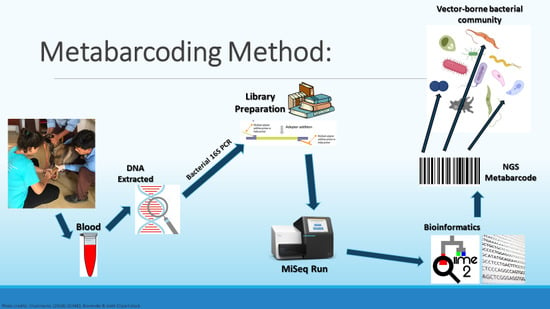
A Host-Specific Blocking Primer Combined with Optimal DNA Extraction Improves the Detection Capability of a Metabarcoding Protocol for Canine Vector-Borne Bacteria
Abstract
1. Introduction
2. Results
2.1. Blocking Primer Performance
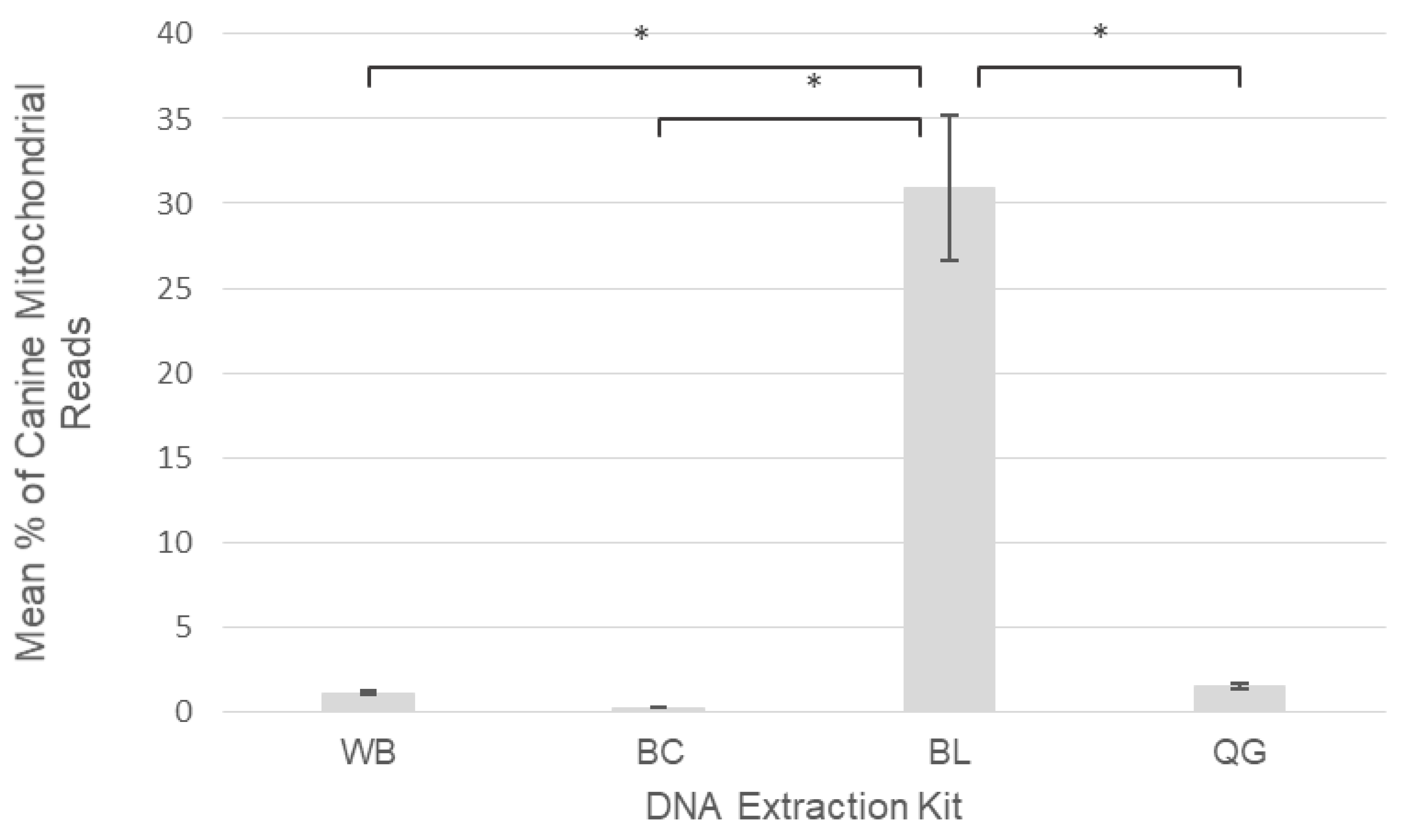
2.2. DNA Extraction Kit Performance
3. Discussion
3.1. Blocking Primer Performance
3.2. DNA Extraction Kit Performance
4. Conclusions
5. Materials and Methods
5.1. Sampling and DNA Extraction
- (1)
- Extraction of 500 µL of whole blood using the Maxwell® RSC Whole Blood DNA Kit (Promega, Madison, WI, USA) on the automated Maxwell® RSC 48 Instrument DNA extraction robot. This was conducted as per the manufacturer’s instructions and eluted in 100 µL of Ambion Nuclease-Free Water (Life Technologies, Carlsbad, CA, USA).
- (2)
- Extraction of 250 µL of Buffy Coat layer using the Maxwell® RSC Buffy Coat DNA Kit (Promega, Madison, WI, USA) on the Maxwell® RSC 48 Instrument, conducted as per the manufacturer’s instructions and eluted in 100 µL of Ambion Nuclease-Free Water.
- (3)
- Manual extraction of 200 µL of whole blood, using the ISOLATE II Genomic DNA Kit (Bioline, Memphis, TN, USA), using the manufacturer’s protocol with a final elution step in 50 µL.
- (4)
- Manual extraction of 100 µL of whole blood, using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) using the manufacturer’s protocol with a final elution step in 50 µL.
5.2. DNA Quantification
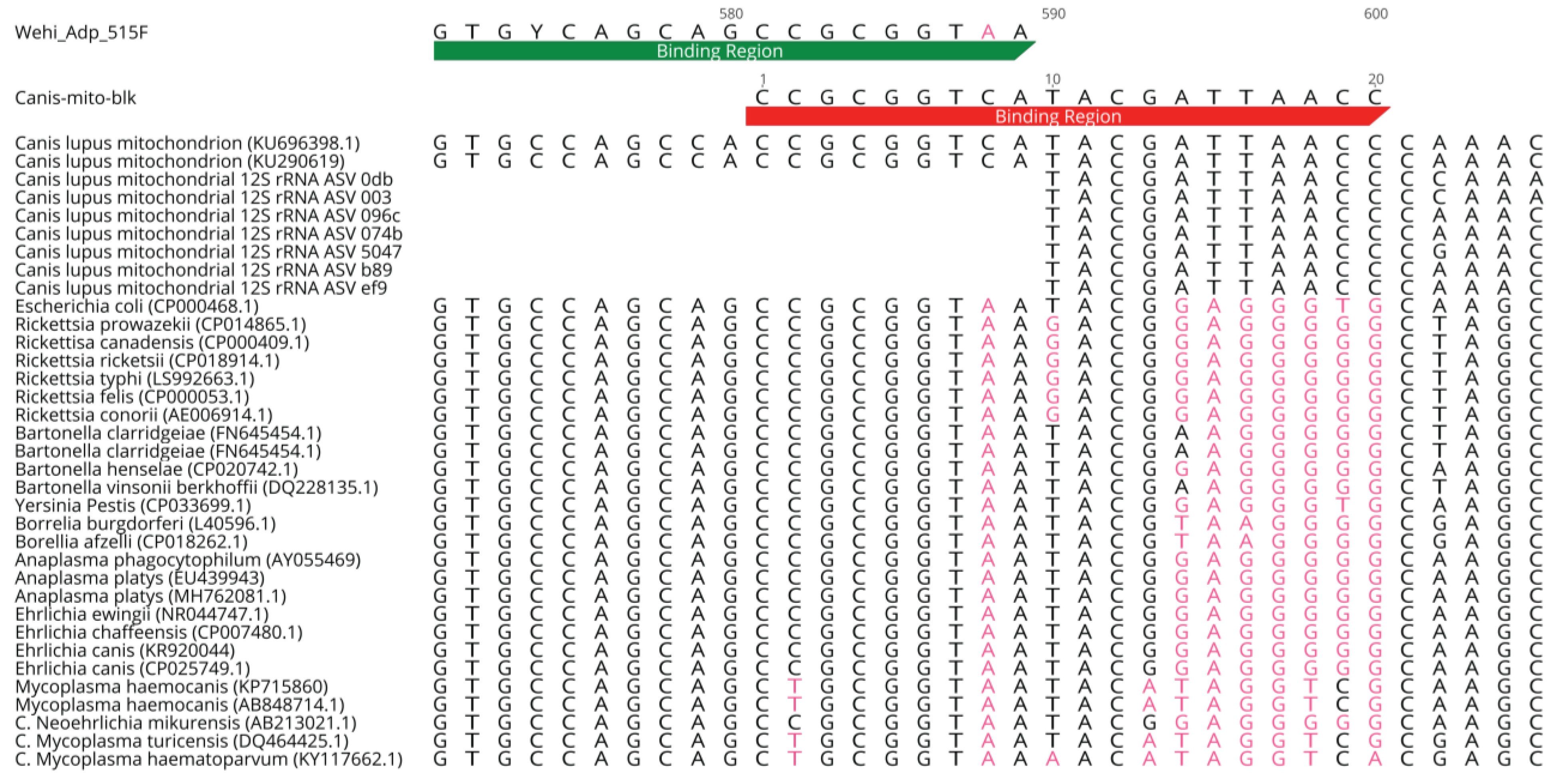
5.3. Design of Blocking Primer
5.4. Validation of Canis-mito-blk
5.5. Bacterial 16S rRNA Metabarcoding Assessment of Canis-mito-blk Efficacy
5.6. Bacterial 16S rRNA Metabarcoding Comparison of DNA Extraction Kit Performance
5.7. Bioinformatics
5.8. Data Analysis of Blocking Primer Performance
5.9. Data Analysis of DNA Extraction Kit Performance
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maggi, R.G.; Krämer, F. A review on the occurrence of companion vector-borne diseases in pet animals in Latin America. Parasit. Vectors 2019, 12, 145. [Google Scholar] [CrossRef]
- Traub, R.J.; Irwin, P.; Dantas-Torres, F.; Tort, G.P.; Labarthe, N.V.; Inpankaew, T.; Gatne, M.; Linh, B.K.; Schwan, V.; Watanabe, M.; et al. Toward the formation of a Companion Animal Parasite Council for the Tropics (CAPCT). Parasit. Vectors 2015, 8, 271. [Google Scholar] [CrossRef]
- Koh, F.X.; Panchadcharam, C.; Tay, S.T. Vector-borne diseases in stray dogs in peninsular Malaysia and molecular detection of Anaplasma and Ehrlichia spp. from Rhipicephalus sanguineus (Acari: Ixodidae) ticks. J. Med. Entomol. 2016, 53, 183–187. [Google Scholar] [CrossRef]
- Rucksaken, R.; Maneeruttanarungroj, C.; Maswanna, T.; Sussadee, M.; Kanbutra, P. Comparison of conventional polymerase chain reaction and routine blood smear for the detection of Babesia canis, Hepatozoon canis, Ehrlichia canis, and Anaplasma platys in Buriram Province, Thailand. Vet. World 2019, 12, 700–705. [Google Scholar] [CrossRef]
- Irwin, P.J.; Jefferies, R. Arthropod-transmitted diseases of companion animals in Southeast Asia. Trends Parasitol. 2004, 20, 27–34. [Google Scholar] [CrossRef]
- Inpankaew, T.; Hii, S.F.; Chimnoi, W.; Traub, R.J. Canine vector-borne pathogens in semi-domesticated dogs residing in northern Cambodia. Parasit. Vectors 2016, 9, 253. [Google Scholar] [CrossRef]
- Mylonakis, M.E.; Harrus, S.; Breitschwerdt, E.B. An update on the treatment of canine monocytic ehrlichiosis (Ehrlichia canis). Vet. J. 2019, 246, 45–53. [Google Scholar] [CrossRef]
- Kaewmongkol, G.; Lukkana, N.; Yangtara, S.; Kaewmongkol, S.; Thengchaisri, N.; Sirinarumitr, T.; Jittapalapong, S.; Fenwick, S.G. Association of Ehrlichia canis, Hemotropic Mycoplasma spp. and Anaplasma platys and severe anemia in dogs in Thailand. Vet. Microbiol. 2017, 201, 195–200. [Google Scholar] [CrossRef]
- Little, S.E. Ehrlichiosis and anaplasmosis in dogs and cats. Vet. Clin. North Am. Small Anim. Pract. 2010, 40, 1121–1140. [Google Scholar] [CrossRef]
- Carrade, D.D.; Foley, J.E.; Borjesson, D.L.; Sykes, J.E. Canine granulocytic anaplasmosis: A review. J. Vet. Intern. Med. 2009, 23, 1129–1141. [Google Scholar] [CrossRef]
- Chomel, B.B.; Mac Donald, K.A.; Kasten, R.W.; Chang, C.C.; Wey, A.C.; Foley, J.E.; Thomas, W.P.; Kittleson, M.D. Aortic valve endocarditis in a dog due to Bartonella clarridgeiae. J. Clin. Microbiol. 2001, 39, 3548–3554. [Google Scholar] [CrossRef]
- Hii, S.F.; Kopp, S.R.; Abdad, M.Y.; Thompson, M.F.; O’Leary, C.A.; Rees, R.L.; Traub, R.J. Molecular evidence supports the role of dogs as potential reservoirs for Rickettsia felis. Vector-Borne Zoonotic Dis. 2011, 11, 1007–1012. [Google Scholar] [CrossRef]
- Ng-Nguyen, D.; Hii, S.-F.; Hoang, M.-T.T.; Nguyen, V.-A.T.; Rees, R.; Stenos, J.; Traub, R.J. Domestic dogs (Canis familiaris) are natural mammalian reservoirs for flea-borne spotted fever caused by Rickettsia felis. Sci. Rep. 2020, 10, 4151. [Google Scholar] [CrossRef]
- Fourie, J.J.; Evans, A.; Labuschagne, M.; Crafford, D.; Madder, M.; Pollmeier, M.; Schunack, B. Transmission of Anaplasma phagocytophilum (Foggie, 1949) by Ixodes ricinus (Linnaeus, 1758) ticks feeding on dogs and artificial membranes. Parasit. Vectors 2019, 12, 136. [Google Scholar] [CrossRef]
- Krämer, F.; Hüsken, R.; Krüdewagen, E.M.; Deuster, K.; Blagburn, B.; Straubinger, R.K.; Butler, J.; Fingerle, V.; Charles, S.; Settje, T.; et al. Prevention of transmission of Borrelia burgdorferi sensu lato and Anaplasma phagocytophilum by Ixodes spp. ticks to dogs treated with the Seresto® collar (imidacloprid 10% + flumethrin 4.5%). Parasitol. Res. 2019, 119, 299–315. [Google Scholar]
- Krupka, I.; Straubinger, R.K. Lyme Borreliosis in dogs and cats: Background, diagnosis, treatment and prevention of infections with Borrelia burgdorferi sensu stricto. Vet. Clin. North Am. Small Anim. Pract. 2010, 40, 1103–1119. [Google Scholar] [CrossRef]
- Bakken, J.S.; Dumler, J.S. Human granulocytic anaplasmosis. Infect. Dis. Clin. North Am. 2015, 29, 341–355. [Google Scholar] [CrossRef]
- Takhampunya, R.; Korkusol, A.; Pongpichit, C.; Yodin, K.; Rungrojn, A.; Chanarat, N.; Promsathaporn, S.; Monkanna, T.; Thaloengsok, S.; Tippayachai, B.; et al. Metagenomic approach to characterizing disease epidemiology in a disease-endemic environment in northern Thailand. Front. Microbiol. 2019, 10, 319. [Google Scholar] [CrossRef]
- Vayssier-Taussat, M.; Moutailler, S.; Michelet, L.; Devillers, E.; Bonnet, S.; Cheval, J.; Hébert, C.; Eloit, M. Next generation sequencing uncovers unexpected bacterial pathogens in ticks in western Europe. PLoS ONE 2013, 8, e81439. [Google Scholar] [CrossRef]
- Greay, T.L.; Gofton, A.W.; Paparini, A.; Ryan, U.M.; Oskam, C.L.; Irwin, P.J. Recent insights into the tick microbiome gained through next-generation sequencing. Parasit. Vectors 2018, 11, 12. [Google Scholar] [CrossRef]
- Huggins, L.G.; Koehler, A.V.; Ng-Nguyen, D.; Wilcox, S.; Schunack, B.; Inpankaew, T.; Traub, R.J. A novel metabarcoding diagnostic tool to explore protozoan haemoparasite diversity in mammals: A proof-of-concept study using canines from the tropics. Sci. Rep. 2019, 9, 12644. [Google Scholar] [CrossRef]
- Huggins, L.G.; Koehler, A.V.; Ng-Nguyen, D.; Wilcox, S.; Schunack, B.; Inpankaew, T.; Traub, R.J. Assessment of a metabarcoding approach for the characterisation of vector-borne bacteria in canines from Bangkok, Thailand. Parasit. Vectors 2019, 12, 394. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ruttayaporn, N.; Saechan, V.; Jirapattharasate, C.; Vudriko, P.; Moumouni, P.F.A.; Cao, S.; Inpankaew, T.; Ybañez, A.P.; Suzuki, H.; et al. Molecular survey of canine vector-borne diseases in stray dogs in Thailand. Parasitol. Int. 2016, 65, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hofstaedter, C.E.; Zhao, C.; Mattei, L.; Tanes, C.; Clarke, E.; Lauder, A.; Sherrill-Mix, S.; Chehoud, C.; Kelsen, J.; et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta-Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Gofton, A.W.; Oskam, C.L.; Lo, N.; Beninati, T.; Wei, H.; McCarl, V.; Murray, D.C.; Paparini, A.; Greay, T.L.; Holmes, A.J.; et al. Inhibition of the endosymbiont “Candidatus Midichloria mitochondrii” during 16S rRNA gene profiling reveals potential pathogens in Ixodes ticks from Australia. Parasit. Vectors 2015, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Liu, H. Unravel the hidden protistan diversity: Application of blocking primers to suppress PCR amplification of metazoan DNA. Appl. Microbiol. Biotechnol. 2018, 102, 389–401. [Google Scholar] [CrossRef]
- Vestheim, H.; Jarman, S.N. Blocking primers to enhance PCR amplification of rare sequences in mixed samples – a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 2008, 5, 12. [Google Scholar] [CrossRef]
- Leray, M.; Agudelo, N.; Mills, S.C.; Meyer, C.P. Effectiveness of annealing blocking primers versus restriction enzymes for characterization of generalist diets: Unexpected prey revealed in the gut contents of two coral reef fish species. PLoS ONE 2013, 8, e58076. [Google Scholar] [CrossRef]
- Boessenkool, S.; Epp, L.S.; Haile, J.; Bellemain, E.; Edwards, M.; Coissac, E.; Willerslev, E.; Brochmann, C. Blocking human contaminant DNA during PCR allows amplification of rare mammal species from sedimentary ancient DNA. Mol. Ecol. 2012, 21, 1806–1815. [Google Scholar] [CrossRef]
- Hino, A.; Maruyama, H.; Kikuchi, T. A novel method to assess the biodiversity of parasites using 18S rDNA Illumina sequencing; parasitome analysis method. Parasitol. Int. 2016, 65, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Glassing, A.; Dowd, S.E.; Galandiuk, S.; Davis, B.; Chiodini, R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Podnecky, N.L.; Elrod, M.G.; Newton, B.R.; Dauphin, L.A.; Shi, J.; Chawalchitiporn, S.; Baggett, H.C.; Hoffmaster, A.R.; Gee, J.E. Comparison of DNA extraction kits for detection of Burkholderia pseudomallei in spiked human whole blood using real-time PCR. PLoS ONE 2013, 8, e58032. [Google Scholar] [CrossRef]
- Yang, G.; Erdman, D.E.; Kodani, M.; Kools, J.; Bowen, M.D.; Fields, B.S. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens. J. Virol. Methods 2011, 171, 195–199. [Google Scholar] [CrossRef]
- Riemann, K.; Adamzik, M.; Frauenrath, S.; Egensperger, R.; Schmid, K.W.; Brockmeyer, N.H.; Siffert, W. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J. Clin. Lab. Anal. 2007, 21, 244–248. [Google Scholar] [CrossRef]
- Laurence, M.; Hatzis, C.; Brash, D.E. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS ONE 2014, 9, e97876. [Google Scholar] [CrossRef]
- Velásquez-Mejía, E.P.; de la Cuesta-Zuluaga, J.; Escobar, J.S. Impact of DNA extraction, sample dilution, and reagent contamination on 16S rRNA gene sequencing of human feces. Appl. Microbiol. Biotechnol. 2018, 102, 403–411. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Alexandre, N.; Santos, A.S.; Núncio, M.S.; de Sousa, R.; Boinas, F.; Bacellar, F. Detection of Ehrlichia canis by polymerase chain reaction in dogs from Portugal. Vet. J. 2009, 181, 343–344. [Google Scholar] [CrossRef]
- Mylonakis, M.E.; Koutinas, A.F.; Billinis, C.; Leontides, L.S.; Kontos, V.; Papadopoulos, O.; Rallis, T.; Fytianou, A. Evaluation of cytology in the diagnosis of acute canine monocytic ehrlichiosis (Ehrlichia canis): A comparison between five methods. Vet. Microbiol. 2003, 91, 197–204. [Google Scholar] [CrossRef]
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D.; et al. Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef] [PubMed]
- Greiner, M.; Gardner, I.A. Epidemiologic issues in the validation of veterinary diagnostic tests. Prev. Vet. Med. 2000, 45, 3–22. [Google Scholar] [CrossRef]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. Miseq: A next generation sequencing platform for genomic analysis. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1706, pp. 223–232. [Google Scholar]
- Vestheim, H.; Deagle, B.E.; Jarman, S.N. Application of blocking oligonucleotides to improve signal-to-noise ratio in a PCR. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2010; Volume 687, pp. 265–274. [Google Scholar]
- Hart, M.L.; Meyer, A.; Johnson, P.J.; Ericsson, A.C. Comparative evaluation of DNA extraction methods from feces of multiple host species for downstream next-generation sequencing. PLoS ONE 2015, 10, e0143334. [Google Scholar] [CrossRef] [PubMed]
- Wiesinger-Mayr, H.; Jordana-Lluch, E.; Martró, E.; Schoenthaler, S.; Noehammer, C. Establishment of a semi-automated pathogen DNA isolation from whole blood and comparison with commercially available kits. J. Microbiol. Methods 2011, 85, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Schiebelhut, L.M.; Abboud, S.S.; Gómez Daglio, L.E.; Swift, H.F.; Dawson, M.N. A comparison of DNA extraction methods for high-throughput DNA analyses. Mol. Ecol. Resour. 2017, 17, 721–729. [Google Scholar] [CrossRef]
- Phillips, K.; McCallum, N.; Welch, L. A comparison of methods for forensic DNA extraction: Chelex-100® and the QIAGEN DNA Investigator Kit (manual and automated). Forensic Sci. Int. Genet. 2012, 6, 282–285. [Google Scholar] [CrossRef]
- Sidstedt, M.; Hedman, J.; Romsos, E.L.; Waitara, L.; Wadsö, L.; Steffen, C.R.; Vallone, P.M.; Rådström, P. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 2018, 410, 2569–2583. [Google Scholar] [CrossRef]
- Hall, C.M.; Jaramillo, S.; Jimenez, R.; Stone, N.E.; Centner, H.; Busch, J.D.; Bratsch, N.; Roe, C.C.; Gee, J.E.; Hoffmaster, A.R.; et al. Burkholderia pseudomallei, the causative agent of melioidosis, is rare but ecologically established and widely dispersed in the environment in Puerto Rico. PLoS Negl. Trop. Dis. 2019, 13, e0007727. [Google Scholar] [CrossRef]
- Zinger, L.; Bonin, A.; Alsos, I.G.; Bálint, M.; Bik, H.; Boyer, F.; Chariton, A.A.; Creer, S.; Coissac, E.; Deagle, B.E.; et al. DNA metabarcoding—Need for robust experimental designs to draw sound ecological conclusions. Mol. Ecol. 2019, 28, 1857–1862. [Google Scholar] [CrossRef]
- Tedersoo, L.; Drenkhan, R.; Anslan, S.; Morales-Rodriguez, C.; Cleary, M. High-throughput identification and diagnostics of pathogens and pests: Overview and practical recommendations. Mol. Ecol. Resour. 2019, 19, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Vijayvargiya, P.; Jeraldo, P.R.; Thoendel, M.J.; Greenwood-Quaintance, K.E.; Esquer Garrigos, Z.; Sohail, M.R.; Chia, N.; Pritt, B.S.; Patel, R. Application of metagenomic shotgun sequencing to detect vector-borne pathogens in clinical blood samples. PLoS ONE 2019, 14, e0222915. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Huber, D.; Antolić, M.; Anzulović, Ž.; Reil, I.; Polkinghorne, A.; Baneth, G.; Beck, R. Retrospective study of canine infectious haemolytic anaemia cases reveals the importance of molecular investigation in accurate postmortal diagnostic protocols. Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Colella, V.; Nguyen, V.L.; Tan, D.Y.; Lu, N.; Fang, F.; Zhijuan, Y.; Wang, J.; Liu, X.; Chen, X.; Dong, J.; et al. Zoonotic vectorborne pathogens and ectoparasites of dogs and cats in Asia. Emerg. Infect. Dis. 2020, 26. [Google Scholar] [CrossRef]
- Ruby, E.G.; Urbanowski, M.; Campbell, J.; Dunn, A.; Faini, M.; Gunsalus, R.; Lostroh, P.; Lupp, C.; McCann, J.; Millikan, D.; et al. Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 2005, 102, 3004–3009. [Google Scholar] [CrossRef]
- Nyholm, S.V.; McFall-Ngai, M.J. The winnowing: Establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2004, 2, 632–642. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Nearing, J.T.; Douglas, G.M.; Comeau, A.M. Denoising the Denoisers: An independent evaluation of microbiome sequence error-correction approaches. PeerJ 2018, 8, e5364. [Google Scholar]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Spellerberg, I.F.; Fedor, P.J. A tribute to Claude-Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon-Wiener” Index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef]
- Gill, C.A.; Joanes, D.N. Bayesian estimation of Shannon’s index of diversity. Biometrika 1979, 66, 81–85. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]
- Ogedengbe, M.E.; El-Sherry, S.; Ogedengbe, J.D.; Chapman, H.D.; Barta, J.R. Phylogenies based on combined mitochondrial and nuclear sequences conflict with morphologically defined genera in the eimeriid coccidia (Apicomplexa). Int. J. Parasitol. 2018, 48, 59–69. [Google Scholar] [CrossRef]
- Allaby, M. A Dictionary of Ecology, 4th ed.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
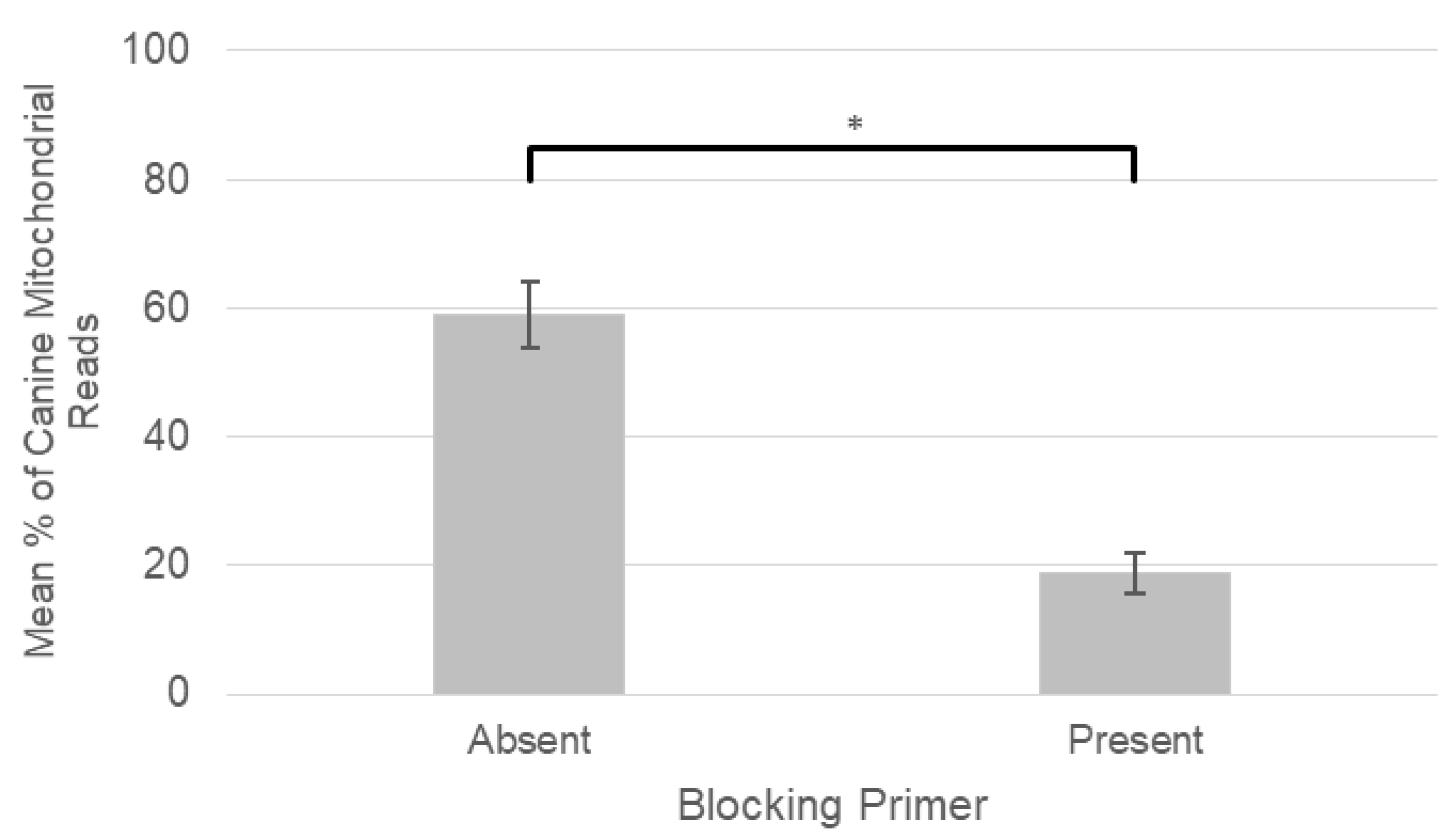
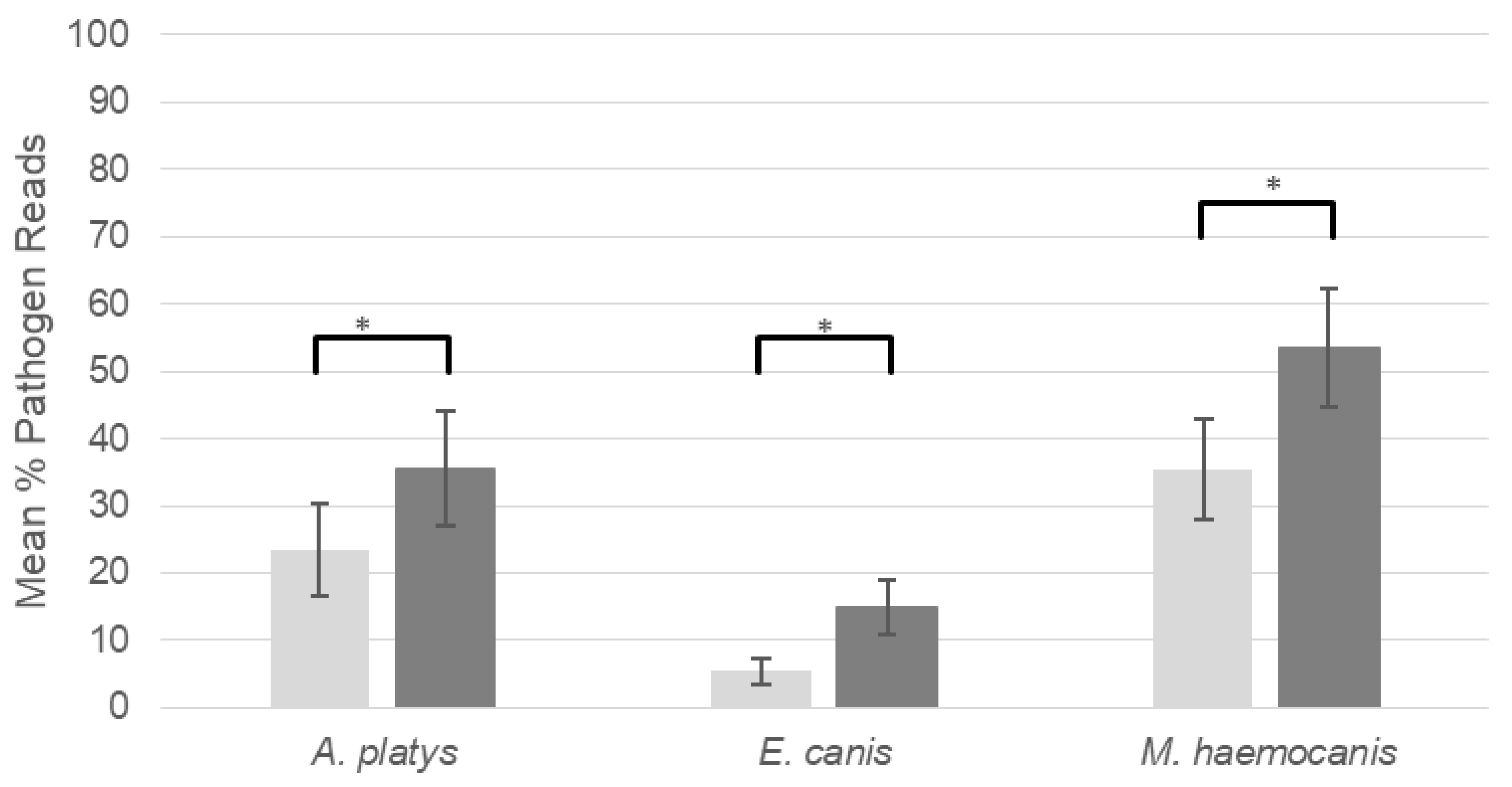
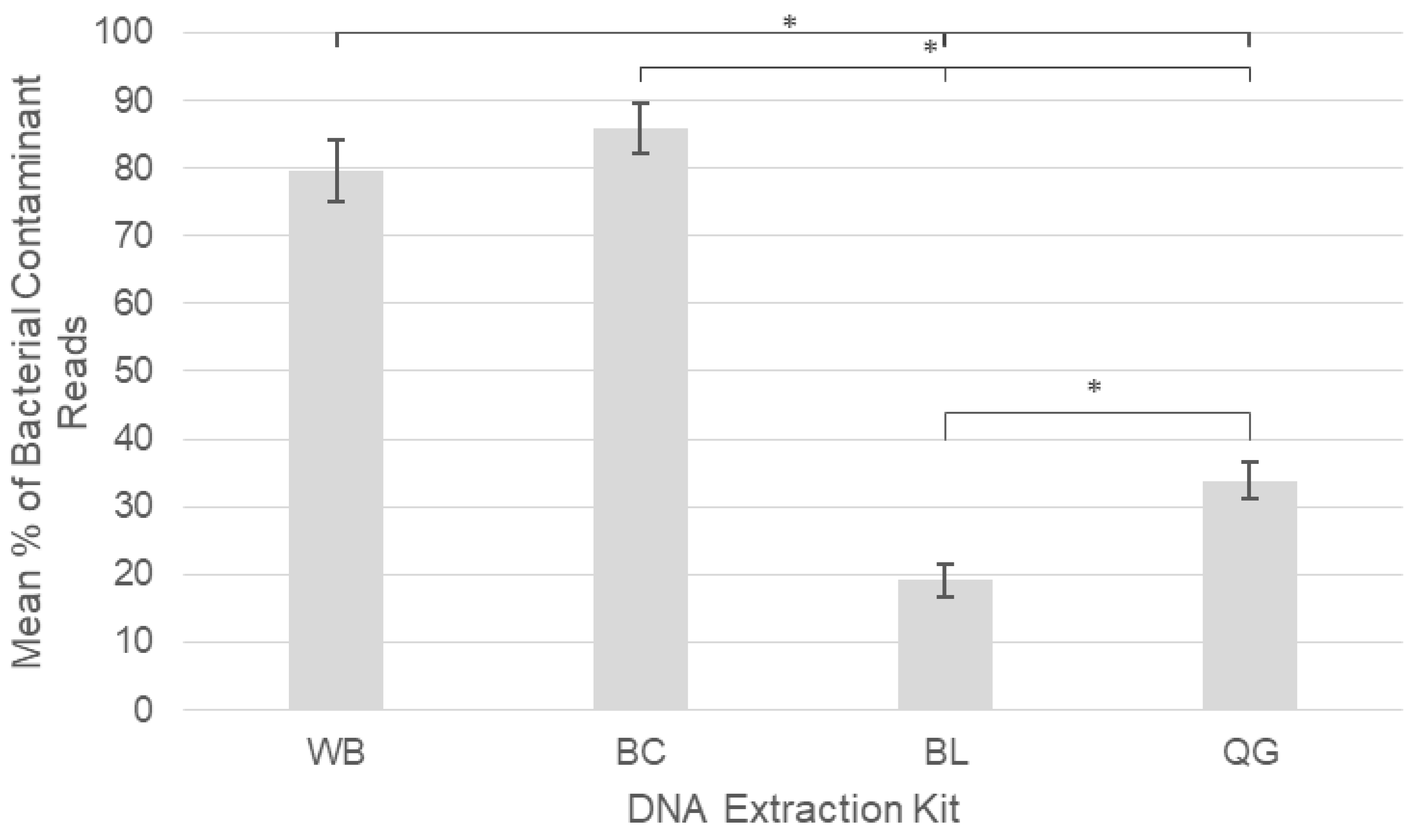
| Vector-Borne Bacteria | Total Infections: Blocking Primer Absent | Total Infections: Blocking Primer Present |
|---|---|---|
| A. platys | 17 | 20 |
| E. canis | 19 | 24 |
| M. haemocanis | 19 | 19 |
| Bacterial Diversity: Shannon–Wiener (H) Index | Bacterial Diversity: Simpson (D) Index | ||
|---|---|---|---|
| BP Absent | BP Present | BP Absent | BP Present |
| 1.81 | 2.3 | 3.15 | 4.07 |
| DNA Extraction Kit | Mean DNA Quantity ± S.E. (ng/µL) | DNA Quantity in ExtractionNegative Controls (ng/µL) | Total Raw Reads | Reads Post-Filtering | Total ASVs |
|---|---|---|---|---|---|
| Promega Whole Blood (WB) | 483.8 (±150.9) | 0.005–0.017 | 5,351,987 | 3,857,213 | 435 |
| Promega Buffy Coat (BC) | 680.2 (±394.7) | 0.012–0.024 | 5,916,458 | 4,716,590 | 428 |
| Bioline Whole Blood (BL) | 3.4 (±0.6) | <0.001 | 5,935,103 | 4,188,285 | 1401 |
| Qiagen Whole Blood (QG) | 13.9 (±1.1) | 0.001–0.014 | 4,462,788 | 3,559,370 | 6683 |
| Vector-Borne Bacteria | Promega Whole Blood (WB) | Promega Buffy Coat (BC) | Bioline Whole Blood (BL) | Qiagen Whole Blood (QG) |
|---|---|---|---|---|
| A. platys | 18 | 17 | 13 | 22 |
| E. canis | 7 | 7 | 6 | 8 |
| M. haemocanis | 3 | 4 | 4 | 5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huggins, L.G.; Koehler, A.V.; Schunack, B.; Inpankaew, T.; Traub, R.J. A Host-Specific Blocking Primer Combined with Optimal DNA Extraction Improves the Detection Capability of a Metabarcoding Protocol for Canine Vector-Borne Bacteria. Pathogens 2020, 9, 258. https://doi.org/10.3390/pathogens9040258
Huggins LG, Koehler AV, Schunack B, Inpankaew T, Traub RJ. A Host-Specific Blocking Primer Combined with Optimal DNA Extraction Improves the Detection Capability of a Metabarcoding Protocol for Canine Vector-Borne Bacteria. Pathogens. 2020; 9(4):258. https://doi.org/10.3390/pathogens9040258
Chicago/Turabian StyleHuggins, Lucas G., Anson V. Koehler, Bettina Schunack, Tawin Inpankaew, and Rebecca J. Traub. 2020. "A Host-Specific Blocking Primer Combined with Optimal DNA Extraction Improves the Detection Capability of a Metabarcoding Protocol for Canine Vector-Borne Bacteria" Pathogens 9, no. 4: 258. https://doi.org/10.3390/pathogens9040258
APA StyleHuggins, L. G., Koehler, A. V., Schunack, B., Inpankaew, T., & Traub, R. J. (2020). A Host-Specific Blocking Primer Combined with Optimal DNA Extraction Improves the Detection Capability of a Metabarcoding Protocol for Canine Vector-Borne Bacteria. Pathogens, 9(4), 258. https://doi.org/10.3390/pathogens9040258