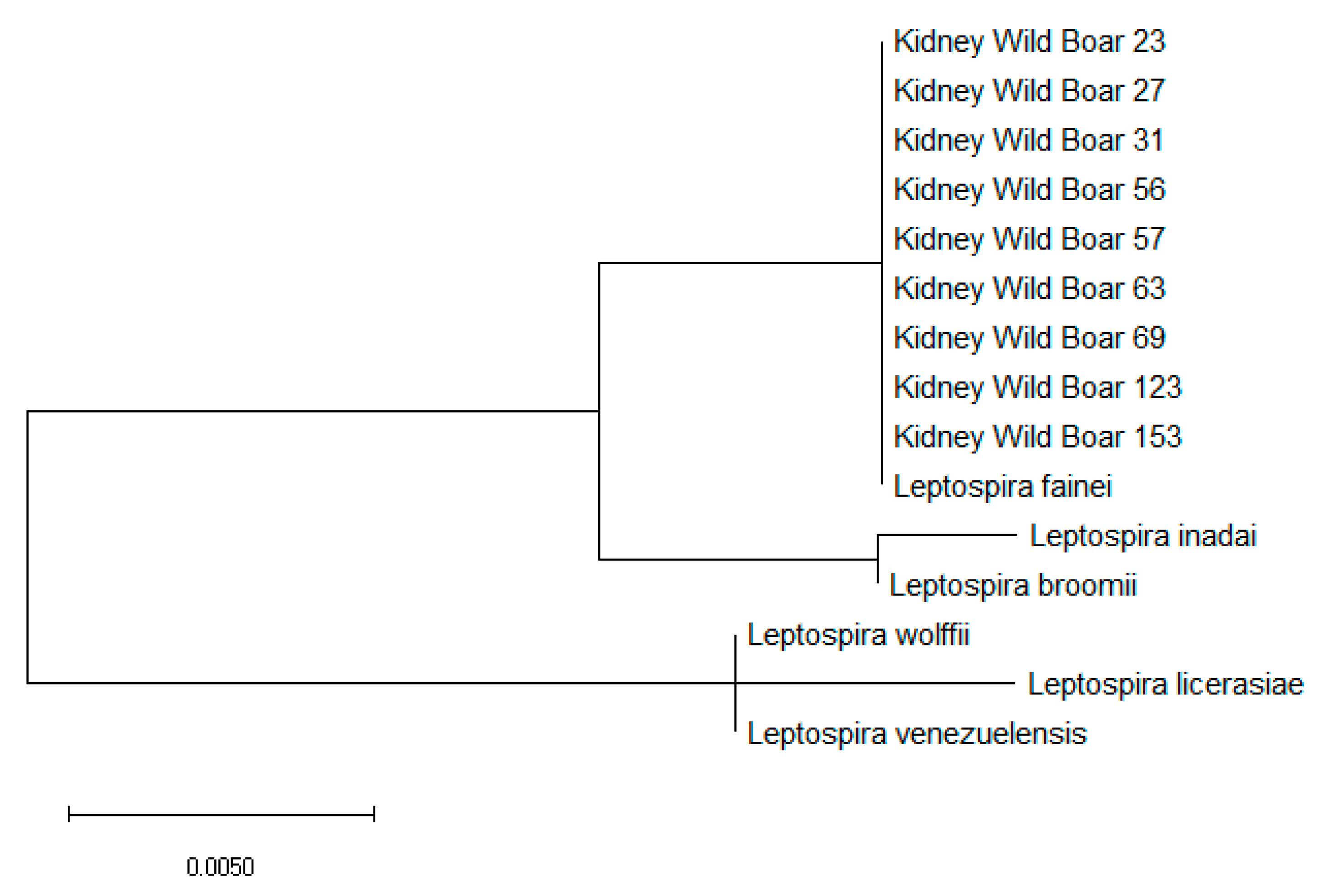
2.3. Leptospira spp. Isolation, Characterization and Genotyping
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Microscopic Agglutination Test (MAT)
4.3. Leptospira spp. Isolation
4.4. Molecular Analysis
4.5. Leptospira spp. Characterization and Genotyping
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Massei, G.; Kindberg, J.; Licoppe, A.; Gačić, D.; Šprem, N.; Kamler, J.; Baubet, E.; Hohmann, U.; Monaco, A.; Ozoliņš, J.; et al. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag. Sci. 2015, 71, 492–500. [Google Scholar] [CrossRef]
- Castillo-Contreras, R.; Carvalho, J.; Serrano, E.; Mentaberre, G.; Fernández-Aguilar, X.; Colom, A.; González-Crespo, C.; Lavín, S.; López-Olvera, J.R. Urban wild boars prefer fragmented areas with food resources near natural corridors. Sci. Total Environ. 2018, 615, 282–288. [Google Scholar] [CrossRef]
- Santilli, F.; Varuzza, P. Factors affecting wild boar (Sus scrofa) abundance in southern Tuscany. Hystrix, Ital. J. Mammal. 2013, 24, 169–173. [Google Scholar]
- Pittiglio, C.; Khomenko, S.; Beltran-Alcrudo, D. Wild boar mapping using population-density statistics: From polygons to high resolution raster maps. PLoS ONE 2018, 13, e0193295. [Google Scholar] [CrossRef]
- Carnevali, L.; Pedrotti, L.; Riga, F.; Toso, S. Banca Dati Ungulati: Status, distribuzione, consistenza, gestione e prelievo venatorio delle popolazioni di Ungulati in Italia. Rapporto 2001–2005. Biol. e Conserv. della Fauna 2009, 117, 1–168. [Google Scholar]
- Lombardini, M.; Meriggi, A.; Fozzi, A. Factors influencing wild boar damage to agricultural crops in Sardinia (Italy). Curr. Zool. 2017, 63, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Bertelloni, F.; Mignone, W.; Spina, S.; Berio, E.; Razzuoli, E.; Vencia, W.; Franco, V.; Cecchi, F.; Bogi, S.; et al. Molecular detection of Leptospira spp. in wild boar (Sus scrofa) hunted in Liguria region (Italy). Comp. Immunol. Microbiol. Infect. Dis. 2020, 68, 101410. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J.; Lindsay, D.S.; Sriranganathan, N. Wild boars as sources for infectious diseases in livestock and humans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009, 364, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Bertelloni, F.; Mazzei, M.; Cilia, G.; Forzan, M.; Felicioli, A.; Sagona, S.; Bandecchi, P.; Turchi, B.; Cerri, D.; Fratini, F. Serological Survey on Bacterial and Viral Pathogens in Wild Boars Hunted in Tuscany. Ecohealth 2020, 17, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef]
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef]
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.A.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus 2013, 2, 362. [Google Scholar] [CrossRef]
- Barragan, V.; Chiriboga, J.; Miller, E.; Olivas, S.; Birdsell, D.; Hepp, C.; Hornstra, H.; Schupp, J.M.; Morales, M.; Gonzalez, M.; et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016, 10, e0004990. [Google Scholar] [CrossRef]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Morales, M.; et al. High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141–2147. [Google Scholar] [CrossRef]
- Ruiz-Fons, F. A Review of the Current Status of Relevant Zoonotic Pathogens in Wild Swine (Sus scrofa) Populations: Changes Modulating the Risk of Transmission to Humans. Transbound. Emerg. Dis. 2017, 64, 68–88. [Google Scholar] [CrossRef]
- Dupouey, J.; Faucher, B.; Edouard, S.; Richet, H.; Kodjo, A.; Drancourt, M.; Davoust, B. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 77–83. [Google Scholar] [CrossRef]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar]
- Ellis, W.A. Animal Leptospirosis. In Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology; Adler, B., Ed.; Springer: Berlin, Germany, 2015; Volume 387, pp. 99–137. [Google Scholar]
- Cerri, D.; Ebani, V.V.; Fratini, F.; Pinzauti, P.; Andreani, E. Epidemiology of leptospirosis: Observations on serological data obtained by a "diagnostic laboratory for leptospirosis" from 1995 to 2001. New Microbiol. 2003, 26, 383–389. [Google Scholar] [PubMed]
- Żmudzki, J.; Jabłoński, A.; Nowak, A.; Zębek, S.; Arent, Z.; Bocian, Ł.; Pejsak, Z. First overall report of Leptospira infections in wild boars in Poland. Acta Vet. Scand. 2015, 58, 3. [Google Scholar] [CrossRef]
- Vale-Goncalves, H.M.; Cabral, J.A.; Faria, M.C.; Nunes-Pereira, M.; Faria, A.S.; Veloso, O.; Vieira, M.L.; Paiva-Cardoso, M.N. Prevalence of Leptospira antibodies in wild boars (Sus scrofa) from Northern Portugal: Risk factor analysis. Epidemiol. Infect. 2015, 143, 2126–2130. [Google Scholar] [CrossRef] [PubMed]
- Boqvist, S.; Bergström, K.; Magnusson, U. Prevalence of Antibody to Six Leptospira Servovars in Swedish Wild Boars. J. Wildl. Dis. 2012, 48, 492–496. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bertelloni, F.; Cilia, G.; Turchi, B.; Pinzauti, P.; Cerri, D.; Fratini, F. Epidemiology of leptospirosis in North-Central Italy: Fifteen years of serological data (2002–2016). Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, S.; Figarolli, B.M.; D’Incau, M.; Foschi, G.; Gennero, M.S.; Giordani, R.; Giordani, R.; Natale, A.; Papa, P.; Ponti, N.; et al. Serological surveillance of Leptospirosis in Italy: Two-year national data (2010-2011). Vet. Ital. 2016, 52, 129–138. [Google Scholar]
- Coppola, F.; Cilia, G.; Bertelloni, F.; Casini, L.; D’Addio, E.; Fratini, F.; Cerri, D.; Felicioli, A. Crested porcupine (Hystrix cristata L.): A new potential host for pathogenic Leptospira among semi-fossorial mammals. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101472. [Google Scholar] [CrossRef]
- Fratini, F.; Turchi, B.; Ebani, V.V.; Bertelloni, F.; Galiero, A.; Cerri, D. The presence of Leptospira in coypus (Myocastor coypus) and rats (Rattus norvegicus) living in a protected wetland in Tuscany (Italy). Vet. Arh. 2015, 85, 407–414. [Google Scholar]
- Bertelloni, F.; Turchi, B.; Vattiata, E.; Viola, P.; Pardini, S.; Cerri, D.; Fratini, F. Serological survey on Leptospira infection in slaughtered swine in North-Central Italy. Epidemiol. Infect. 2018, 1–6. [Google Scholar] [CrossRef]
- Ebani, V.V.; Cerri, D.; Poli, A.; Andreani, E. Prevalence of Leptospira and Brucella antibodies in wild boars (Sus scrofa) in Tuscany, Italy. J. Wildl. Dis. 2003, 39, 718–722. [Google Scholar] [CrossRef][Green Version]
- Verin, R.; Varuzza, P.; Mazzei, M.; Poli, A. Serologic, molecular, and pathologic survey of pseudorabies virus infection in hunted wild boars (Sus scrofa) in Italy. J. Wildl. Dis. 2014, 50, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Ebani, V.V.; Bertelloni, F.; Pinzauti, P.; Cerri, D. Seroprevalence of Leptospira spp. and Borrelia burgdorferi sensu lato in Italian horses. Ann. Agric. Environ. Med. 2012, 19, 237–240. [Google Scholar] [PubMed]
- Hartskeerl, R.; Collares-Pereira, M.; Ellis, W.A. Emergence, control and re-emerging leptospirosis: Dynamics of infection in the changing world. Clin. Microbiol. Infect. 2011, 17, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Montagnaro, S.; Sasso, S.; De Martino, L.; Longo, M.; Iovane, V.; Ghiurmino, G.; Pisanelli, G.; Nava, D.; Baldi, L.; Pagnini, U. Prevalence of Antibodies to Selected Viral and Bacterial Pathogens in Wild Boar (Sus scrofa) in Campania Region, Italy. J. Wildl. Dis. 2010, 46, 316–319. [Google Scholar] [CrossRef]
- Piredda, I.; Palmas, B.; Noworol, M.; Canu, M.; Fiori, E.; Picardeau, M.; Tola, A.; Pintore, A.; Ponti, N. Indagine sulla prevalenza della leptospirosi nei cinghiali del centro-nord Sardegna. In Proceedings of the XIII Congresso Nazionale S.I.Di.L.V., Trani, Italy, 12–14 October 2011; pp. 63–64. [Google Scholar]
- Figarolli, B.M.; Gaffuri, A.; Alborali, G.L.; D’Incau, M.; Tagliabue, S. Survey on leptospirosis in wild boars (Sus scrofa) in Lombardy, Northern Italy. In Proceedings of the 1°ELS meeting, Dubrovnik, Croatian, 30 April –1 May 2012; pp. 63–64. [Google Scholar]
- Chiari, M.; Figarolli, B.M.; Tagliabue, S.; Alborali, G.L.; Bertoletti, M.; Papetti, A.; D’Incau, M.; Zanoni, M.; Boniotti, M.B. Seroprevalence and risk factors of leptospirosis in wild boars ( Sus scrofa ) in northern Italy. Hystrix, Ital. J. Mammal. 2016, 27. [Google Scholar] [CrossRef]
- Tagliabue, S.; Raffo, A.; Foni, E.; Candotti, R.; Barigazzi, G. Anticorpi per Leptospira interrogans in sieri di cinghiale selvatico (Sus scrofa) nell’Appennino parmense. In Proceedings of the Convegno Nazionale di Ecopatologia della fauna selvatica, Bologna, Italy, 9–11 February 1995. [Google Scholar]
- Espí, A.; Prieto, J.M.; Alzaga, V. Leptospiral antibodies in Iberian red deer (Cervus elaphus hispanicus), fallow deer (Dama dama) and European wild boar (Sus scrofa) in Asturias, Northern Spain. Vet. J. 2010, 183, 226–227. [Google Scholar] [CrossRef][Green Version]
- Jansen, A.; Nöckler, K.; Schönberg, A.; Luge, E.; Ehlert, D.; Schneider, T. Wild boars as possible source of hemorrhagic leptospirosis in Berlin, Germany. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 544–546. [Google Scholar] [CrossRef]
- Vicente, J.; León-Vizcaíno, L.; Gortázar, C.; Cubero, M.J.; González, M.; Martín-Atance, P. Antibodies to selected viral and bacterial pathogens in European wild boars from southcentral Spain. J. Wildl. Dis. 2002, 38, 649–652. [Google Scholar] [CrossRef]
- Vengust, G.; Lindtner-Knific, R.; Zele, D.; Bidovec, A. Leptospira antibodies in wild boars (Sus scrofa) in Slovenia. Eur. J. Wildl. Res. 2008, 54, 749–752. [Google Scholar] [CrossRef]
- Treml, F.; Pikula, J.; Holešovská, Z. Prevalence of antibodies against leptospires in the wild boar (Sus scrofa L., 1758). Vet. Med. (Praha). 2012, 48, 66–70. [Google Scholar] [CrossRef]
- Arent, Z.; Gilmore, C.; Brem, S.; Ellis, W.A. Molecular studies on European equine isolates of Leptospira interrogans serovars Bratislava and Muenchen. Infect. Genet. Evol. 2015, 34, 26–31. [Google Scholar] [CrossRef]
- Rocha, T.; Ellis, W.A.; Montgomery, J.; Gilmore, C.; Regalla, J.; Brem, S. Microbiological and serological study of leptospirosis in horses at slaughter: First isolations. Res. Vet. Sci. 2004, 76, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A.; McParland, P.J.; Bryson, D.G.; Cassells, J.A. Boars as carriers of leptospires of the Australis serogroup on farms with an abortion problem. Vet. Rec. 1986, 118, 563. [Google Scholar] [CrossRef] [PubMed]
- Slavica, A.; Cvetnić, Z.; Konjević, D.; Janicki, Z.; Severin, K.; Dežđek, D.; Starešina, V.; Sindičić, M.; Antić, J. Detection of Leptospira spp. serovars in wild boars (Sus scrofa) from continental Croatia. Vet. Arh. 2010, 80, 247–257. [Google Scholar]
- Agampodi, S.B.; Matthias, M.A.; Moreno, A.C.; Vinetz, J.M. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: Association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin. Infect. Dis. 2012, 54, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Merien, F.; Baranton, G.; Perolat, P. Comparison of Polymerase Chain Reaction with Microagglutination Test and Culture for Diagnosis of Leptospirosis. J. Infect. Dis. 1995, 172, 281–285. [Google Scholar] [CrossRef]
- Hall, C.; Lambourne, J. The Challenges of Diagnosing Leptospirosis. J. Travel Med. 2014, 21, 139–140. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fornazari, F.; Langoni, H.; Marson, P.M.; Nóbrega, D.B.; Teixeira, C.R. Leptospira reservoirs among wildlife in Brazil: Beyond rodents. Acta Trop. 2018, 178, 205–212. [Google Scholar] [PubMed]
- Koizumi, N.; Muto, M.; Yamada, A.; Watanabe, H. Prevalence of Leptospira spp. in the Kidneys of Wild Boars and Deer in Japan. J. Vet. Med. Sci. 2009, 71, 797–799. [Google Scholar] [CrossRef][Green Version]
- Koizumi, N.; Uchida, M.; Makino, T.; Taguri, T.; Kuroki, T.; Muto, M.; Kato, Y.; Watanabe, H. Isolation and characterization of Leptospira spp. from raccoons in Japan. J. Vet. Med. Sci. 2009, 71, 425–429. [Google Scholar] [CrossRef]
- Pedersen, K.; Anderson, T.D.; Bevins, S.N.; Pabilonia, K.L.; Whitley, P.N.; Virchow, D.R.; Gidlewski, T. Evidence of leptospirosis in the kidneys and serum of feral swine (Sus scrofa) in the United States. Epidemiol. Infect. 2017, 145, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Scanziani, E.; Origgi, F.; Giusti, A.M.; Iacchia, G.; Vasino, A.; Pirovano, G.; Scarpa, P.; Tagliabue, S. Serological survey of leptospiral infection in kennelled dogs in Italy. J. Small Anim. Pract. 2002, 43, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, E.; Radaelli, E.; Bertoletti, I.; Bianchi, A.; Scanziani, E.; Tagliabue, S.; Mattiello, S. Leptospira spp. infection in wild ruminants: A survey in Central Italian Alps. Vet. Ital. 2014, 50, 285–291. [Google Scholar] [PubMed]
- Zanzani, S.A.; Cerbo, A.D.; Gazzonis, A.L.; Epis, S.; Invernizzi, A.; Tagliabue, S.; Manfredi, M.T. Parasitic and Bacterial Infections of Myocastor coypus in a Metropolitan Area of Northwestern Italy. J. Wildl. Dis. 2016, 52, 126–130. [Google Scholar] [CrossRef]
- Vitale, M.; Agnello, S.; Chetta, M.; Amato, B.; Vitale, G.; Bella, C.D.; Vicari, D.; Presti, V.D.M.L. Human leptospirosis cases in Palermo Italy. The role of rodents and climate. J. Infect. Public Health 2018, 11, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Ciceroni, L.; Lombardo, D.; Pinto, A.; Ciarrocchi, S.; Simeoni, J. Prevalence of antibodies to Leptospira serovars in sheep and goats in Alto Adige-South Tyrol. J. Vet. Med. B. Infect. Dis. Vet. Public Health 2000, 47, 217–223. [Google Scholar] [CrossRef]
- Russo, L.; Massei, G.; Genov, P.V. Daily home range and activity of wild boar in a mediterranean area free from hunting. Ethol. Ecol. Evol. 1997, 9, 287–294. [Google Scholar] [CrossRef]
- Boitani, L.; Mattei, L.; Nonis, D.; Corsi, F. Spatial and Activity Patterns of Wild Boars in Tuscany, Italy. J. Mammal. 1994, 75, 600–612. [Google Scholar] [CrossRef]
- Massolo, A.; Della Stella, R.M. Population structure variations of wild boar Sus scrofa in central Italy. Ital. J. Zool. 2006, 73, 137–144. [Google Scholar] [CrossRef]
- Perolat, P.; Chappel, R.J.; Adler, B.; Baranton, G.; Bulach, D.M.; Billinghurst, M.L.; Letocart, M.; Merien, F.; Serrano, M.S. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. 1998, 48, 851–858. [Google Scholar] [CrossRef]
- Chappel, R.J.; Khalik, D.A.; Adler, B.; Bulach, D.M.; Faine, S.; Perolat, P.; Vallance, V. Serological titres to Leptospira fainei serovar hurstbridge in human sera in Australia. Epidemiol. Infect. 1998, 121, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Arzouni, J.P.; Parola, P.; Scola, B.L.; Postic, D.; Brouqui, P.; Raoult, D. Human infection caused by Leptospira fainei. Emerg. Infect. Dis. 2002, 8, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.M.; Boye, K.; Blom, J.; Schlichting, P.; Krogfelt, K.A. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J. Med. Microbiol. 2001, 50, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Soares, T.S.M.; do Rosário Dias de Oliveira Latorre, M.; Laporta, G.Z.; Buzzar, M.R. Spatial and seasonal analysis on leptospirosis in the municipality of São Paulo, Southeastern Brazil, 1998 to 2006. Rev. Saude Publica 2010, 44, 283–291. [Google Scholar] [CrossRef]
- Miller, D.A.; Wilson, M.A.; Beran, G.W. Relationships between prevalence of Leptospira interrogans in cattle, and regional, climatic, and seasonal factors. Am. J. Vet. Res. 1991, 52, 1766–1768. [Google Scholar]
- Chadsuthi, S.; Modchang, C.; Lenbury, Y.; Iamsirithaworn, S.; Triampo, W. Modeling seasonal leptospirosis transmission and its association with rainfall and temperature in Thailand using time-series and ARIMAX analyses. Asian Pac. J. Trop. Med. 2012, 5, 539–546. [Google Scholar] [CrossRef]
- Ward, M.P. Seasonality of canine leptospirosis in the United States and Canada and its association with rainfall. Prev. Vet. Med. 2002, 56, 203–213. [Google Scholar] [CrossRef]
- Stoddard, R.A.; Bui, D.; Haberling, D.L.; Wuthiekanun, V.; Thaipadungpanit, J.; Hoffmaster, A.R. Viability of Leptospira isolates from a human outbreak in Thailand in various water types, pH, and temperature conditions. Am. J. Trop. Med. Hyg. 2014, 91, 1020–1022. [Google Scholar] [CrossRef]
- Filho, J.G.; Nazário, N.O.; Freitas, P.F.; Pinto, G.A.; Schlindwein, A.D. Temporal analysis of the relationship between leptospirosis, rainfall levels and seasonality, Santa Catarina, Brazil, 2005-2015. Rev. Inst. Med. Trop. Sao Paulo 2018, 60, e39. [Google Scholar]
- Kupek, E.; de Sousa Santos Faversani, M.C.; de Souza Philippi, J.M. The relationship between rainfall and human leptospirosis in Florianópolis, Brazil, 1991–1996. Braz. J. Infect. Dis. 2000, 4, 131–134. [Google Scholar]
- Hacker, K.P.; Sacramento, G.A.; Cruz, J.S.; De Oliveira, D.; Nery, N.; Lindow, J.C.; Carvalho, M.; Hagan, J.; Diggle, P.J.; Begon, M.; et al. Influence of rainfall on leptospira infection and disease in a tropical urban setting, Brazil. Emerg. Infect. Dis. 2020, 26, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Regione Toscana - Settore Idrologico Regionale. Analisi dei Dati Termometrici Report Anno 2018. 2018. Available online: https://www.sir.toscana.it/supports/download/report/2018_situazione_termometrica.pdf (accessed on 10 May 2020).
- Regione Toscana - Settore Idrologico Regionale. Report Pluviometrico Anno 2018. 2018. Available online: https://www.sir.toscana.it/supports/download/report/2018_situazione_idrologica.pdf (accessed on 10 May 2020).
- Regione Toscana - Settore Idrologico Regionale. Analisi dei Dati Termometrici Report Anno 2019. 2019. Available online: https://www.sir.toscana.it/supports/download/report/2019_situazione_termometrica.pdf (accessed on 10 May 2020).
- Regione Toscana - Settore Idrologico Regionale. Report Pluviometrico Anno 2019. 2019. Available online: https://www.sir.toscana.it/supports/download/report/2019_situazione_idrologica.pdf (accessed on 10 May 2020).
- Regione Toscana - Settore Idrologico Regionale. Report Pluviometrico del Mese di Gennaio 2020. 2020. Available online: https://www.sir.toscana.it/supports/download/report/2020-01_report_cumulate_mensili.pdf (accessed on 10 May 2020).
- Regione Toscana - Settore Idrologico Regionale. Analisi Dati Termometrici Report Gennaio 2020. 2020. Available online: https://www.sir.toscana.it/supports/download/report/report_termometria_2020-01.pdf (accessed on 10 May 2020).
- Arenas-Montes, A.; García-Bocanegra, I.; Paniagua, J.; Franco, J.J.; Miró, F.; Fernández-Morente, M.; Carbonero, A.; Arenas, A. Blood sampling by puncture in the cavernous sinus from hunted wild boar. Eur. J. Wildl. Res. 2013, 59, 299–303. [Google Scholar] [CrossRef]
- Sáez-Royuela, C.; Gomariz, R.P.; Luis Tellería, J. Age Determination of European Wild Boar; Wiley: Hoboken, NJ, USA, 1989; Volume 17. [Google Scholar]
- OIE. Leptospirosis. Man. Diagnostic Tests Vaccines Terr. Anim. 2014. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.12_LEPTO.pdf (accessed on 10 May 2020).
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Bedir, O.; Kilic, A.; Atabek, E.; Kuskucu, A.M.; Turhan, V.; Basustaoglu, A.C. Simultaneous detection and differentiation of pathogenic and nonpathogenic Leptospira spp. by multiplex real-time PCR (TaqMan) assay. Polish J. Microbiol. 2010, 59, 167–173. [Google Scholar] [CrossRef]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.G.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A single Multilocus Sequence Typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Devi, S.M.; de los Á Valverde, M.; Vijayachari, P.; Machang’u, R.S.; Ellis, W.A.; Hartskeerl, R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Varni, V.; Ruybal, P.; Lauthier, J.J.; Tomasini, N.; Brihuega, B.; Koval, A.; Caimi, K. Reassessment of MLST schemes for Leptospira spp. typing worldwide. Infect. Genet. Evol. 2014, 22, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- R. R Development Core Team. A language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria 2015. [Google Scholar] [CrossRef]
- Nuti, M.; Amaddeo, D.; Crovatto, M.; Ghionni, A.; Polato, D.; Lillini, E.; Pitzus, E.; Santini, G.F. Infections in an alpine environment: Antibodies to hantaviruses, leptospira, rickettsiae, and Borrelia burgdorferi in defined Italian populations. Am. J. Trop. Med. Hyg. 1993, 48, 20–25. [Google Scholar] [CrossRef]
- Nuti, M.; Amaddeo, D.; Autorino, G.L.; Crovatto, M.; Crucil, C.; Ghionni, A.; Giommi, M.; Salvati, F.; Santini, G.F. Seroprevalence of antibodies to hantaviruses and leptospires in selected Italian population groups. Eur. J. Epidemiol. 1992, 8, 98–102. [Google Scholar] [PubMed]
- Istisan, R.; Graziani, C.; Duranti, A.; Morelli, A.; Busani, L.; Pezzotti, P. Rapoorti ISTISAN Istituto Superiore di Sanità 16/1. In Proceedings of the I Simposio internazionale Nuove strategie per gli interventi di prevenzione dello stress da lavoro, Sassari-Alghero, Italy, 8–10 July 2015. [Google Scholar]
- Ngugi, J.N.; Fèvre, E.M.; Mgode, G.F.; Obonyo, M.; Mhamphi, G.G.; Otieno, C.A.; Cook, E.A.J. Seroprevalence and associated risk factors of leptospirosis in slaughter pigs; A neglected public health risk, western Kenya. BMC Vet. Res. 2019, 15, 403. [Google Scholar] [CrossRef] [PubMed]
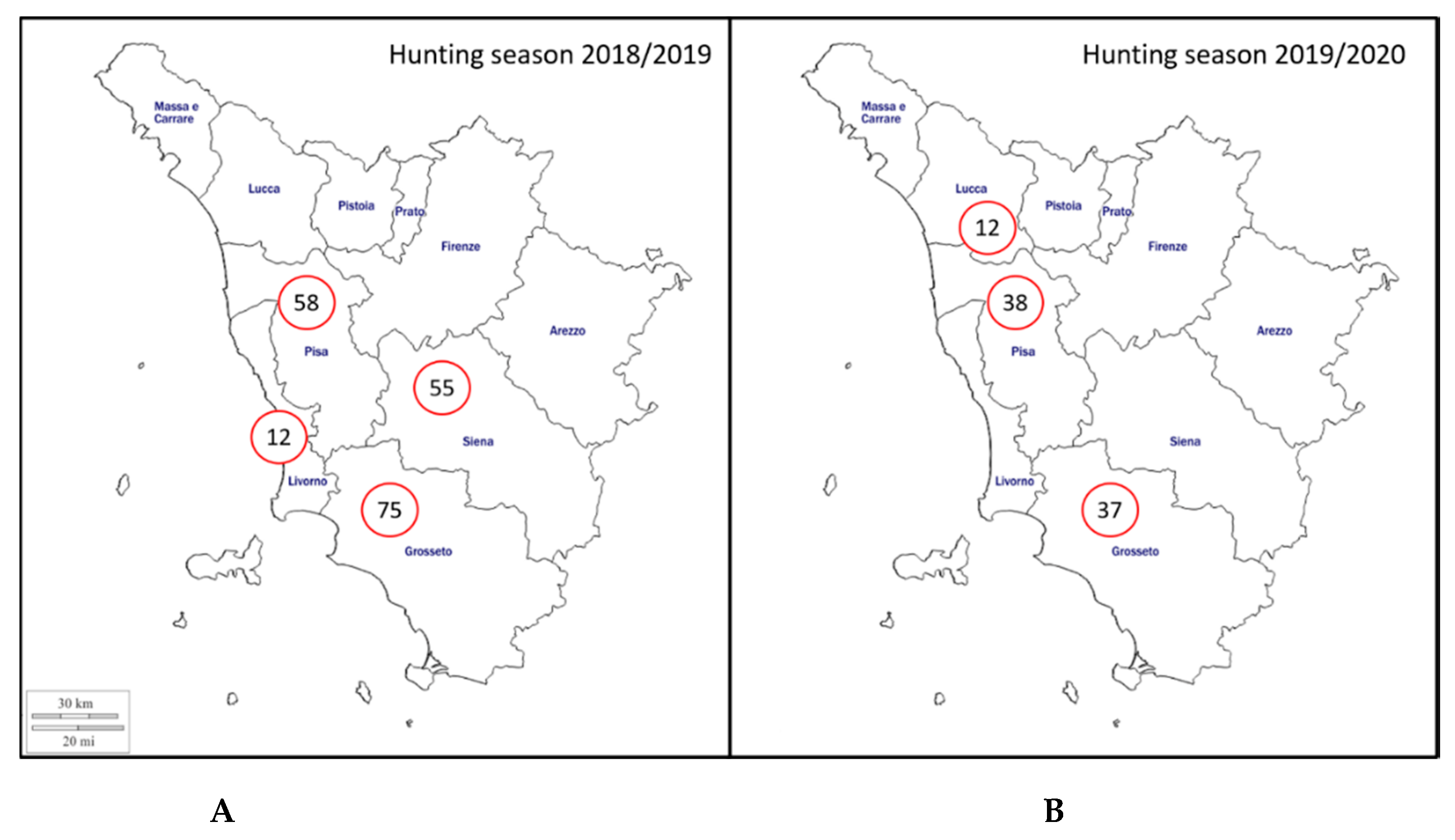
| Hunting Season | Province | Sex | Age Class | Examined Wild Boar | MAT-Positive Sera (%) | PCR-Positive Kidneys (%) |
|---|---|---|---|---|---|---|
| 2018/2019 | Pisa | Male | Adult | 9 | 2 (22.2) | 1 (11.1) |
| (n = 200) | (n = 58) | (n = 30) | Subadult | 10 | 2 (20.0) | 4 (40.0) |
| Young | 11 | 3 (27.3) | 0 | |||
| Female | Adult | 14 | 2 (14.3) | 2 (14.3) | ||
| (n = 28) | Subadult | 5 | 1 (20.0) | 2 (40.0) | ||
| Young | 9 | 1 (11.1) | 2 (22.2) | |||
| Grosseto | Male | Adult | 10 | 2 (20.0) | 2 (20.0) | |
| (n = 75) | (n = 29) | Subadult | 5 | 1 (20.0) | 0 | |
| Young | 14 | 1 (7.1) | 3 (21.4) | |||
| Female | Adult | 22 | 2 (9.09) | 1 (4.6) | ||
| (n = 46) | Subadult | 5 | 0 | 0 | ||
| Young | 19 | 2 (10.5) | 4 (21.1) | |||
| Siena | Male | Adult | 10 | 2 (20.0) | 0 | |
| (n = 55) | (n = 22) | Subadult | 4 | 1 (25.0) | 0 | |
| Young | 8 | 0 | 1 (12.5) | |||
| Female | Adult | 21 | 5 (23.8) | 3 (14.3) | ||
| (n = 33) | Subadult | 2 | 0 | 0 | ||
| Young | 10 | 1 (10.0) | 3 (30.0) | |||
| Livorno | Male | Adult | 2 | 0 | 0 | |
| (n = 12) | (n = 4) | Subadult | 0 | 0 | 0 | |
| Young | 2 | 1 (50.0) | 1 (50.0) | |||
| Female | Adult | 4 | 1 (25.0) | 1 (25.0) | ||
| (n=8) | Subadult | 0 | 0 | 0 | ||
| Young | 4 | 2 (50.0) | 1 (25.0) | |||
| 2019/2020 | Pisa | Male | Adult | 6 | 0 | 0 |
| (n = 87) | (n = 38) | (n = 13) | Subadult | 4 | 0 | 0 |
| Young | 3 | 0 | 0 | |||
| Female | Adult | 21 | 2 (9.52) | 0 | ||
| (n = 25) | Subadult | 1 | 1 (100) | 0 | ||
| Young | 3 | 0 | 1 (33.3) | |||
| Grosseto | Male | Adult | 11 | 1 (9.09) | 0 | |
| (n = 37) | (n = 16) | Subadult | 1 | 0 | 0 | |
| Young | 4 | 0 | 0 | |||
| Female | Adult | 10 | 1 (10.0) | 0 | ||
| (n = 21) | Subadult | 5 | 1 (20.0) | 0 | ||
| Young | 6 | 1 (16.7) | 0 | |||
| Lucca | Male | Adult | 1 | 0 | 0 | |
| (n=12) | (n = 4) | Subadult | 0 | 0 | 0 | |
| Young | 3 | 0 | 0 | |||
| Female | Adult | 4 | 0 | 0 | ||
| (n = 8) | Subadult | 0 | 0 | 0 | ||
| Young | 4 | 0 | 0 |
| Leptospira Serogroup | Titer | Total (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12800 | ||
| Icterohaemorrhagiae | 1 | 1 (2.56%) | |||||||
| Canicola | 1 | 1 | 2 (5.13%) | ||||||
| Pomona | 8 | 1 | 3 | 12 (30.8%) | |||||
| Grippotyphosa | |||||||||
| Tarassovi | 4 | 1 | 1 | 1 | 1 | 1 | 9 (23.1%) | ||
| Australis | 5 | 5 | 2 | 1 | 1 | 14 (35.9%) | |||
| Sejroe | |||||||||
| Ballum | 1 | 1 (2.56%) | |||||||
| Total | 17 | 3 | 10 | 2 | 2 | 2 | 2 | 1 | 39 (100%) |
| Sample | Wild Boar | Isolates Characterization | |||
|---|---|---|---|---|---|
| Sex | Age Class | Province | Anti-Serum MAT Serogroup | MLST (Sequence Type) | |
| Kidney 5 | Male | Subadult | Pisa | Tarassovi | Tarassovi (ST 153) |
| Kidney 14 | Male | Subadult | Pisa | Australis | Bratislava (ST 24) |
| Kidney 15 | Male | Subadult | Pisa | Australis | Bratislava (ST 24) |
| Kidney 22 | Female | Adult | Livorno | Australis | Bratislava (ST 24) |
| Sample | Wild Boar | Isolate Characterization | ||
|---|---|---|---|---|
| Sex | Age Class | Province | Leptospira Species | |
| Kidney 1 | Female | Young | Pisa | L. borgpetersenii |
| Kidney 2 | Female | Subadult | Pisa | L. interrogans |
| Kidney 3 | Male | Adult | Pisa | L. borgpetersenii |
| Kidney 4 | Female | Young | Pisa | L. interrogans |
| Kidney 6 | Male | Young | Siena | L. interrogans |
| Kidney 7 | Female | Young | Siena | L. interrogans |
| Kidney 12 | Female | Young | Siena | L. interrogans |
| Kidney13 | Female | Adult | Siena | L. interrogans |
| Kidney 20 | Male | Young | Grosseto | L. interrogans |
| Kidney 24 | Female | Young | Livorno | L. interrogans |
| Kidney 36 | Female | Adult | Grosseto | L. interrogans |
| Kidney39 | Female | Young | Grosseto | L. interrogans |
| Kidney 42 | Female | Young | Siena | L. interrogans |
| Kidney 53 | Female | Young | Grosseto | L. interrogans |
| Kidney 54 | Male | Young | Grosseto | L. kirschneri |
| Kidney 55 | Male | Young | Grosseto | L. interrogans |
| Kidney 58 | Female | Adult | Pisa | L. borgpetersenii |
| Kidney 64 | Female | Adult | Siena | L. interrogans |
| Kidney 65 | Female | Adult | Siena | L. interrogans |
| Kidney 67 | Female | Adult | Pisa | L. interrogans |
| Kidney 68 | Female | Subadult | Pisa | L. interrogans |
| Kidney 71 | Male | Adult | Grosseto | L. interrogans |
| Kidney 72 | Female | Young | Grosseto | L. borgpetersenii |
| Kidney 75 | Male | Subadult | Pisa | L. interrogans |
| Kidney 78 | Male | Young | Livorno | L. interrogans |
| Kidney 97 | Male | Adult | Grosseto | L. interrogans |
| Kidney 165 | Female | Young | Grosseto | L. interrogans |
| Kidney 208 | Female | Young | Pisa | L. interrogans |
| Sample | Wild Boar | Isolate Characterization | ||
|---|---|---|---|---|
| Sex | Age Class | Province | Leptospira Species | |
| Kidney 23 | Male | Young | Livorno | L. fainei |
| Kidney 27 | Male | Adult | Pisa | L. fainei |
| Kidney 31 | Female | Adult | Pisa | L. fainei |
| Kidney 56 | Male | Young | Grosseto | L. fainei |
| Kidney 57 | Male | Adult | Pisa | L. fainei |
| Kidney 63 | Male | Adult | Siena | L. fainei |
| Kidney 69 | Female | Subadult | Pisa | L. fainei |
| Kidney123 | Male | Adult | Livorno | L. fainei |
| Kidney 153 | Male | Adult | Siena | L. fainei |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilia, G.; Bertelloni, F.; Angelini, M.; Cerri, D.; Fratini, F. Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy. Pathogens 2020, 9, 377. https://doi.org/10.3390/pathogens9050377
Cilia G, Bertelloni F, Angelini M, Cerri D, Fratini F. Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy. Pathogens. 2020; 9(5):377. https://doi.org/10.3390/pathogens9050377
Chicago/Turabian StyleCilia, Giovanni, Fabrizio Bertelloni, Marta Angelini, Domenico Cerri, and Filippo Fratini. 2020. "Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy" Pathogens 9, no. 5: 377. https://doi.org/10.3390/pathogens9050377
APA StyleCilia, G., Bertelloni, F., Angelini, M., Cerri, D., & Fratini, F. (2020). Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy. Pathogens, 9(5), 377. https://doi.org/10.3390/pathogens9050377







