Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster
Abstract
:1. Introduction
2. Results
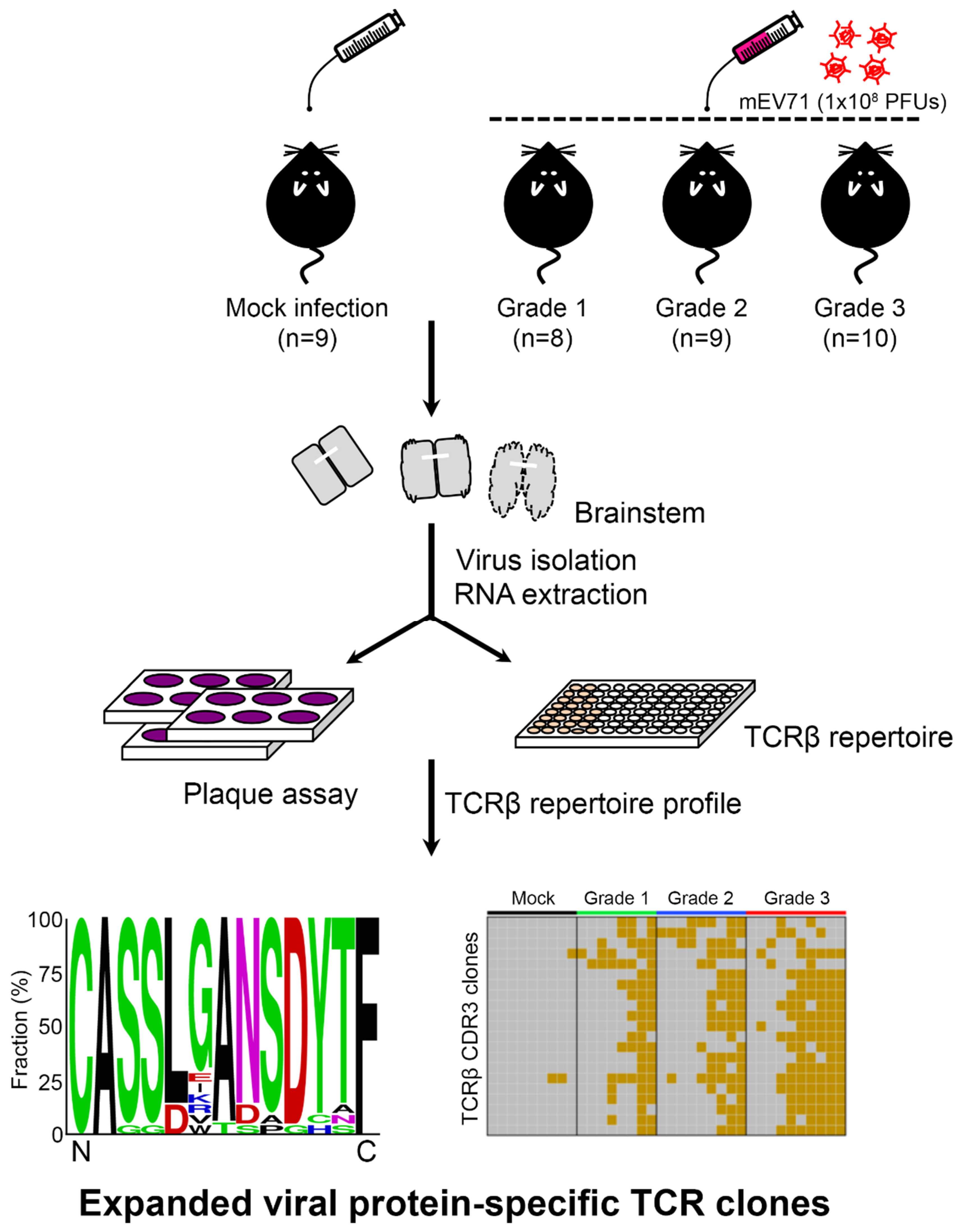
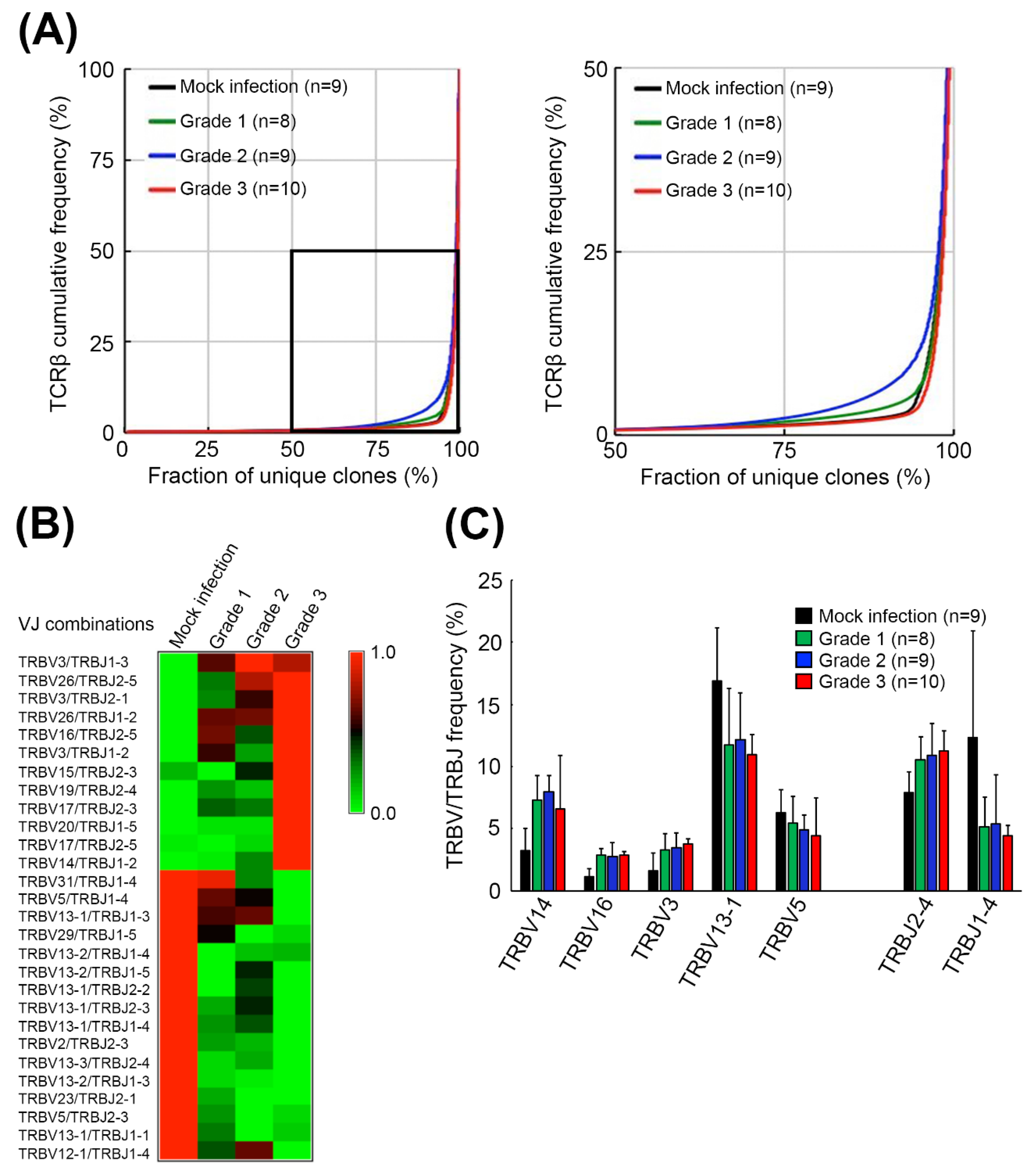
2.1. EV71 Infection Skewed the Host TCRβ Repertoire
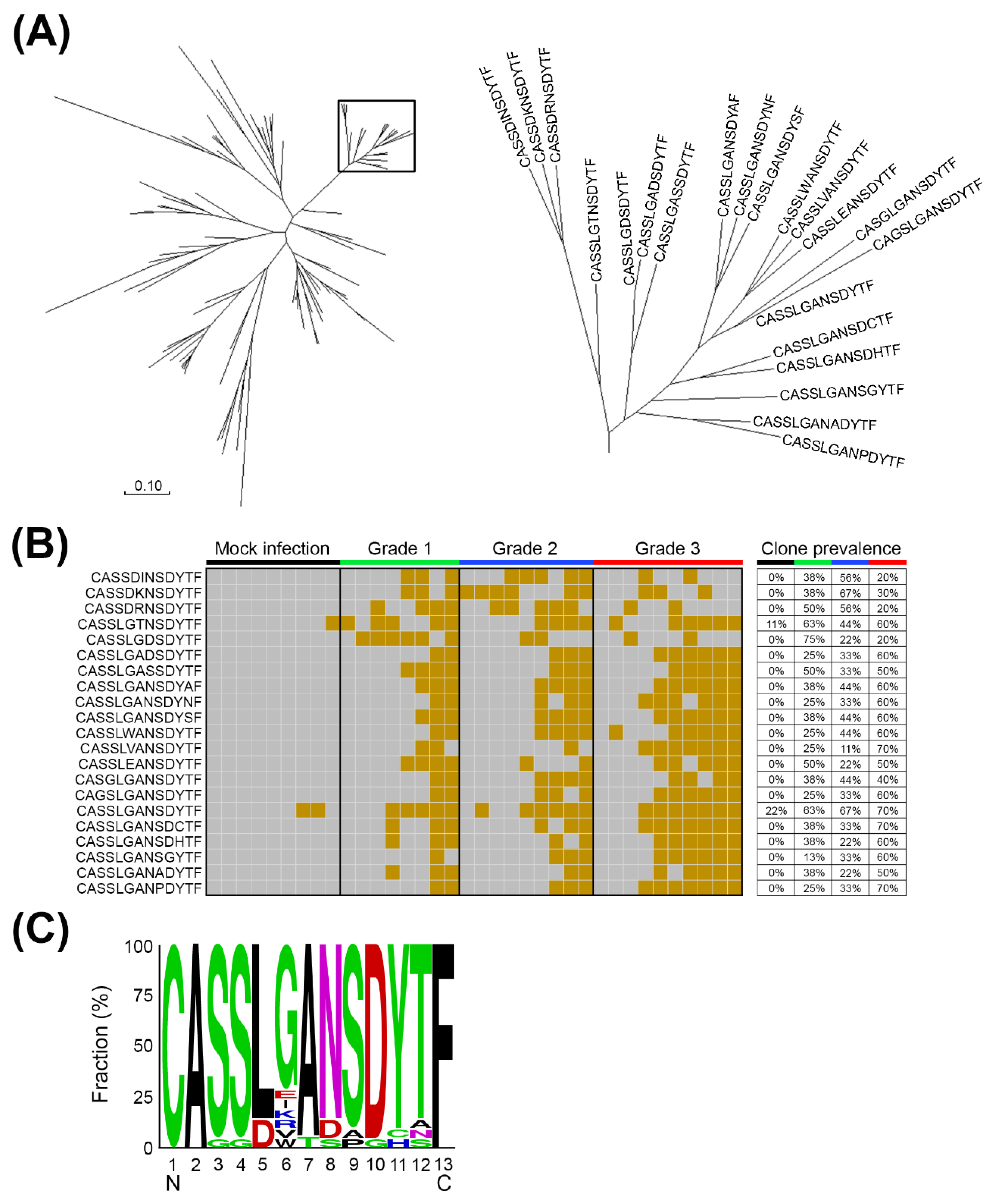
2.2. The Expanded TCRβ CDR3 Clones Prevailed in mEV71-Infected Mouse Brainstems
2.3. The Expanded TCRβ CDR3 Clones Were Elicited by EV71 Viral Protein 1
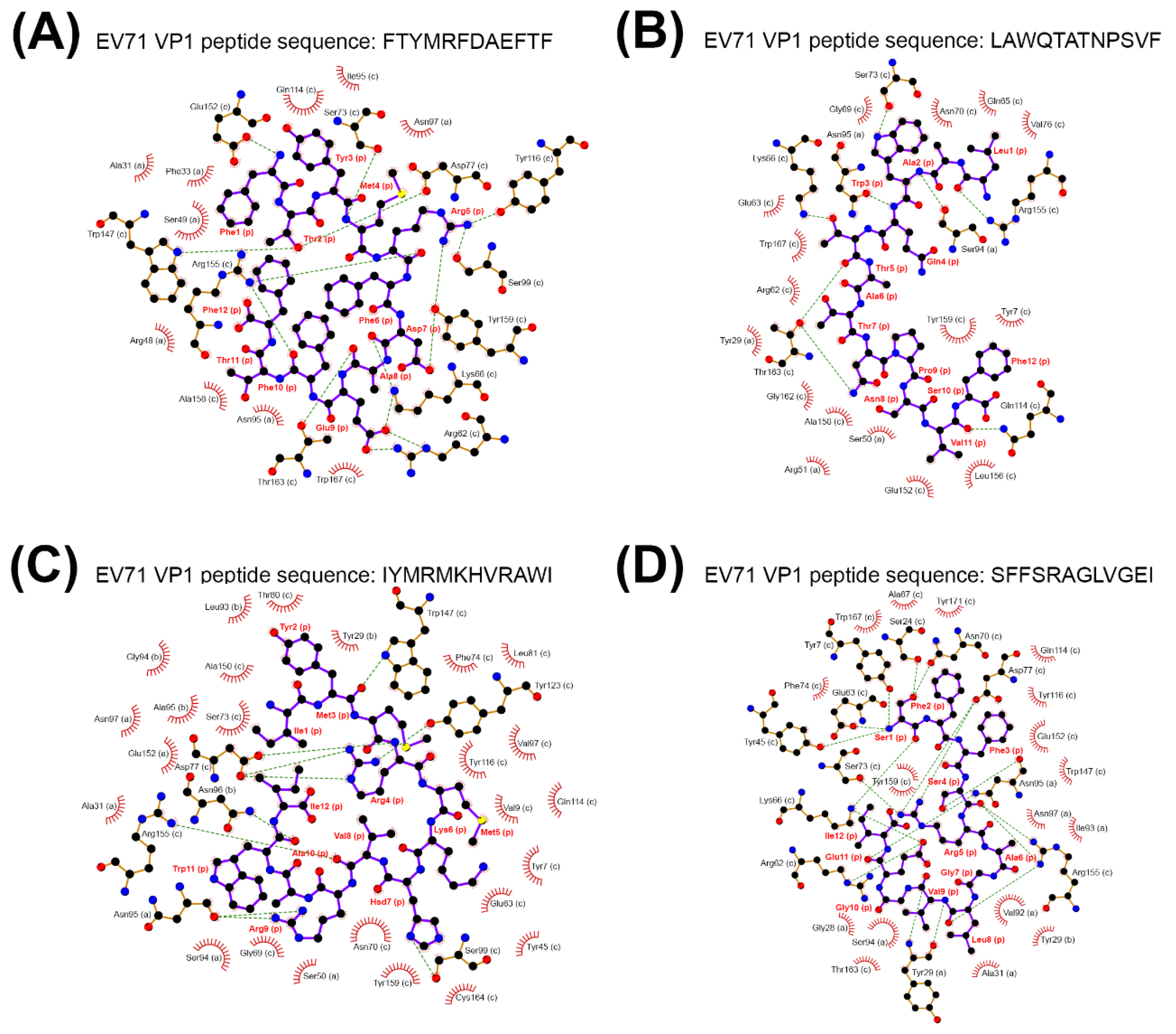
2.4. Scrambled CDR3 Sequences Disturbed the Stable Binding of TCRβ-MHC-Viral Peptide Complex
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. mEV71 Propagation
4.3. RNA Extraction
4.4. Plaque Assay
4.5. mEV71 Infection
4.6. Severity Assessment
4.7. TCRβ Library Preparation and NGS
4.8. NGS Data Processing
4.9. Identification of mEV71 Infection-Related CDR3 Clones
4.10. Ligand-Binding Prediction
4.11. Molecular Dynamics Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yi, E.J.; Shin, Y.J.; Kim, J.H.; Kim, T.G.; Chang, S.Y. Enterovirus 71 infection and vaccines. Clin. Exp. Vaccine Res. 2017, 6, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Sivanandy, P.; Zi Xien, F.; Woon Kit, L.; Tze Wei, Y.; Hui En, K.; Chia Lynn, L. A review on current trends in the treatment of human infection with H7N9-avian influenza A. J. Infect. Public Health 2018. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Cordon, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Chang, L.Y.; Huang, L.M.; Gau, S.S.; Wu, Y.Y.; Hsia, S.H.; Fan, T.Y.; Lin, K.L.; Huang, Y.C.; Lu, C.Y.; Lin, T.Y. Neurodevelopment and cognition in children after enterovirus 71 infection. N. Engl. J. Med. 2007, 356, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Huang, C.C.; Liu, C.C.; Chang, Y.C.; Chen, C.Y.; Wang, S.T.; Yeh, T.F. Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 1999, 341, 936–942. [Google Scholar] [CrossRef]
- Chang, L.Y.; Huang, Y.C.; Lin, T.Y. Fulminant neurogenic pulmonary oedema with hand, foot, and mouth disease. Lancet 1998, 352, 367–368. [Google Scholar]
- Chang, Y.L.; Ho, B.C.; Sher, S.; Yu, S.L.; Yang, P.C. miR-146a and miR-370 coordinate enterovirus 71-induced cell apoptosis through targeting SOS1 and GADD45beta. Cell. Microbiol. 2015, 17, 802–818. [Google Scholar] [CrossRef]
- Ho, B.C.; Yu, I.S.; Lu, L.F.; Rudensky, A.; Chen, H.Y.; Tsai, C.W.; Chang, Y.L.; Wu, C.T.; Chang, L.Y.; Shih, S.R.; et al. Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon. Nat. Commun. 2014, 5, 3344. [Google Scholar] [CrossRef] [Green Version]
- Ho, B.C.; Yu, S.L.; Chen, J.J.; Chang, S.Y.; Yan, B.S.; Hong, Q.S.; Singh, S.; Kao, C.L.; Chen, H.Y.; Su, K.Y.; et al. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 2011, 9, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Xu, W.; Xia, J.; Liang, Z.; Liu, Y.; Zhang, X.; Tan, X.; Wang, L.; Mao, Q.; Wu, J.; et al. Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N. Engl. J. Med. 2014, 370, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, L.; Mo, Z.; Wang, X.; Xia, J.; Liang, Z.; Zhang, Y.; Li, Y.; Mao, Q.; Wang, J.; et al. An inactivated enterovirus 71 vaccine in healthy children. N. Engl. J. Med. 2014, 370, 829–837. [Google Scholar] [CrossRef]
- Mao, Q.Y.; Wang, Y.; Bian, L.; Xu, M.; Liang, Z. EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev. Vaccines 2016, 15, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.; Akkaya, M.; Pierce, S.K. B cell signaling in context. Nat. Immunol. 2019, 20, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Courtney, A.H.; Lo, W.L.; Weiss, A. TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem. Sci. 2018, 43, 108–123. [Google Scholar] [CrossRef]
- Chang, L.Y.; Hsiung, C.A.; Lu, C.Y.; Lin, T.Y.; Huang, F.Y.; Lai, Y.H.; Chiang, Y.P.; Chiang, B.L.; Lee, C.Y.; Huang, L.M. Status of cellular rather than humoral immunity is correlated with clinical outcome of enterovirus 71. Pediatric Res. 2006, 60, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Hsia, S.H.; Huang, Y.C.; Wu, C.T.; Chang, L.Y. Proinflammatory cytokine reactions in enterovirus 71 infections of the central nervous system. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 36, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.M.; Lei, H.Y.; Huang, K.J.; Wu, J.M.; Wang, J.R.; Yu, C.K.; Su, I.J.; Liu, C.C. Pathogenesis of enterovirus 71 brainstem encephalitis in pediatric patients: Roles of cytokines and cellular immune activation in patients with pulmonary edema. J. Infect. Dis. 2003, 188, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Nakagawa, H.; Fujita, M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018, 109, 513–522. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Y.; Zhang, C.; Wu, Q.; Zhang, J.; Ye, Z.; Qian, Q. Application of immune repertoire sequencing in cancer immunotherapy. Int. Immunopharmacol. 2019, 74, 105688. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.A.; Ober, B.; Malchiodi, E.L.; Lebedeva, M.I.; Braden, B.C.; Ysern, X.; Kim, J.K.; Shao, X.; Ward, E.S.; Mariuzza, R.A. Crystal structure of the V alpha domain of a T cell antigen receptor. Science 1995, 270, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Bentley, G.A.; Boulot, G.; Karjalainen, K.; Mariuzza, R.A. Crystal structure of the beta chain of a T cell antigen receptor. Science 1995, 267, 1984–1987. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.J.; Doherty, P.C.; McCluskey, J.; Rossjohn, J. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 2006, 6, 883–894. [Google Scholar] [CrossRef]
- Miconnet, I. Probing the T-cell receptor repertoire with deep sequencing. Curr. Opin. HIV AIDS 2012, 7, 64–70. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT--the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Lefranc, M.P.; Giudicelli, V.; Duroux, P.; Jabado-Michaloud, J.; Folch, G.; Aouinti, S.; Carillon, E.; Duvergey, H.; Houles, A.; Paysan-Lafosse, T.; et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015, 43, D413–D422. [Google Scholar] [CrossRef] [Green Version]
- Shifrut, E.; Baruch, K.; Gal, H.; Ndifon, W.; Deczkowska, A.; Schwartz, M.; Friedman, N. CD4(+) T Cell-Receptor Repertoire Diversity is Compromised in the Spleen but Not in the Bone Marrow of Aged Mice Due to Private and Sporadic Clonal Expansions. Front. Immunol. 2013, 4, 379. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.L.; Ying, X.L.; Han, X.; Sun, X.X.; Jin, Q.; Yang, F. Characterization of the enterovirus 71 VP1 protein as a vaccine candidate. J. Med. Virol. 2015, 87, 256–262. [Google Scholar] [CrossRef]
- Tan, S.; Tan, X.; Sun, X.; Lu, G.; Chen, C.C.; Yan, J.; Liu, J.; Xu, W.; Gao, G.F. VP2 dominated CD4+ T cell responses against enterovirus 71 and cross-reactivity against coxsackievirus A16 and polioviruses in a healthy population. J. Immunol. 2013, 191, 1637–1647. [Google Scholar] [CrossRef] [Green Version]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.; Lundegaard, C.; Blicher, T.; Lamberth, K.; Harndahl, M.; Justesen, S.; Roder, G.; Peters, B.; Sette, A.; Lund, O.; et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE 2007, 2, e796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karosiene, E.; Rasmussen, M.; Blicher, T.; Lund, O.; Buus, S.; Nielsen, M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 2013, 65, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004, 32, W96–W99. [Google Scholar] [CrossRef] [Green Version]
- Gohlke, H.; Hendlich, M.; Klebe, G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000, 295, 337–356. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- La Gruta, N.L.; Turner, S.J. T cell mediated immunity to influenza: Mechanisms of viral control. Trends Immunol. 2014, 35, 396–402. [Google Scholar] [CrossRef]
- Trautmann, L.; Rimbert, M.; Echasserieau, K.; Saulquin, X.; Neveu, B.; Dechanet, J.; Cerundolo, V.; Bonneville, M. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 2005, 175, 6123–6132. [Google Scholar] [CrossRef]
- Lehner, P.J.; Wang, E.C.; Moss, P.A.; Williams, S.; Platt, K.; Friedman, S.M.; Bell, J.I.; Borysiewicz, L.K. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J. Exp. Med. 1995, 181, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Argaet, V.P.; Schmidt, C.W.; Burrows, S.R.; Silins, S.L.; Kurilla, M.G.; Doolan, D.L.; Suhrbier, A.; Moss, D.J.; Kieff, E.; Sculley, T.B.; et al. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 1994, 180, 2335–2340. [Google Scholar] [CrossRef]
- Kapasi, Z.F.; Murali-Krishna, K.; McRae, M.L.; Ahmed, R. Defective generation but normal maintenance of memory T cells in old mice. Eur. J. Immunol. 2002, 32, 1567–1573. [Google Scholar] [CrossRef]
- Dong, C.; Wang, J.; Liu, L.; Zhao, H.; Shi, H.; Zhang, Y.; Jiang, L.; Li, Q. Optimized development of a candidate strain of inactivated EV71 vaccine and analysis of its immunogenicity in rhesus monkeys. Hum. Vaccines 2010, 6, 1028–1037. [Google Scholar] [CrossRef] [Green Version]
- Ong, K.C.; Devi, S.; Cardosa, M.J.; Wong, K.T. Formaldehyde-inactivated whole-virus vaccine protects a murine model of enterovirus 71 encephalomyelitis against disease. J. Virol. 2010, 84, 661–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foo, D.G.; Alonso, S.; Phoon, M.C.; Ramachandran, N.P.; Chow, V.T.; Poh, C.L. Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res. 2007, 125, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Tung, W.S.; Bakar, S.A.; Sekawi, Z.; Rosli, R. DNA vaccine constructs against enterovirus 71 elicit immune response in mice. Genet. Vaccines Ther. 2007, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.C.; Ho, M.S.; Wu, J.C.; Chen, W.J.; Huang, J.H.; Chou, S.T.; Hu, Y.C. Immunization with virus-like particles of enterovirus 71 elicits potent immune responses and protects mice against lethal challenge. Vaccine 2008, 26, 1855–1862. [Google Scholar] [CrossRef]
- Liang, Z.; Wang, J. EV71 vaccine, an invaluable gift for children. Clin. Transl. Immunol. 2014, 3, e28. [Google Scholar] [CrossRef]
- Mao, Q.; Dong, C.; Li, X.; Gao, Q.; Guo, Z.; Yao, X.; Wang, Y.; Gao, F.; Li, F.; Xu, M.; et al. Comparative analysis of the immunogenicity and protective effects of inactivated EV71 vaccines in mice. PLoS ONE 2012, 7, e46043. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mo, Z.; Liang, Z.; Zhang, Y.; Li, R.; Ong, K.C.; Wong, K.T.; Yang, E.; Che, Y.; Wang, J.; et al. Immunity and clinical efficacy of an inactivated enterovirus 71 vaccine in healthy Chinese children: A report of further observations. BMC Med. 2015, 13, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.Y.; Lin, Y.W.; Kuo, C.H.; Liu, W.H.; Tai, H.F.; Pan, C.H.; Chen, Y.T.; Hsiao, P.W.; Chan, C.H.; Chang, C.C.; et al. Inactivated Enterovirus 71 Vaccine Produced by 200-L Scale Serum-Free Microcarrier Bioreactor System Provides Cross-Protective Efficacy in Human SCARB2 Transgenic Mouse. PLoS ONE 2015, 10, e0136420. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ju, Y.; Chen, M.; Xie, Z.; Zhou, K.; Tan, Y.; Mo, J. Epidemiological and genetic characteristics of EV71 in hand, foot, and mouth disease in Guangxi, southern China, from 2010 to 2015. PLoS ONE 2017, 12, e0188640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noisumdaeng, P.; Sangsiriwut, K.; Prasertsopon, J.; Klinmalai, C.; Payungporn, S.; Mungaomklang, A.; Chokephaibulkit, K.; Buathong, R.; Thitithanyanont, A.; Puthavathana, P. Complete genome analysis demonstrates multiple introductions of enterovirus 71 and coxsackievirus A16 recombinant strains into Thailand during the past decade. Emerg Microbes Infect 2018, 7, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esfandiarei, M.; McManus, B.M. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathol. 2008, 3, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Karussis, D. The diagnosis of multiple sclerosis and the various related demyelinating syndromes: A critical review. J. Autoimmun. 2014, 48–49, 134–142. [Google Scholar]
- Polman, C.H.; O’Connor, P.W.; Havrdova, E.; Hutchinson, M.; Kappos, L.; Miller, D.H.; Phillips, J.T.; Lublin, F.D.; Giovannoni, G.; Wajgt, A.; et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2006, 354, 899–910. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Malave, E.; Stark-Aroeira, L. A natural anti-T-cell receptor monoclonal antibody protects against experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2011, 234, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Walters, G.; Knight, J.F.; Alexander, S.I. DNA vaccination against specific pathogenic TCRs reduces proteinuria in active Heymann nephritis by inducing specific autoantibodies. J. Immunol. 2003, 171, 4824–4829. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Liang, C.T.; Jiang, S.T.; Chen, K.H.; Yang, C.C.; Cheng, M.L.; Ho, H.Y. A Novel Murine Model Expressing a Chimeric mSCARB2/hSCARB2 Receptor Is Highly Susceptible to Oral Infection with Clinical Isolates of Enterovirus 71. J. Virol. 2019, 93. [Google Scholar]
- Chang, C.S.; Liao, C.C.; Liou, A.T.; Chang, Y.S.; Chang, Y.T.; Tzeng, B.H.; Chen, C.C.; Shih, C. Enterovirus 71 targets the cardiopulmonary system in a robust oral infection mouse model. Sci. Rep. 2019, 9, 11108. [Google Scholar] [CrossRef] [Green Version]
- Shih, C.; Liao, C.C.; Chang, Y.S.; Wu, S.Y.; Chang, C.S.; Liou, A.T. Immunocompetent and Immunodeficient Mouse Models for Enterovirus 71 Pathogenesis and Therapy. Viruses 2018, 10, 674. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.F.; Chou, C.T.; Lei, H.Y.; Liu, C.C.; Wang, S.M.; Yan, J.J.; Su, I.J.; Wang, J.R.; Yeh, T.M.; Chen, S.H.; et al. A mouse-adapted enterovirus 71 strain causes neurological disease in mice after oral infection. J. Virol. 2004, 78, 7916–7924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousfi Monod, M.; Giudicelli, V.; Chaume, D.; Lefranc, M.P. IMGT/JunctionAnalysis: The first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics 2004, 20, i379–i385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Chen, M.H.; Sandberg, D.J.; Babu, K.R.; Bubis, J.; Surya, A.; Ramos, L.S.; Zapata, H.J.; Galan, J.F.; Sandberg, M.N.; Birge, R.R.; et al. Conserved residues in the extracellular loops of short-wavelength cone visual pigments. Biochemistry 2011, 50, 6763–6773. [Google Scholar] [CrossRef] [Green Version]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Kiefer, F.; Arnold, K.; Kunzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. Dbiol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
| Severity Grade | p Value of Trend Test | ||||
|---|---|---|---|---|---|
| Mock Infection | Grade 1 | Grade 2 | Grade 3 | ||
| Mice No. | 9 | 8 | 9 | 10 | |
| Averaged viral titer (log10 PFUs/g) | − | 3.28 ± 1.68 | 5.83 ± 0.34 | 6.41 ± 0.71 | <0.001 |
| Viral titer significance (p value) a | − | − | <0.001 | <0.001 | |
| Average body weight (g) | 4.87 ± 0.55 | 4.16 ± 1.01 | 3.89 ± 0.90 | 3.52 ± 0.60 | <0.001 |
| Body weight significance (p value) b | − | 0.2489 | 0.0532 | 0.0032 | |
| MHC Presenting EV71 VP1 Peptide | Binding Energy |
|---|---|
| MHC Class I-EV71 VP1 Peptide | |
| FTYMRFDAEFTF | −1193.3 a |
| LAWQTATNPSVF | −924 |
| IYMRMKHVRAWI | −872.3 |
| SFFSRAGLVGEI | −881.4 |
| MHC Class II-EV71 VP1 Peptide | |
| VSRALTRALPAPTGQ | − |
| Cleaved EV71 VP1 Peptide a | ||||
|---|---|---|---|---|
| FTYMRFDAEFTF | LAWQTATNPSVF | IYMRMKHVRAWI | SFFSRAGLVGEI | |
| HA_RMSDm | 0.39 | 0.17 | 0.93 | 0.60 |
| Grid score | −180.63 | −164.19 | −118.01 | −155.11 |
| Grid_vdw | −151.80 | −153.65 | −96.85 | −117.20 |
| Grid_es | −28.83 | −10.54 | −21.16 | −37.91 |
| Int. energy | 89.50 | 63.30 | 120.48 | 86.70 |
| Cleaved EV71 VP1 Peptide | CASSLGANSDYTF CDR3 Scramble | HA_RMSDm | Grid Score | Grid_vdw | Grid_es | Int. Energy b |
|---|---|---|---|---|---|---|
| FTYMRFDAEFTF | CASSLGANSDYTF | 0.39 | −180.63 | −151.80 | −28.83 | 89.50 |
| CANSGLSSDAYTF | 1.45 | 483.15 | 486.10 | −2.96 | 260.33 | |
| CGAYLANSSSDTF | 0.51 | −117.71 | −80.91 | −36.81 | 97.31 | |
| CGSNALYSTDASF | 5.88 | −14.21 | −20.92 | 6.71 | 86.97 | |
| LAWQTATNPSVF | CASSLGANSDYTF a | 0.17 | −164.19 | −153.65 | −10.54 | 63.30 |
| CLATGNSAYDSSF | 1.37 | 9095.05 | 9114.54 | −19.49 | 1037.36 | |
| CLSYATASNGSDF | 0.42 | −137.52 | −130.43 | −7.09 | 84.10 | |
| CTSNGAASSDLYF | 2.80 | 58013.92 | 58035.42 | −21.50 | 1905.18 | |
| IYMRMKHVRAWI | CASSLGANSDYTF | 0.93 | −118.01 | −96.85 | −21.16 | 120.48 |
| CAALYTDSSGSNF | 4.30 | 1423.73 | 1409.09 | 14.63 | 157.11 | |
| CLTAAGNDYSSSF | 0.60 | −65.99 | −41.48 | −24.51 | 142.61 | |
| CSTSNDSAYLGAF | 0.50 | 166.10 | 199.76 | −33.66 | 114.02 | |
| SFFSRAGLVGEI | CASSLGANSDYTF | 0.60 | −155.11 | −117.20 | −37.91 | 86.70 |
| CAADGLNSSSTYF | 0.46 | 124.89 | 162.56 | −37.67 | 85.43 | |
| CASNGSTASLDYF | 0.46 | −43.34 | 3.89 | −47.22 | 82.62 | |
| CNSYSALTGDASF | 0.54 | 750.47 | 778.18 | −27.71 | 81.88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, Y.-W.; Ho, B.-C.; Chen, M.-H.; Yu, S.-L. Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster. Pathogens 2020, 9, 121. https://doi.org/10.3390/pathogens9020121
Liao Y-W, Ho B-C, Chen M-H, Yu S-L. Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster. Pathogens. 2020; 9(2):121. https://doi.org/10.3390/pathogens9020121
Chicago/Turabian StyleLiao, Yu-Wen, Bing-Ching Ho, Min-Hsuan Chen, and Sung-Liang Yu. 2020. "Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster" Pathogens 9, no. 2: 121. https://doi.org/10.3390/pathogens9020121
APA StyleLiao, Y.-W., Ho, B.-C., Chen, M.-H., & Yu, S.-L. (2020). Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster. Pathogens, 9(2), 121. https://doi.org/10.3390/pathogens9020121




