Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia
Abstract
1. Introduction
2. Results
2.1. Clinical Specimens
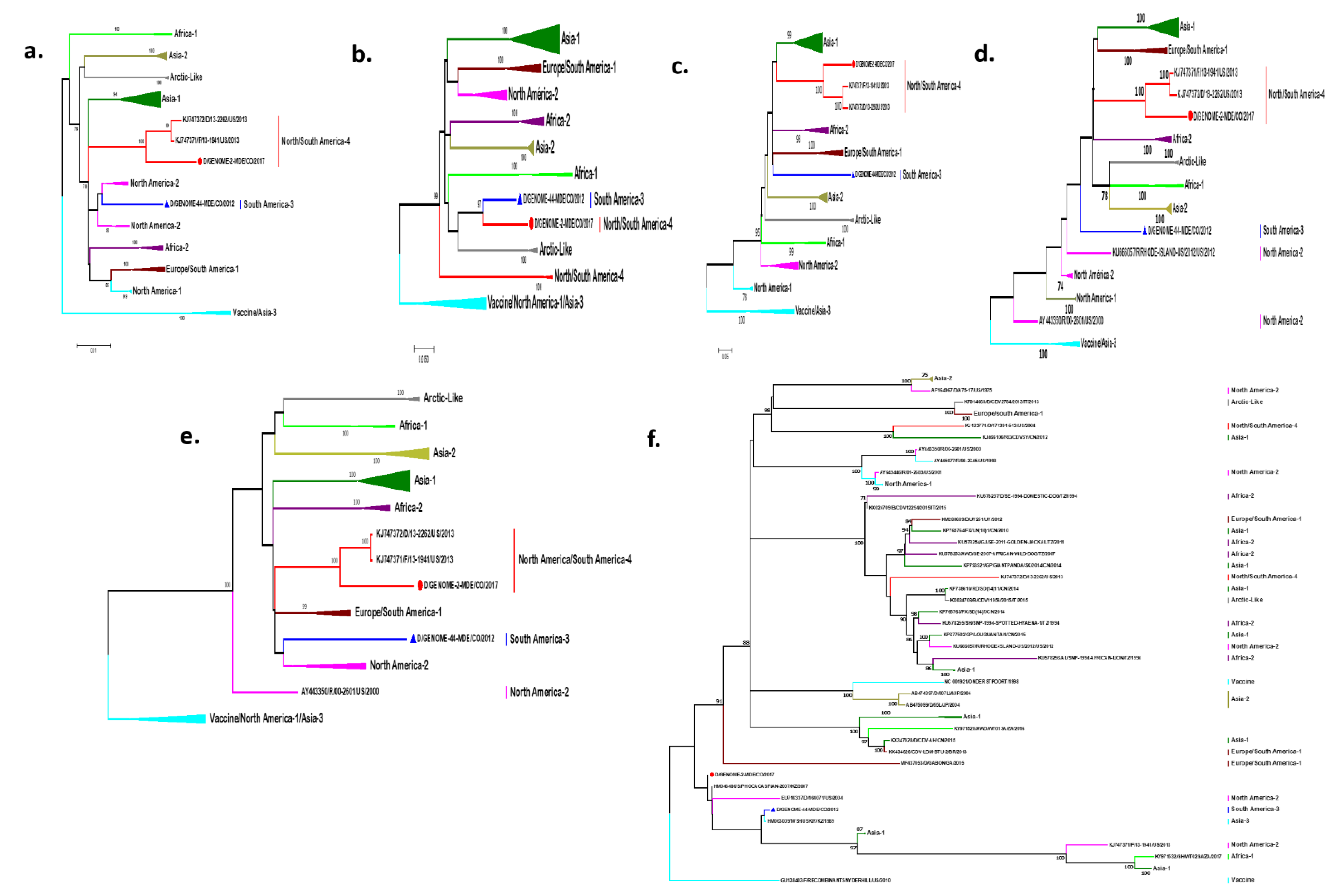
2.2. Phylogenetic Analysis of the Entire Genome
2.3. Recombination Analysis
2.4. Analysis of the Amino Acids of CDV
2.5. Analysis of Positive Selection
2.6. The CDV Molecular Clock
3. Discussion
4. Materials and Methods
4.1. Type of Study and Ethical Considerations
4.2. Clinical Specimens
4.3. RNA Extraction
4.4. Synthesis of Complementary DNA (cDNA)
4.5. PCR and Sequencing
4.6. Phylogenetic Analysis
4.7. Recombination Analysis
4.8. Analysis of the CDV Amino Acids
4.9. Analysis of Selection Pressure
4.10. Molecular Clocks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- MacLachlan, N.; Dubovi, E.; Fenner, F. Fenner’s Veterinary Virology, 4th ed.; Academic Press: Boston, MA, USA, 2011; pp. 299–325. [Google Scholar]
- Martinez-Gutierrez, M.; Ruiz-Saenz, J. Diversity of susceptible hosts in canine distemper virus infection: A systematic review and data synthesis. BMC Vet. Res. 2016, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses (ICTV). 2019. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 10 December 2019).
- Lunardi, M.; Darold, G.M.; Amude, A.M.; Headley, S.A.; Sonne, L.; Yamauchi, K.C.I.; Boabaid, F.M.; Alfieri, A.F.; Alfieri, A.A. Canine distemper virus active infection in order pilosa, family myrmecophagidae, species tamandua tetradactyla. Vet. Microbiol. 2018, 220, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Chinnakannan, S.K.; Nanda, S.K.; Baron, M.D. Morbillivirus v proteins exhibit multiple mechanisms to block type 1 and type 2 interferon signalling pathways. PLoS ONE 2013, 8, e57063. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, K.M.; Pfaller, C.K.; Conzelmann, K.K. The measles virus v protein binds to p65 (rela) to suppress nf-kappab activity. J. Virol. 2011, 85, 3162–3171. [Google Scholar] [CrossRef]
- Duque-Valencia, J.; Sarute, N.; Olarte-Castillo, X.A.; Ruiz-Saenz, J. Evolution and interspecies transmission of canine distemper virus-an outlook of the diverse evolutionary landscapes of a multi-host virus. Viruses 2019, 11, 582. [Google Scholar] [CrossRef]
- Martella, V.; Bianchi, A.; Bertoletti, I.; Pedrotti, L.; Gugiatti, A.; Catella, A.; Cordioli, P.; Lucente, M.S.; Elia, G.; Buonavoglia, C. Canine distemper epizootic among red foxes, Italy, 2009. Emerg. Infect. Dis. 2010, 16, 2007. [Google Scholar] [CrossRef]
- Espinal, M.A.; Diaz, F.J.; Ruiz-Saenz, J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet. Microbiol. 2014, 172, 168–176. [Google Scholar] [CrossRef]
- Pope, J.P.; Miller, D.L.; Riley, M.C.; Anis, E.; Wilkes, R.P. Characterization of a novel canine distemper virus causing disease in wildlife. J. Vet. Diagn. Investig. 2016, 28, 506–513. [Google Scholar] [CrossRef]
- Nikolin, V.M.; Olarte-Castillo, X.A.; Osterrieder, N.; Hofer, H.; Dubovi, E.; Mazzoni, C.J.; Brunner, E.; Goller, K.V.; Fyumagwa, R.D.; Moehlman, P.D.; et al. Canine distemper virus in the serengeti ecosystem: Molecular adaptation to different carnivore species. Mol. Ecol. 2017, 26, 2111–2130. [Google Scholar] [CrossRef]
- Anis, E.; Newell, T.K.; Dyer, N.; Wilkes, R.P. Phylogenetic analysis of the wild-type strains of canine distemper virus circulating in the United States. Virol. J. 2018, 15, 118. [Google Scholar] [CrossRef]
- Bolt, G.; Jensen, T.D.; Gottschalck, E.; Arctander, P.; Appel, M.J.; Buckland, R.; Blixenkrone-Moller, M. Genetic diversity of the attachment (h) protein gene of current field isolates of canine distemper virus. J. Gen. Virol. 1997, 78 Pt 2, 367–372. [Google Scholar] [CrossRef]
- Duque-Valencia, J.; Forero-Munoz, N.R.; Diaz, F.J.; Martins, E.; Barato, P.; Ruiz-Saenz, J. Phylogenetic evidence of the intercontinental circulation of a canine distemper virus lineage in the Americas. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Ke, G.M.; Ho, C.H.; Chiang, M.J.; Sanno-Duanda, B.; Chung, C.S.; Lin, M.Y.; Shi, Y.Y.; Yang, M.H.; Tyan, Y.C.; Liao, P.C.; et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet. Res. 2015, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Cirone, F.; Elia, G.; Lorusso, E.; Decaro, N.; Campolo, M.; Desario, C.; Lucente, M.; Bellacicco, A.; Blixenkrone-Møller, M. Heterogeneity within the hemagglutinin genes of canine distemper virus (cdv) strains detected in Italy. Vet. Microbiol. 2006, 116, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Brinkmann, H.; Philippe, H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005, 6, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Budaszewski Rda, F.; Pinto, L.D.; Weber, M.N.; Caldart, E.T.; Alves, C.D.; Martella, V.; Ikuta, N.; Lunge, V.R.; Canal, C.W. Genotyping of canine distemper virus strains circulating in Brazil from 2008 to 2012. Virus Res. 2014, 180, 76–83. [Google Scholar] [CrossRef]
- Yuan, C.; Liu, W.; Wang, Y.; Hou, J.; Zhang, L.; Wang, G. Homologous recombination is a force in the evolution of canine distemper virus. PLoS ONE 2017, 12, e0175416. [Google Scholar] [CrossRef]
- Otsuki, N.; Nakatsu, Y.; Kubota, T.; Sekizuka, T.; Seki, F.; Sakai, K.; Kuroda, M.; Yamaguchi, R.; Takeda, M. The v protein of canine distemper virus is required for virus replication in human epithelial cells. PLoS ONE 2013, 8, e82343. [Google Scholar] [CrossRef]
- Otsuki, N.; Sekizuka, T.; Seki, F.; Sakai, K.; Kubota, T.; Nakatsu, Y.; Chen, S.; Fukuhara, H.; Maenaka, K.; Yamaguchi, R. Canine distemper virus with the intact c protein has the potential to replicate in human epithelial cells by using human nectin4 as a receptor. Virology 2013, 435, 485–492. [Google Scholar] [CrossRef]
- Panzera, Y.; Sarute, N.; Iraola, G.; Hernandez, M.; Perez, R. Molecular phylogeography of canine distemper virus: Geographic origin and global spreading. Mol. Phylogenet. Evol. 2015, 92, 147–154. [Google Scholar] [CrossRef]
- Yi, L.; Cao, Z.; Tong, M.; Cheng, Y.; Yang, Y.; Li, S.; Wang, J.; Lin, P.; Sun, Y.; Zhang, M. Identification of a novel linear b-cell epitope using a monoclonal antibody against the carboxy terminus of the canine distemper virus nucleoprotein and sequence analysis of the identified epitope in different cdv isolates. Virol. J. 2017, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yi, L.; Cao, Z.; Cheng, Y.; Tong, M.; Wang, J.; Lin, P.; Cheng, S. Identification of linear b-cell epitopes on the phosphoprotein of canine distemper virus using four monoclonal antibodies. Virus Res. 2018, 257, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Anis, E.; Holford, A.L.; Galyon, G.D.; Wilkes, R.P. Antigenic analysis of genetic variants of canine distemper virus. Vet. Microbiol. 2018, 219, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, H.; Bai, X.; Martella, V.; Hu, B.; Sun, Y.; Zhu, C.; Zhang, L.; Liu, H.; Xu, S.; et al. Emergence of canine distemper virus strains with two amino acid substitutions in the haemagglutinin protein, detected from vaccinated carnivores in north-eastern China in 2012–2013. Vet. J. 2014, 200, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Svitek, N.; Gerhauser, I.; Goncalves, C.; Grabski, E.; Döring, M.; Kalinke, U.; Anderson, D.E.; Cattaneo, R.; Von Messling, V. Morbillivirus control of the interferon response: Relevance of stat2 and mda5 but not stat1 for canine distemper virus virulence in ferrets. J. Virol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Okabayashi, T.; Fujii, N. Measles virus c protein suppresses gamma-activated factor formation and virus-induced cell growth arrest. Virology 2011, 414, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kalbermatter, D.; Shrestha, N.; Ader-Ebert, N.; Herren, M.; Moll, P.; Plemper, R.K.; Altmann, K.H.; Langedijk, J.P.; Gall, F.; Lindenmann, U.; et al. Primary resistance mechanism of the canine distemper virus fusion protein against a small-molecule membrane fusion inhibitor. Virus Res. 2019, 259, 28–37. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Shaw, M.A.; Goodman, S.J. Pathogen evolution and disease emergence in carnivores. Proc. Biol. Sci. 2007, 274, 3165–3174. [Google Scholar] [CrossRef]
- Sawatsky, B.; Cattaneo, R.; von Messling, V. Canine distemper virus spread and transmission to naive ferrets: Selective pressure on slam-dependent entry. J. Virol. 2018. [Google Scholar] [CrossRef]
- Sanjuan, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef]
- Riley, M.C.; Wilkes, R.P. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the united states associated with disease in wildlife and domestic canine populations. Virol. J. 2015, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. Rdp4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Jung, E.; Brunak, S. Netnglyc 1.0 server. Center for Biological Sequence Analysis, Technical University of Denmark. 2004. Available online: http://www.Cbs.Dtudk/services/NetNGlyc (accessed on 10 December 2019).
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. Fubar: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with beauti and the beast 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 10. [Google Scholar] [CrossRef]

| SA-4 | NA-4 | SA-3 | VAC | NA-1 | ASIA-3 | NA-2 | EU/SA-1 | ARC-L | AFR-1 | AFR-2 | ASIA-1 | ASIA-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SA-4 | 0.003 | 0.004 | 0.005 | 0.005 | 0.005 | 0.004 | 0.004 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | |
| 0.007 | 0.009 | 0.01 | 0.011 | 0.011 | 0.007 | 0.008 | 0.01 | 0.009 | 0.009 | 0.009 | 0.01 | ||
| NA-4 | 0.061 | 0.004 | 0.005 | 0.004 | 0.005 | 0.004 | 0.004 | 0.005 | 0.004 | 0.005 | 0.004 | 0.005 | |
| 0.031 | 0.009 | 0.011 | 0.011 | 0.011 | 0.008 | 0.008 | 0.01 | 0.009 | 0.009 | 0.009 | 0.009 | ||
| SA-3 | 0.123 | 0.121 | 0.005 | 0.004 | 0.005 | 0.004 | 0.004 | 0.005 | 0.004 | 0.005 | 0.004 | 0.004 | |
| 0.06 | 0.064 | 0.011 | 0.012 | 0.011 | 0.007 | 0.008 | 0.011 | 0.009 | 0.01 | 0.009 | 0.011 | ||
| VAC | 0.177 | 0.172 | 0.174 | 0.004 | 0.003 | 0.005 | 0.005 | 0.004 | 0.005 | 0.005 | 0.004 | 0.005 | |
| 0.093 | 0.095 | 0.104 | 0.006 | 0.006 | 0.009 | 0.009 | 0.011 | 0.009 | 0.01 | 0.01 | 0.011 | ||
| NA-1 | 0.124 | 0.12 | 0.112 | 0.109 | 0.004 | 0.003 | 0.004 | 0.004 | 0.004 | 0.005 | 0.004 | 0.005 | |
| 0.085 | 0.088 | 0.096 | 0.038 | 0.007 | 0.009 | 0.01 | 0.011 | 0.01 | 0.01 | 0.01 | 0.011 | ||
| ASIA-3 | 0.173 | 0.166 | 0.167 | 0.057 | 0.106 | 0.005 | 0.005 | 0.004 | 0.004 | 0.005 | 0.005 | 0.005 | |
| 0.084 | 0.088 | 0.096 | 0.039 | 0.032 | 0.01 | 0.01 | 0.011 | 0.01 | 0.01 | 0.01 | 0.011 | ||
| NA-2 | 0.099 | 0.1 | 0.088 | 0.149 | 0.073 | 0.147 | 0.003 | 0.004 | 0.004 | 0.004 | 0.004 | 0.004 | |
| 0.055 | 0.057 | 0.056 | 0.092 | 0.084 | 0.083 | 0.005 | 0.009 | 0.008 | 0.007 | 0.007 | 0.009 | ||
| EU/SA-1 | 0.13 | 0.119 | 0.12 | 0.17 | 0.114 | 0.166 | 0.097 | 0.004 | 0.004 | 0.004 | 0.004 | 0.005 | |
| 0.055 | 0.053 | 0.056 | 0.087 | 0.08 | 0.077 | 0.043 | 0.009 | 0.008 | 0.007 | 0.007 | 0.009 | ||
| ARC-L | 0.14 | 0.131 | 0.135 | 0.175 | 0.126 | 0.167 | 0.112 | 0.129 | 0.004 | 0.004 | 0.004 | 0.005 | |
| 0.077 | 0.071 | 0.085 | 0.101 | 0.092 | 0.091 | 0.07 | 0.066 | 0.009 | 0.01 | 0.01 | 0.01 | ||
| AFR-1 | 0.137 | 0.129 | 0.129 | 0.166 | 0.126 | 0.163 | 0.11 | 0.13 | 0.128 | 0.004 | 0.004 | 0.004 | |
| 0.062 | 0.064 | 0.069 | 0.092 | 0.085 | 0.083 | 0.062 | 0.057 | 0.064 | 0.009 | 0.008 | 0.009 | ||
| AFR-2 | 0.132 | 0.12 | 0.123 | 0.169 | 0.122 | 0.165 | 0.102 | 0.121 | 0.13 | 0.129 | 0.004 | 0.005 | |
| 0.066 | 0.064 | 0.073 | 0.095 | 0.09 | 0.089 | 0.06 | 0.053 | 0.08 | 0.066 | 0.008 | 0.01 | ||
| ASIA-1 | 0.131 | 0.118 | 0.122 | 0.172 | 0.122 | 0.167 | 0.102 | 0.118 | 0.134 | 0.133 | 0.123 | 0.005 | |
| 0.065 | 0.068 | 0.075 | 0.097 | 0.088 | 0.088 | 0.061 | 0.053 | 0.079 | 0.067 | 0.066 | 0.009 | ||
| ASIA-2 | 0.132 | 0.123 | 0.126 | 0.165 | 0.118 | 0.161 | 0.105 | 0.125 | 0.121 | 0.123 | 0.124 | 0.128 | |
| 0.071 | 0.071 | 0.081 | 0.102 | 0.096 | 0.096 | 0.07 | 0.064 | 0.08 | 0.068 | 0.078 | 0.069 |
| Gene | Substitutions |
|---|---|
| N | T410A, A428T, P448S, H450N, L456F, L456V |
| P | C40H, C40R, T49A, G50S, N59D, T67A, P97L, P97A, E99D, G106E, E124A, T135A, G137S, R221G, R221E, E233G, L256P, E264G, G270E, N278S, L287P, T426A |
| M | Not found |
| F | K3N, K3E, K7E, P23H, R34Q, A35T, Y48H, D49G, T60I, L74S, N82D, Q88H, K96Q, E103K, P107S, I110T, I110V, S616I, A646T |
| H | H30Q, T56A, D238Y, R241E, R241G, E247K, D329N, H330Q, M342V, K370Q, G376N, N446D, I506T, D531N, N572D, A586T |
| L | G139D, T149A, N1705K, S1707P, T1708I, S1712L, H2010Q, H2017Y, S2076F |
| Protein | Substitutions |
|---|---|
| N | L229V, I296M, E467K |
| P/V | V133A, M267V |
| V | Y267C |
| C | L6W, A27V, T30I, T33A, C44S, R47K, R74L, L89P, T93M, A109V, K146R, K154R, Q172R, P173L |
| M | T84P, F178L, N206D, L329Q |
| F | C116Y, R331P |
| H | D540G, M548T |
| L | D1748N |
| Gene | # Amino Acids | # Positive Selection Sites | # Negative Selection Sites | dN/dS |
|---|---|---|---|---|
| N | 524 | 2 | 278 | 0.0249 |
| P | 508 | 9 | 90 | 0.05 |
| V | 299 | 9 | 33 | 0.177 |
| C | 175 | 8 | 12 | 0.128 |
| M | 336 | 2 | 155 | 0.005 |
| F | 663 | 7 | 262 | 0.02 |
| H | 605 | 1 | 207 | 0.006 |
| L | 2185 | Not found | Not found | Not estimated |
| Gene | Substitution Model | Evolution Rate | tMRCA | ||
|---|---|---|---|---|---|
| Mean | 95% Confidence Interval | Median | 95% Confidence Interval | ||
| N | T92 + g | 3.73 × 10−4 | 2.71 × 10−4–4.72 × 10−4 | 1931 | 1905–1954 |
| P | Tn93 + g | 2.65 × 10−4 | 1.69 × 10−4–3.61 × 10−4 | 1888 | 1838–1927 |
| M | T92 + g | 3.29 × 10−4 | 1.91 × 10−4–4.72 × 10−4 | 1892 | 1835–1938 |
| F | T92 + g | 3.25 × 10−4 | 2.06 × 10−4–4.41 × 10−4 | 1835 | 1766–1891 |
| H | T92 + g | 4.99 × 10−4 | 3.87 × 10−4–6.13 × 10−4 | 1903 | 1876–1928 |
| L | Gtr + g | Not calculated | Not calculated | Not calculated | Not calculated |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duque-Valencia, J.; Diaz, F.J.; Ruiz-Saenz, J. Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia. Pathogens 2020, 9, 26. https://doi.org/10.3390/pathogens9010026
Duque-Valencia J, Diaz FJ, Ruiz-Saenz J. Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia. Pathogens. 2020; 9(1):26. https://doi.org/10.3390/pathogens9010026
Chicago/Turabian StyleDuque-Valencia, July, Francisco J. Diaz, and Julian Ruiz-Saenz. 2020. "Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia" Pathogens 9, no. 1: 26. https://doi.org/10.3390/pathogens9010026
APA StyleDuque-Valencia, J., Diaz, F. J., & Ruiz-Saenz, J. (2020). Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia. Pathogens, 9(1), 26. https://doi.org/10.3390/pathogens9010026






