Piscirickettsia salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors
Abstract
1. Introduction
2. Results
2.1. Identification and Visualization of Extrachromosomal Elements in P. Salmonis LF-89
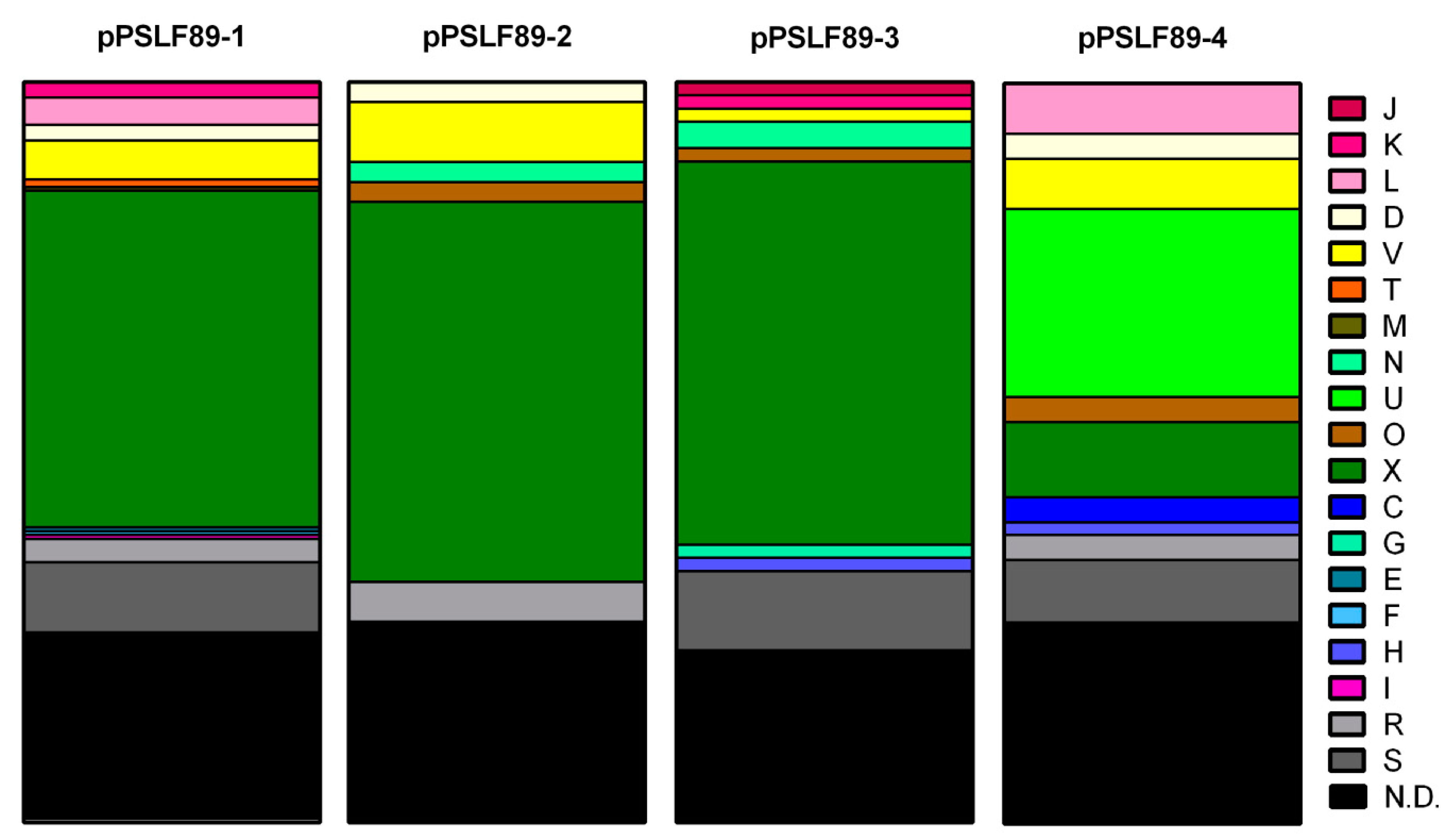
2.2. Categorization of Plasmidial Proteins in COGs
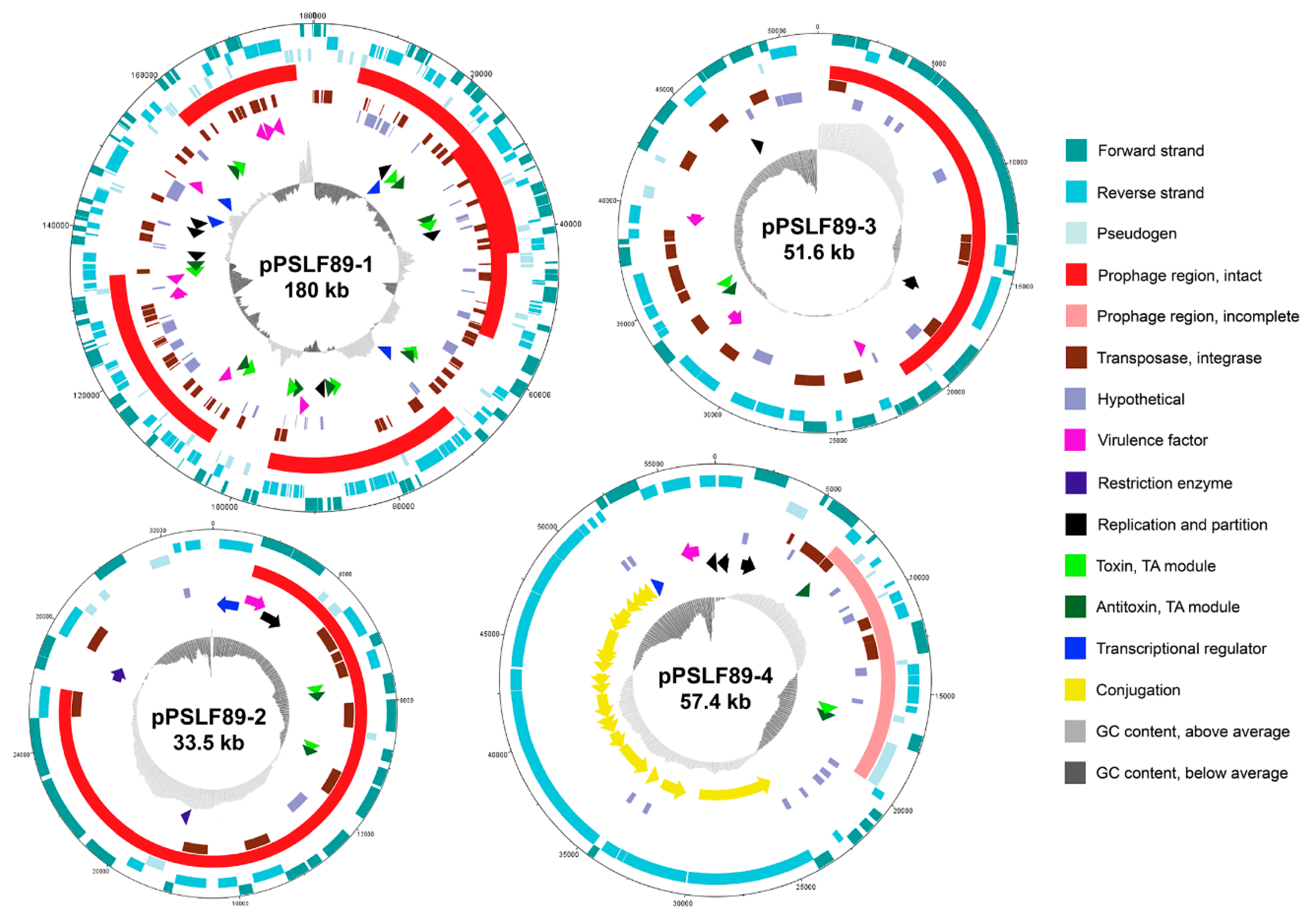
2.3. Annotation and Sequence Analysis of P. salmonis LF-89 Plasmids
2.3.1. Replication and Plasmid Stability Elements
2.3.2. DNA Mobilization and Gene Transfer in Plasmids
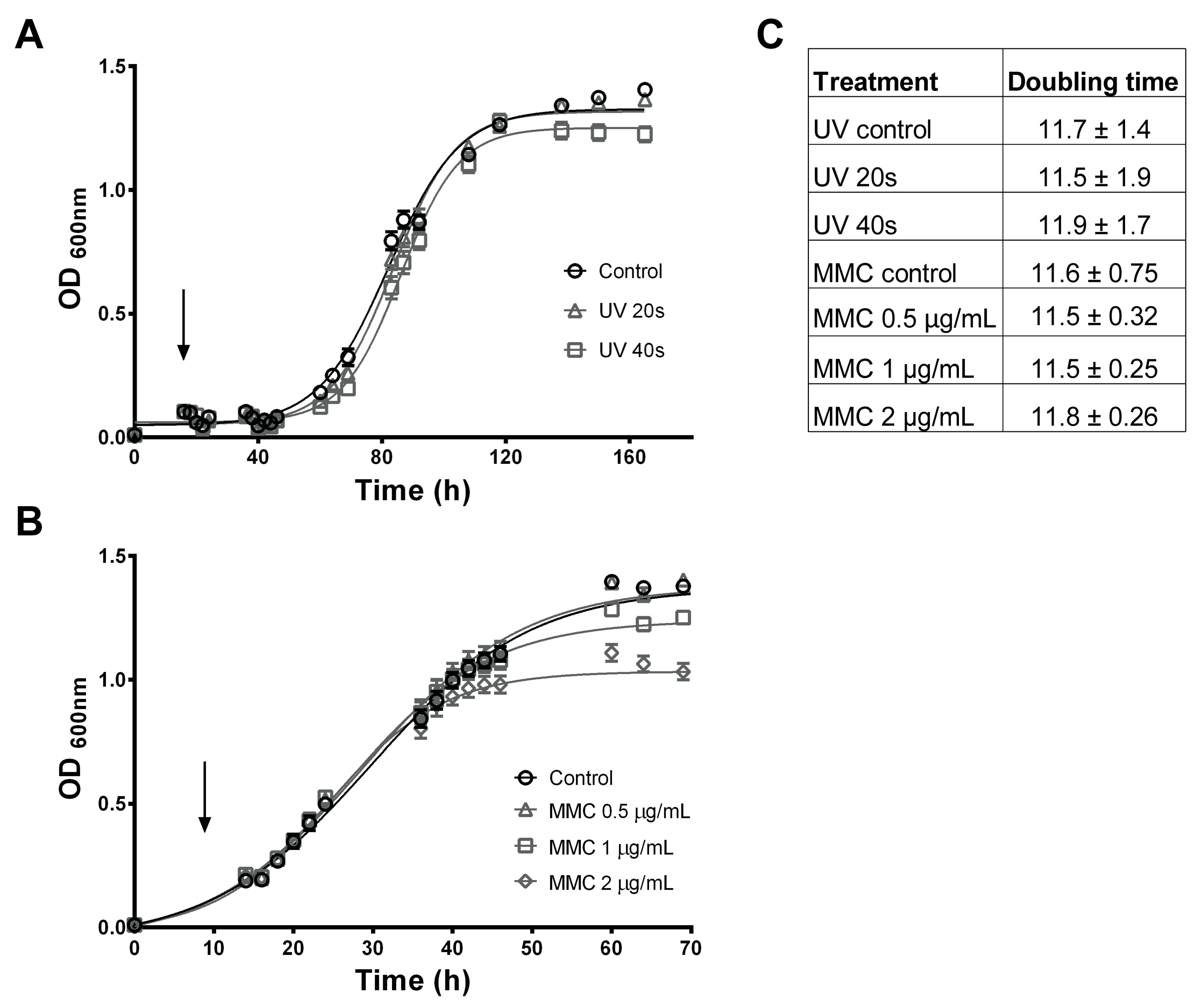
2.3.3. Prophage Sequences and Prophage Induction in P. salmonis LF-89
2.3.4. Plasmid Virulence Factors
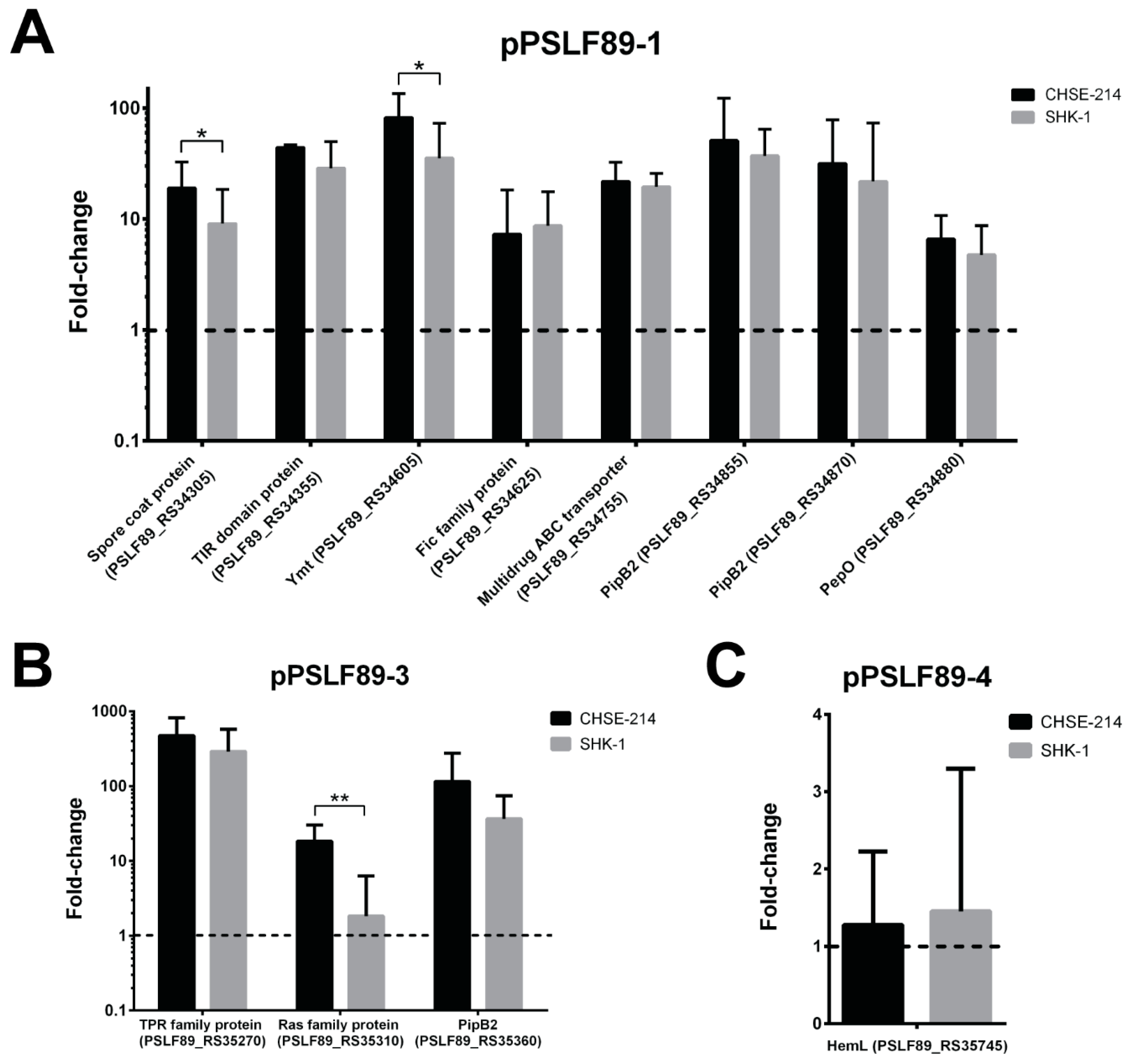
2.4. Plasmid Genes Predicted as Virulence Factors Were Overexpressed during Infection of Salmon Macrophages
2.5. Plasmid Virulence Factors are Shared and Highly Conserved among Other P. salmonis Strains
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Purification of P. salmonis Plasmids
4.3. Plasmid Sequencing and Annotation
4.4. Design and Synthesis of Digoxigenin-Labeled Oligonucleotides
4.5. Southern Blot Analysis
4.6. Plasmid Copy Number
4.7. Prophage Inductions
4.8. Cell Cultures
4.9. P. salmonis Infection in Cell Cultures
4.10. Nucleic Acids Purification and Transcripts Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in fish: A review. J. Fish Dis. 2013, 37, 1–26. [Google Scholar]
- Bravo, S.; Campos, M. Coho salmon syndrome in Chile. Am. Fish. Soc. Newsl. 1989, 17, 3. [Google Scholar]
- Cvitanich, J.D.; Garate, N.O.; Smith, C.E. The isolation of a rickettsia-like organism causing disease and mortality in Chilean salmonids and its confirmation by Koch’s postulate. J. Fish Dis. 1991, 14, 121–145. [Google Scholar] [CrossRef]
- Fryer, J.L.; Lannan, C.N.; Giovannoni, S.J.; Wood, N.D. Piscirickettsia salmonis gen. nov., sp. nov., the causative agent of an epizootic disease in salmonid fishes. Int. J. Syst. Bacteriol. 1992, 42, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Pulgar, R.; Travisany, D.; Zuñiga, A.; Maass, A.; Cambiazo, V. Complete genome sequence of Piscirickettsia salmonis LF-89 (ATCC VR-1361) a major pathogen of farmed salmonid fish. J. Biotechnol. 2015, 212, 30–31. [Google Scholar] [CrossRef]
- Nourdin-Galindo, G.; Sánchez, P.; Molina, C.F.; Espinoza-Rojas, D.A.; Oliver, C.; Ruiz, P.; Vargas-Chacoff, L.; Cárcamo, J.G.; Figueroa, J.E.; Mancilla, M.; et al. Comparative pan-genome analysis of Piscirickettsia salmonis reveals genomic divergences within genogroups. Front. Cell. Infect. Microbiol. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Chaconas, G.; Norris, S.J. Peaceful coexistence amongst Borrelia plasmids: Getting by with a little help from their friends? Plasmid 2014, 70, 161–167. [Google Scholar] [CrossRef][Green Version]
- Carneiro, L.C.; Mendes, P.V.C.; Silva, S.P.; Souza, G.R.L.; Bataus, L.A.M. Characterization of a cryptic and intriguing low molecular weight plasmid. Curr. Microbiol. 2016, 72, 351–356. [Google Scholar] [CrossRef]
- Wang, Y.; Zhuang, X.; Zhong, Y.; Zhang, C.; Zhang, Y.; Zeng, L. Distribution of plasmids in distinct Leptospira pathogenic species. PLoS Negl. Trop. Dis. 2015, 9, e0004220. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Int. J. Med. Microbiol. 2011, 301, 654–658. [Google Scholar] [CrossRef]
- Del Solar, G.; Giraldo, R.; Ruiz-Echevarría, M.J.; Espinosa, M.; Díaz-Orejas, R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998, 62, 434–464. [Google Scholar] [PubMed]
- Davison, J. Genetic exchange between bacteria in the environment. Plasmid 1999, 42, 73–91. [Google Scholar] [CrossRef]
- Kubasova, T.; Cejkova, D.; Matiasovicova, J.; Sekelova, Z.; Polansky, O.; Medvecky, M.; Rychlik, I.; Juricova, H. Antibiotic resistance, core-genome and protein expression in incHI1 plasmids in Salmonella Typhimurium. Genome Biol. Evol. 2016, 8, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Villa, L.; Poirel, L.; Nordmann, P.; Carattoli, A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16s RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013, 68, 34–39. [Google Scholar] [CrossRef]
- Gulig, P.A.; Danbara, H.; Guiney, D.G.; Lax, A.J.; Norel, F.; Rhen, M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 1993, 7, 825–830. [Google Scholar] [CrossRef]
- Fricke, W.F.; McDermott, P.F.; Mammel, M.K.; Zhao, S.; Johnson, T.J.; Rasko, D.A.; Fedorka-Cray, P.J.; Pedroso, A.; Whichard, J.M.; LeClerc, J.E.; et al. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 2009, 75, 5963–5971. [Google Scholar] [CrossRef]
- Monchy, S.; Benotmane, M.A.; Janssen, P.; Vallaeys, T.; Taghavi, S.; Van Der Lelie, D.; Mergeay, M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 2007, 189, 7417–7425. [Google Scholar] [CrossRef]
- Aminov, R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502. [Google Scholar] [CrossRef]
- López-Guerrero, M.G.; Ormeño-Orrillo, E.; Acosta, J.L.; Mendoza-Vargas, A.; Rogel, M.A.; Ramírez, M.A.; Rosenblueth, M.; Martínez-Romero, J.; Martínez-Romero, E. Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid 2012, 68, 149–158. [Google Scholar] [CrossRef]
- Di Lorenzo, M.; Stork, M. Plasmid-encoded iron uptake systems. Microbiol. Spectr. 2015, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Voth, D.E.; Beare, P.A.; Howe, D.; Sharma, U.M.; Samoilis, G.; Cockrell, D.C.; Omsland, A.; Heinzen, R. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J. Bacteriol. 2011, 193, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Amy, J.; Bulach, D.; Knight, D.; Riley, T.; Johanesen, P.; Lyras, D. Identification of large cryptic plasmids in Clostridioides (Clostridium) difficile. Plasmid 2018, 96–97, 25–38. [Google Scholar] [CrossRef]
- Guler, H.I.; Ceylan, E.; Canakci, S.; Belduz, A.O. A novel cryptic and theta type plasmid (pHIG22) from Thermus scodotuctus sp. K6. Gene 2018, 679, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Zaleski, P.; Wawrzyniak, P.; Sobolewska, A.; Łukasiewicz, N.; Baran, P.; Romańczuk, K.; Daniszewska, K.; Kierył, P.; Płucienniczak, G.; Płucienniczak, A. PIGWZ12—A cryptic plasmid with a modular structure. Plasmid 2015, 79, 37–47. [Google Scholar] [CrossRef]
- Harris, L.; van Zyl, L.J.; Kirby-McCullough, B.M.; Damelin, L.H.; Tiemessen, C.T.; Trindade, M. Identification and sequence analysis of two novel cryptic plasmids isolated from the vaginal mucosa of South African women. Plasmid 2018, 98, 56–62. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Reznikoff, W.S.; Bay, J. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 2005, 3, 688–699. [Google Scholar] [CrossRef]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Jackson, R.W.; Vinatzer, B.; Arnold, D.L.; Dorus, S.; Murillo, J. The influence of the accessory genome on bacterial pathogen evolution. Mob. Genet. Elements 2011, 1, 55–65. [Google Scholar] [CrossRef]
- Sobecky, P.A. Approaches to investigating the ecology of plasmids in marine bacterial communities. Plasmid 2002, 48, 213–221. [Google Scholar] [CrossRef]
- Halary, S.; Leigh, J.W.; Cheaib, B.; Lopez, P.; Bapteste, E. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. USA 2010, 107, 127–132. [Google Scholar] [CrossRef]
- Gómez, F.; Henríquez, V.; Marshall, S.H. Additional evidence of the facultative intracellular nature of the fish bacterial pathogen Piscirickettsia salmonis. Arch. Med. Vet. 2009, 267, 261–267. [Google Scholar] [CrossRef]
- Saavedra, J.; Grandón, M.; Villalobos-González, J.; Bohle, H.; Bustos, P.; Mancilla, M. Isolation, functional characterization and transmissibility of p3PS10, a multidrug resistance plasmid of the fish pathogen Piscirickettsia salmonis. Front. Microbiol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Oliver, C.; Hernández, M.A.; Tandberg, J.I.; Valenzuela, K.N.; Lagos, L.X.; Haro, R.E.; Sánchez, P.; Ruiz, P.A.; Sanhueza-Oyarzún, C.; Cortés, M.A.; et al. The proteome of biologically active membrane vesicles from Piscirickettsia salmonis LF-89 type strain identifies plasmid-encoded putative toxins. Front. Cell. Infect. Microbiol. 2017, 7, 420. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Dong, N.; Lin, D.; Yang, X.; Wong, M.H.Y.; Chan, E.W.C.; Chen, S. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Yanagiya, K.; Maejima, Y.; Nakata, H.; Tokuda, M.; Moriuchi, R.; Dohra, H.; Inoue, K.; Ohkuma, M.; Kimbara, K.; Shintani, M. Novel self-transmissible and broad-host-range plasmids exogenously captured from anaerobic granules or cow manure. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Knodler, L.A.; Steele-Mortimer, O. The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate sif extension. Mol. Biol. Cell 2005, 16, 4108–4123. [Google Scholar] [CrossRef]
- Balado, M.; Benzekri, H.; Labella, A.M.; Claros, M.G.; Manchado, M.; Borrego, J.J.; Osorio, C.R.; Lemos, M.L. Genomic analysis of the marine fish pathogen Photobacterium damselae subsp. piscicida: Insertion sequences proliferation is associated with chromosomal reorganisations and rampant gene decay. Infect. Genet. Evol. 2017, 54, 221–229. [Google Scholar]
- Backert, S.; Grohmann Editors, E. Current Topics in Microbiology and Immunology Type IV Secretion in Gram-Negative and Gram-Positive Bacteria; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; ISBN 9783319752402. [Google Scholar]
- Li, P.L.; Hwang, I.; Miyagi, H.; True, H.; Farrand, S.K. Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J. Bacteriol. 1999, 181, 5033–5041. [Google Scholar]
- Farrand, S.K.; Hwang, I.; Cook, D.M. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 1996, 178, 4233–4247. [Google Scholar] [CrossRef]
- Molofsky, A.B.; Swanson, M.S. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 2003, 50, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Otsuka, Y. Prokaryotic toxin—Antitoxin systems: Novel regulations of the toxins. Curr. Genet. 2016, 62, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Machuca, A.; Martinez, V. Transcriptome analysis of the intracellular facultative pathogen Piscirickettsia salmonis: Expression of putative groups of genes associated with virulence and iron metabolism. PLoS ONE 2016, 11, e0168855. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Origin of Group Identity: Viruses, Addiction and Cooperation; Springer US: Berlin/Heidelberg, Germany, 2009; ISBN 9780387779973. [Google Scholar]
- Rana, R.R.; Zhang, M.; Spear, A.M.; Atkins, H.S.; Byrne, B. Bacterial TIR-containing proteins and host innate immune system evasion. Med. Microbiol. Immunol. 2013, 202, 1–10. [Google Scholar] [CrossRef]
- Roy, C.R.; Cherfils, J. Structure and function of Fic proteins. Nat. Rev. Microbiol. 2015, 13, 631–640. [Google Scholar] [CrossRef]
- Agarwal, V.; Kuchipudi, A.; Fulde, M.; Riesbeck, K.; Bergmann, S.; Blom, A.M. Streptococcus pneumoniae Endopeptidase O (PepO) is a multifunctional plasminogen-and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 2013, 288, 6849–6863. [Google Scholar] [CrossRef]
- Zakrzewska, A.; Ágoston, V.C.; Ordas, A.; Mink, M.; Spaink, H.P.; Meijer, A.H. Deep sequencing of the zebrafish transcriptome response to Mycobacterium infection. Mol. Immunol. 2009, 46, 2918–2930. [Google Scholar]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-Binding Proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef]
- Kondo, Y.; Ohara, N.; Sato, K.; Yoshimura, M.; Yukitake, H.; Naito, M.; Fujiwara, T.; Nakayama, K. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 2010, 78, 2846–2856. [Google Scholar] [CrossRef]
- Bang, S.; Min, C.K.; Ha, N.Y.; Choi, M.S.; Kim, I.S.; Kim, Y.S.; Cho, N.H. Inhibition of eukaryotic translation by tetratricopeptide-repeat proteins of Orientia tsutsugamushi. J. Microbiol. 2016, 54, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Million-Weaver, S.; Camps, M. Mechanisms of plasmid segregation: Have multicopy plasmids been overlooked? Plasmid 2014, 75, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Yuksel, S.A.; Thompson, K.D.; Ellis, A.E.; Adams, A. Purification of Piscirickettsia salmonis and associated phage particles. Dis. Aquat. Organ. 2001, 44, 231–235. [Google Scholar] [CrossRef]
- Gómez, F.A.; Cárdenas, C.; Henríquez, V.; Marshall, S.H. Characterization of a functional toxin-antitoxin module in the genome of the fish pathogen Piscirickettsia salmonis. FEMS Microbiol. Lett. 2011, 317, 83–92. [Google Scholar] [CrossRef]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef]
- Dy, R.L.; Richter, C.; Salmond, G.P.C.; Fineran, P.C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef]
- Casjens, S.R. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 2005, 8, 451–458. [Google Scholar] [CrossRef]
- Romeo, T.; Gong, M. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J. Bacteriol. 1993, 175, 5740–5741. [Google Scholar] [CrossRef]
- Agaras, B.; Sobrero, P.; Valverde, C. A CsrA/RsmA translational regulator gene encoded in the replication region of a Sinorhizobium meliloti cryptic plasmid complements Pseudomonas fluorescens rsmA/E mutants. Microbiology (United Kingdom) 2013, 159, 230–242. [Google Scholar] [CrossRef]
- Glöckner, G.; Albert-Weissenberger, C.; Weinmann, E.; Jacobi, S.; Schunder, E.; Steinert, M.; Hacker, J.; Heuner, K. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 2008, 298, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Guan, Z.; Cao, S.; Peng, D.; Ruan, L.; Jiang, D.; Sun, M. Plasmids are vectors for redundant chromosomal genes in the group. BMC Genom. 2015, 16, 6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yañez, A.J.; Valenzuela, K.; Silva, H.; Retamales, J.; Romero, A.; Enriquez, R.; Figueroa, J.; Claude, A.; Gonzalez, J.; Carcamo, J.G. Broth medium for the successful culture of the fish pathogen Piscirickettsia salmonis. Dis. Aquat. Organ. 2012, 97, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Mandakovic, D.; Glasner, B.; Maldonado, J.; Aravena, P.; Gonzalez, M.; Cambiazo, V.; Pulgar, R. Genomic-based restriction enzyme selection for specific detection of Piscirickettsia salmonis by 16S rDNA PCR-RFLP. Front. Microbiol. 2016, 7, 643. [Google Scholar] [CrossRef][Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, 1–6. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, 347–352. [Google Scholar] [CrossRef]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, 606–611. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Škulj, M.; Okršlar, V.; Jalen, Š.; Jevševar, S.; Slanc, P.; Štrukelj, B.; Menart, V. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb. Cell Fact. 2008, 7, 1–12. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Raya, R.; Hébert, E.M. Isolation of Phage via Induction of Lysogens. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 23–32. ISBN 9781603271646. [Google Scholar]
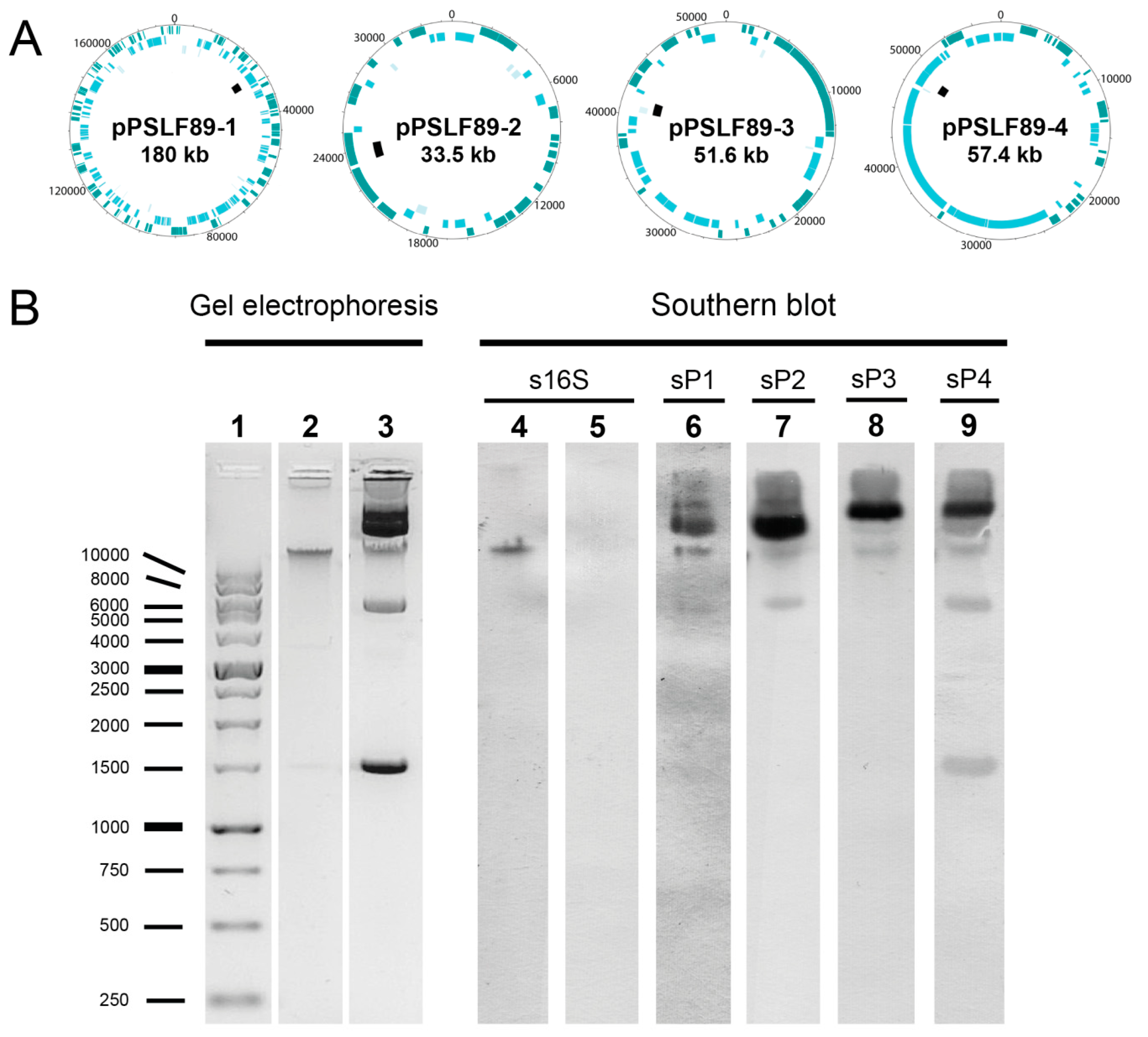

| P. salmonis LF-89 | |||||
|---|---|---|---|---|---|
| Plasmid | Size (bp) | %GC | Proteins | Genes | Pseudogenes |
| pPSLF89-1 | 180,124 | 38.90 | 189 | 233 | 42 |
| pPSLF89-2 | 33,516 | 40.61 | 37 | 44 | 7 |
| pPSLF89-3 | 51,573 | 39.06 | 56 | 60 | 4 |
| pPSLF89-4 | 57,445 | 37.28 | 59 | 66 | 7 |
| Replicon | Prophage Region | Size (kb) | Position in the Replicon | % GC | Percent of Phage Proteins | Percent of Hypothetical Proteins | Number of Phage Species |
|---|---|---|---|---|---|---|---|
| Chromosome | 1 (intact) | 38.9 | 2,079,639–2,118,283 | 36.9 | 41.2 | 58.8 | 11 |
| pPSLF89-1 | 1 (intact) | 36.4 | 6546–42,955 | 37.9 | 63.3 | 36.7 | 9 |
| 2 (intact) | 30.8 | 25,095–55,982 | 39.1 | 65.4 | 34.6 | 7 | |
| 3 (intact) | 28.1 | 68,521–96,644 | 39.6 | 83.3 | 16.7 | 6 | |
| 4 (intact) | 28.5 | 105,452–134,001 | 38.4 | 46.4 | 53.6 | 3 | |
| 5 (intact) | 18.2 | 159,247–177,535 | 38.2 | 61.9 | 38.1 | 4 | |
| pPSLF89-2 | 1 (intact) | 24.5 | 1476–25,982 | 40.56 | 62.9 | 37.1 | 14 |
| pPSLF89-3 | 1 (intact) | 20.6 | 609–21,232 | 41.2 | 65.4 | 34.6 | 11 |
| pPSLF89-4 | 1 (incomplete) | 12.9 | 6733–19,700 | 39.6 | 52.2 | 47.8 | 11 |
| Strain | Year of Isolation | Assembly | Replicons | TIR Domain Protein (PSLF89_RS34355) | Multidrug ABC Transporterpermease (PSLF89_RS34755) | Spore Coat Protein (PSLF89_RS34305) | Ymt Toxin (PSLF89_RS34605) | Fic/DOC Family Protein (PSLF89_RS34625) | PipB2 (PSLF89_RS34855) | Pentapeptiderepeats Family Protein PipB2 (PSLF89_RS34870) | Endopeptidase O (PSLF89_RS34880) | Tetratricopeptide Repeat Family Protein (PSLF89_RS35270) | Ras Family Protein (PSLF89_RS35310) | Pentapeptide Repeats Family Protein PipB2 (PSLF89_RS35360) | HemL Glutamate-1-semialdehyde Aminotransferase (PSLF89_RS35745) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genogroup LF89-like | LF-89 ATCC VR-1361 | 1989 | GCA_000300295.4 | chromosome | ||||||||||||
| pPSLF89-1 | ||||||||||||||||
| pPSLF89-2 | ||||||||||||||||
| pPSLF89-3 | ||||||||||||||||
| pPSLF89-4 | ||||||||||||||||
| PSCGR01 | 2010 | GCA_001514395.1 | chromosome | |||||||||||||
| pPsCRG01-1 | ||||||||||||||||
| pPsCRG01-2 | ||||||||||||||||
| pPsCRG01-3 | ||||||||||||||||
| pPSCRG01-4 | ||||||||||||||||
| PM22180B | 2011 | GCA_001932895.1 | chromosome | |||||||||||||
| p1PS13 | ||||||||||||||||
| p2PS13 | ||||||||||||||||
| p3PS13 | ||||||||||||||||
| p4PS13 | ||||||||||||||||
| PM25344B | 2011 | GCA_001932955.1 | chromosome | |||||||||||||
| p1PS14 | ||||||||||||||||
| p2PS14 | ||||||||||||||||
| p3PS14 | ||||||||||||||||
| p4PS14 | ||||||||||||||||
| PM31429B | 2012 | GCA_001932915.1 | chromosome | |||||||||||||
| p1PS12 | ||||||||||||||||
| p2PS12 | ||||||||||||||||
| p3PS12 | ||||||||||||||||
| p4PS12 | ||||||||||||||||
| PM32597B1 | 2012 | GCA_000756415.3 | chromosome | |||||||||||||
| pPSB1-1 | ||||||||||||||||
| pPSB1-2 | ||||||||||||||||
| pPSB1-3 | ||||||||||||||||
| pPSB1-4 | ||||||||||||||||
| CGR02 | 2013 | GCA_001534725.1 | chromosome | |||||||||||||
| pPSCRG02-1 | ||||||||||||||||
| pPSCRG02-2 | ||||||||||||||||
| pPSCRG02-4 | ||||||||||||||||
| AY3800B | 2013 | GCA_001746795.1 | chromosome | |||||||||||||
| p1PS10 | ||||||||||||||||
| p2PS10 | ||||||||||||||||
| p3PS10 | ||||||||||||||||
| p4PS10 | ||||||||||||||||
| AY3864B | 2013 | GCA_001932935.1 | chromosome | |||||||||||||
| p1PS11 | ||||||||||||||||
| p2PS11 | ||||||||||||||||
| p3PS11 | ||||||||||||||||
| p4PS11 | ||||||||||||||||
| PM49811B | 2014 | GCA_001932815.1 | chromosome | |||||||||||||
| p1PS6 | ||||||||||||||||
| p2PS6 | ||||||||||||||||
| p3PS6 | ||||||||||||||||
| p4PS6 | ||||||||||||||||
| PM58386B | 2015 | GCA_001932835.1 | chromosome | |||||||||||||
| p1PS7 | ||||||||||||||||
| p2PS7 | ||||||||||||||||
| p3PS7 | ||||||||||||||||
| p4PS7 | ||||||||||||||||
| AY6297B | 2015 | GCA_001932855.1 | chromosome | |||||||||||||
| p1PS8 | ||||||||||||||||
| p2PS8 | ||||||||||||||||
| p3PS8 | ||||||||||||||||
| p4PS8 | ||||||||||||||||
| AY6532B | 2015 | GCA_001932875.1 | chromosome | |||||||||||||
| p1PS9 | ||||||||||||||||
| p2PS9 | ||||||||||||||||
| p3PS9 | ||||||||||||||||
| p4PS9 | ||||||||||||||||
| Genogroup EM90-like | EM-90 | 1990 | GCA_003850185.1 | chromosome | ||||||||||||
| pPSEM90-1 | ||||||||||||||||
| pPSEM90-2 | ||||||||||||||||
| pPSEM90-3 | ||||||||||||||||
| pPSEM90-4 | ||||||||||||||||
| pPSEM90-5 | ||||||||||||||||
| pPSEM90-6 | ||||||||||||||||
| pPSEM90-7 | ||||||||||||||||
| pPSEM90-8 | ||||||||||||||||
| PM15972A1 | 2010 | GCA_000756435.3 | chromosome | |||||||||||||
| pPSA1-1 | ||||||||||||||||
| pPSA1-2 | ||||||||||||||||
| pPSA1-3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Severín, J.; Travisany, D.; Maass, A.; P. Chávez, F.; Cambiazo, V. Piscirickettsia salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors. Pathogens 2019, 8, 269. https://doi.org/10.3390/pathogens8040269
Ortiz-Severín J, Travisany D, Maass A, P. Chávez F, Cambiazo V. Piscirickettsia salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors. Pathogens. 2019; 8(4):269. https://doi.org/10.3390/pathogens8040269
Chicago/Turabian StyleOrtiz-Severín, Javiera, Dante Travisany, Alejandro Maass, Francisco P. Chávez, and Verónica Cambiazo. 2019. "Piscirickettsia salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors" Pathogens 8, no. 4: 269. https://doi.org/10.3390/pathogens8040269
APA StyleOrtiz-Severín, J., Travisany, D., Maass, A., P. Chávez, F., & Cambiazo, V. (2019). Piscirickettsia salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors. Pathogens, 8(4), 269. https://doi.org/10.3390/pathogens8040269






