Meningitic Escherichia coli Induction of ANGPTL4 in Brain Microvascular Endothelial Cells Contributes to Blood–Brain Barrier Disruption via ARHGAP5/RhoA/MYL5 Signaling Cascade
Abstract
1. Introduction
2. Results
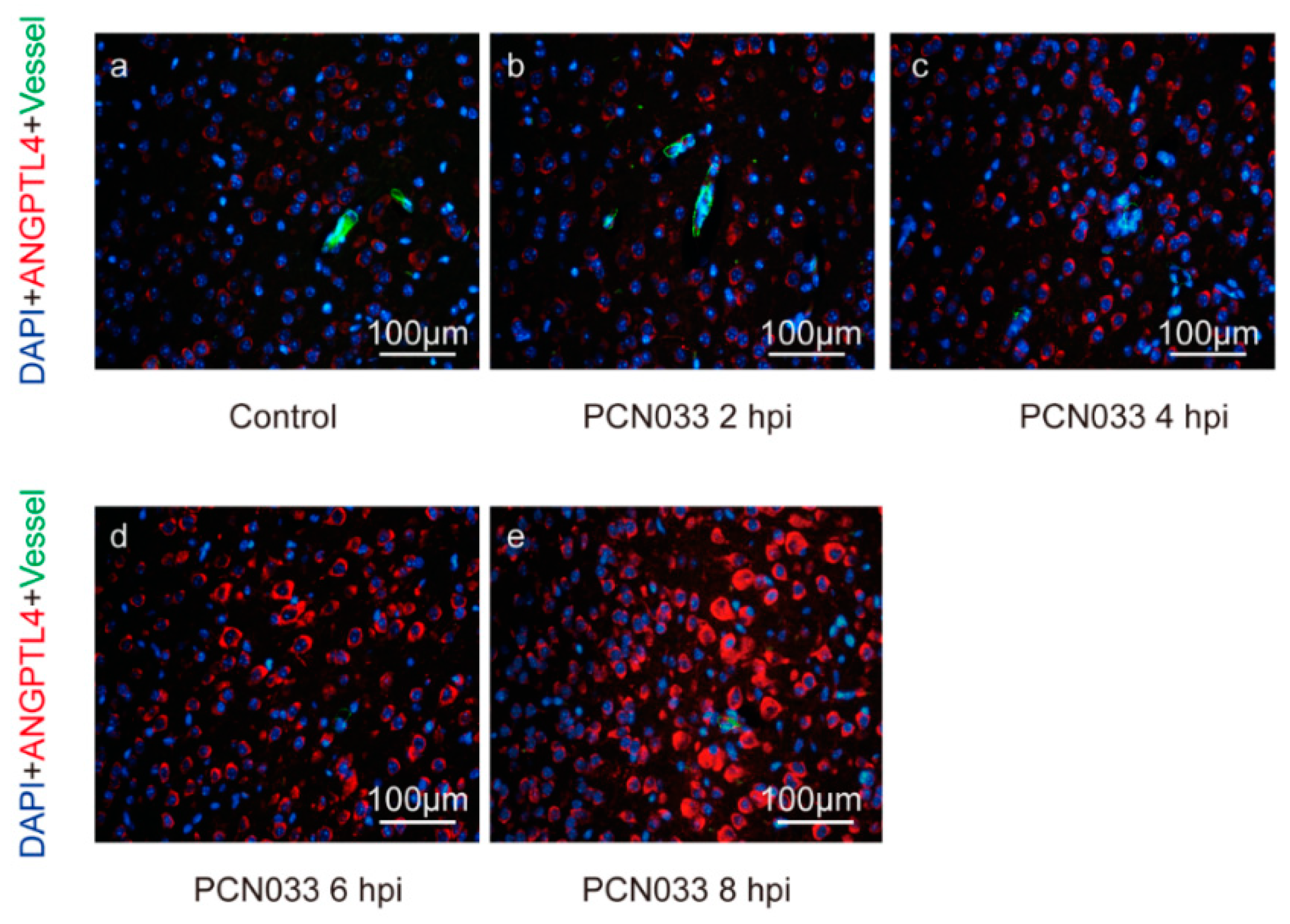
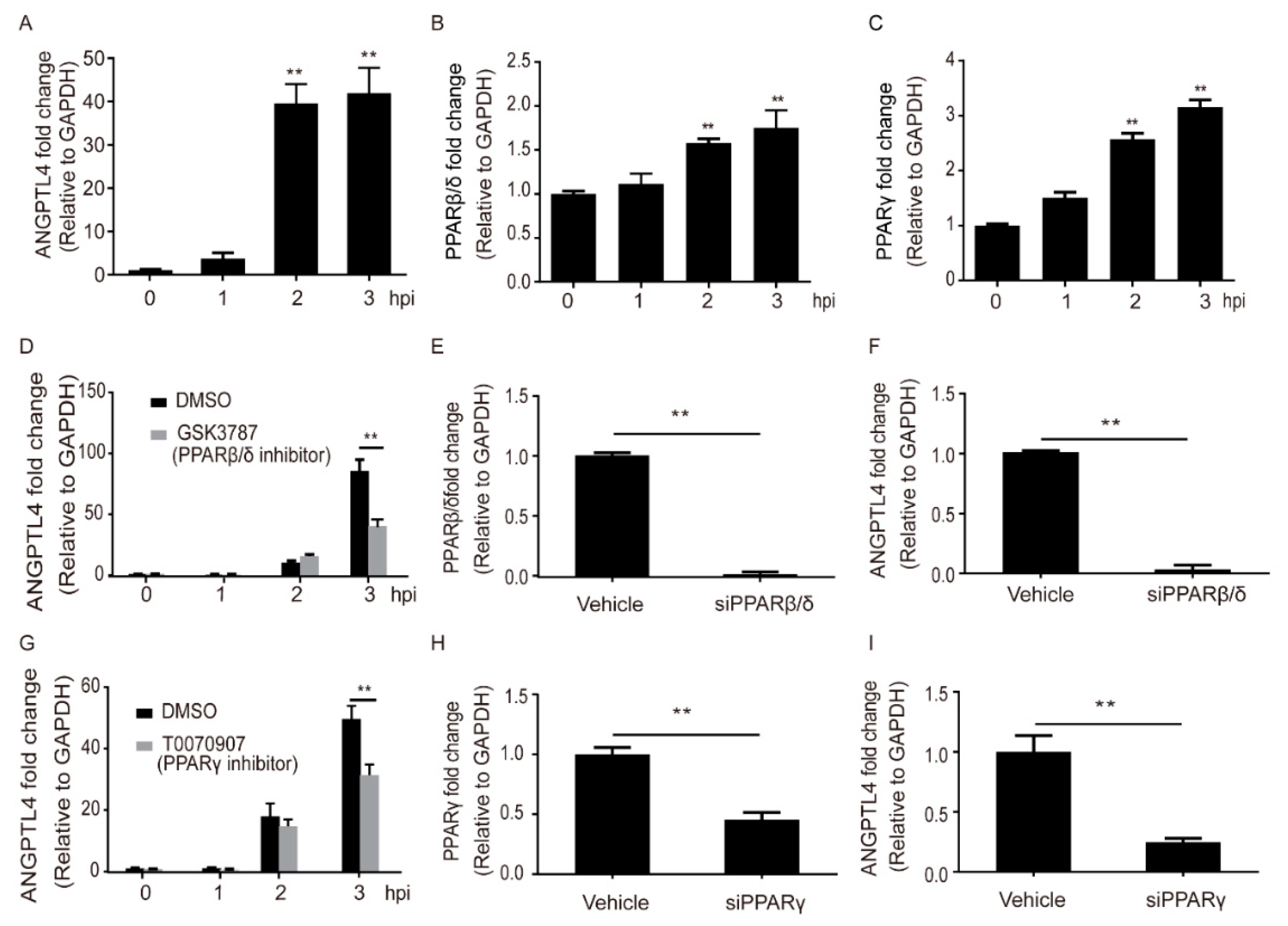
2.1. Meningitic E. coli Induced the Expression of ANGPTL4 through the Activation of PPAR Signaling
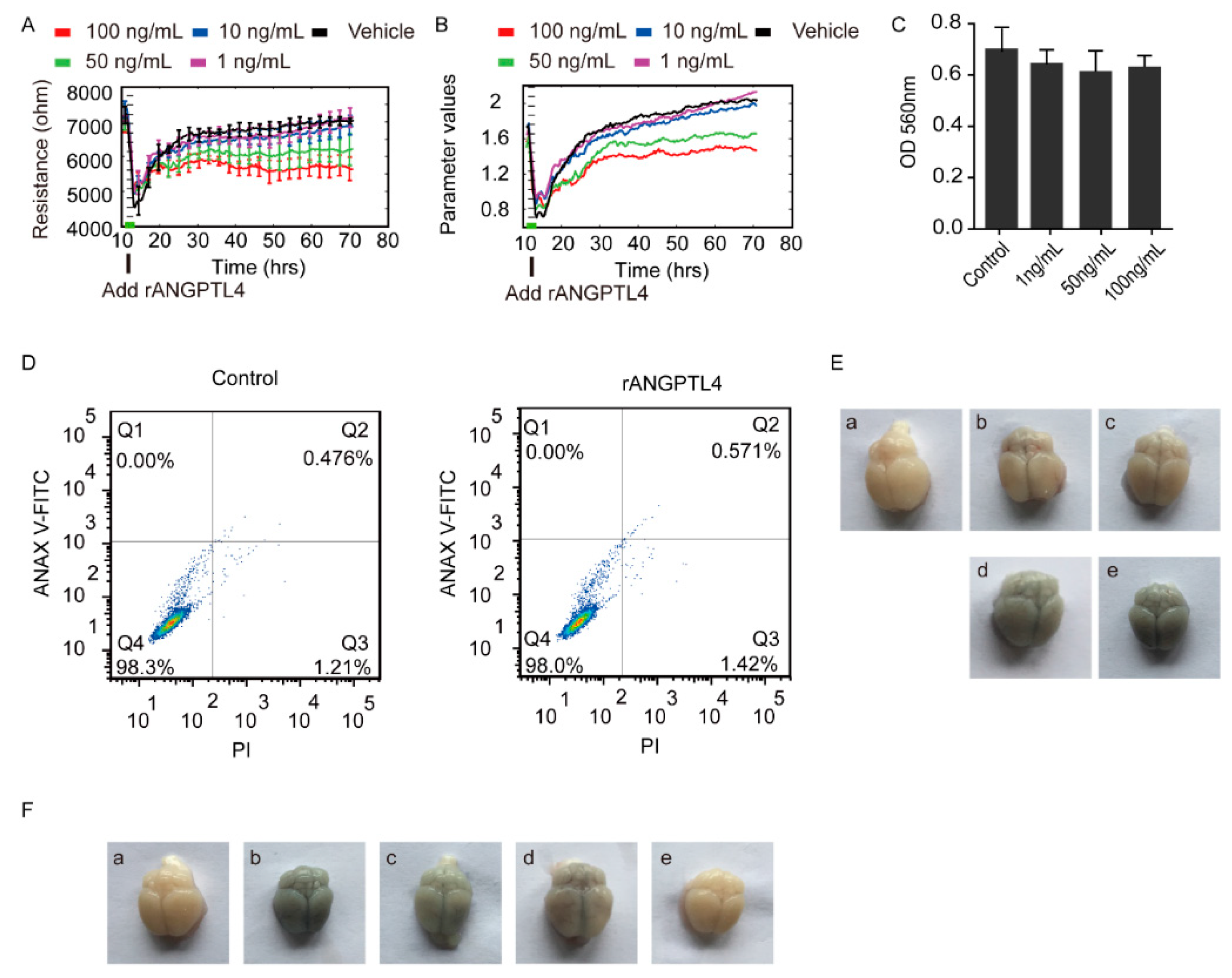
2.2. ANGPTL4 Aggravated the Disruption of BBB without Affecting Vitality of the hBMECs
2.3. ANGPTL4 Did Not Affect Expression of the Tight Junction Proteins as Well as Cytokines Production
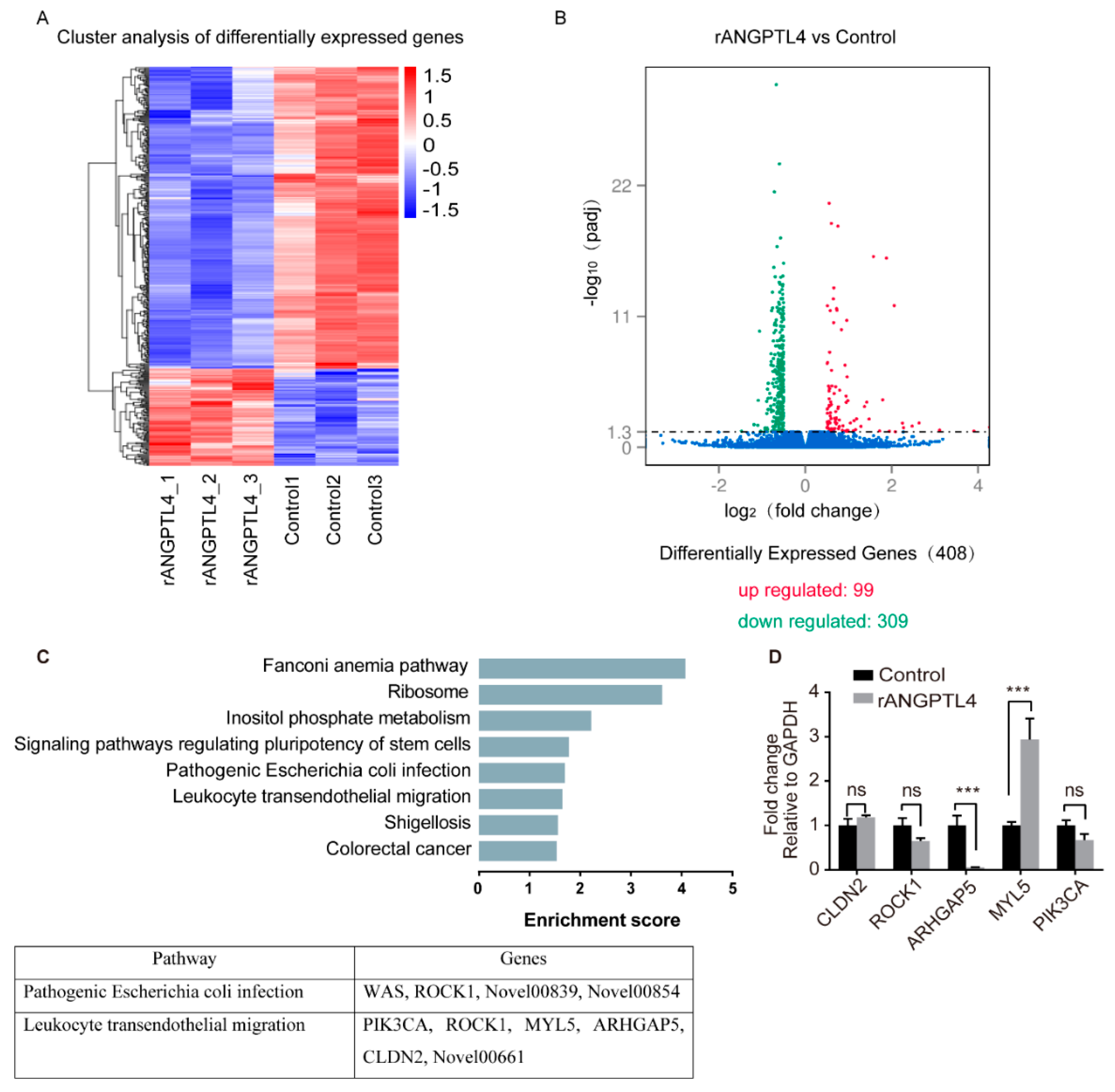
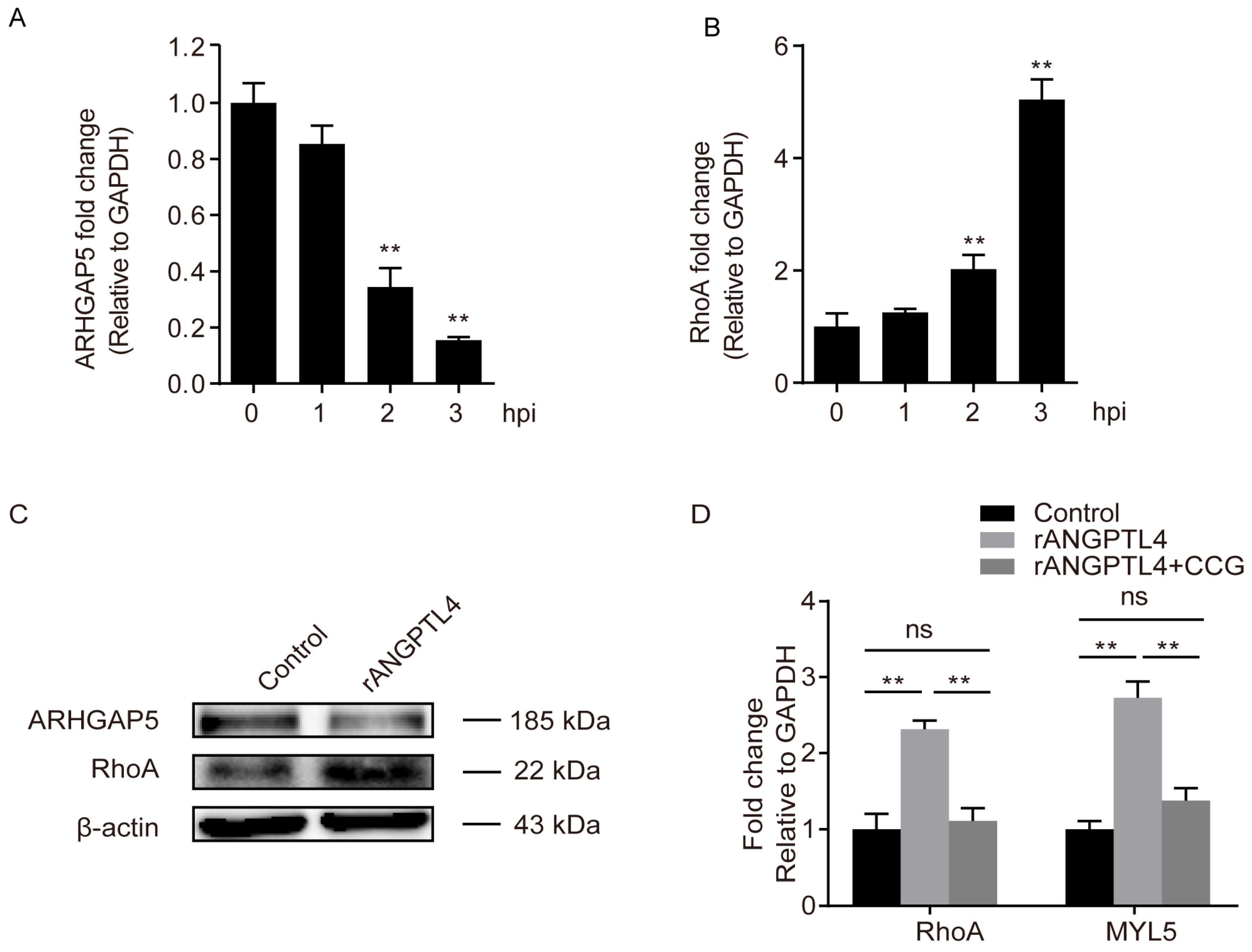
2.4. MYL5 and ARHGAP5 Were Differentially Expressed in Response to rANGPTL4 Treatment
2.5. MYL5 Contributed to ANGPTL4-Induced Barrier Disruption of the hBMECs
2.6. ARHGAP5/RhoA Signaling Facilitated the ANGPTL4 Regulation of MYL5 Expression in hBMECs
3. Discussion
4. Materials and Methods
4.1. Bacteria, Cells, and Culture Conditions
4.2. Protein Isolation and Western Blotting
4.3. siRNA/shRNA Knocking-Down and Gene Overexpression
4.4. Cell Infection and Treatments
4.5. MTT Assay
4.6. Electric Cell-Substrate Impedance Sensing (ECIS)
4.7. Flow Cytometry
4.8. Electrochemiluminescence (ECL) Assay
4.9. Immunofluorescence
4.10. RNA-Sequencing and Analyses
4.11. Animal Tests
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rodrigues, C.M.C.; Maiden, M.C.J. A world without bacterial meningitis: How genomic epidemiology can inform vaccination strategy. F1000Research 2018, 7, 401. [Google Scholar] [CrossRef]
- McGill, F.; Heyderman, R.S.; Panagiotou, S.; Tunkel, A.R.; Solomon, T. Acute bacterial meningitis in adults. Lancet 2016, 388, 3036–3047. [Google Scholar] [CrossRef]
- Kim, K.S. Human meningitis-associated Escherichia coli. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, A.; Thomsen, L.B.; Thomsen, M.S.; Lichota, J.; Fazakas, C.; Krizbai, I.; Moos, T. Transfection of brain capillary endothelial cells in primary culture with defined blood-brain barrier properties. Fluids Barriers CNS 2015, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Palmer, A.M. The role of the blood-CNS barrier in CNS disorders and their treatment. Neurobiol. Dis. 2010, 37, 3–12. [Google Scholar] [CrossRef]
- Edwards, V.L.; Wang, L.C.; Dawson, V.; Stein, D.C.; Song, W. Neisseria gonorrhoeae breaches the apical junction of polarized epithelial cells for transmigration by activating EGFR. Cell. Microbiol. 2013, 15, 1042–1057. [Google Scholar] [CrossRef]
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell 2009, 16, 209–221. [Google Scholar] [CrossRef]
- Ring, A.; Weiser, J.N.; Tuomanen, E.I. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 1998, 102, 347–360. [Google Scholar] [CrossRef]
- Schubert-Unkmeir, A.; Konrad, C.; Slanina, H.; Czapek, F.; Hebling, S.; Frosch, M. Neisseria meningitidis induces brain microvascular endothelial cell detachment from the matrix and cleavage of occludin: A role for MMP-8. PLoS Pathog. 2010, 6, e1000874. [Google Scholar] [CrossRef]
- Goluszko, P.; Popov, V.; Wen, J.; Jones, A.; Yallampalli, C. Group B streptococcus exploits lipid rafts and phosphoinositide 3-kinase/Akt signaling pathway to invade human endometrial cells. Am. J. Obstet. Gynecol. 2008, 199, 548-e1. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.K.; Prasadarao, N.V. Escherichia coli K1 invasion increases human brain microvascular endothelial cell monolayer permeability by disassembling vascular-endothelial cadherins at tight junctions. J. Infect. Dis. 2003, 188, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Patrick, D.; Betts, J.; Frey, E.A.; Prameya, R.; Dorovini-Zis, K.; Finlay, B.B. Haemophilus influenzae lipopolysaccharide disrupts confluent monolayers of bovine brain endothelial cells via a serum-dependent cytotoxic pathway. J. Infect. Dis. 1992, 165, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Kim, K.S. Treatment of bacterial meningitis: An update. Expert Opin. Pharm. 2012, 13, 2189–2206. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S. Mechanisms of microbial traversal of the blood–brain barrier. Nat. Rev. Microbiol. 2008, 6, 625–634. [Google Scholar] [CrossRef]
- Yang, R.; Liu, W.; Miao, L.; Yang, X.; Fu, J.; Dou, B.; Cai, A.; Zong, X.; Tan, C.; Chen, H.; et al. Induction of VEGFA and Snail-1 by meningitic Escherichia coli mediates disruption of the blood-brain barrier. Oncotarget 2016, 7, 63839–63855. [Google Scholar] [CrossRef][Green Version]
- Yang, R.C.; Qu, X.Y.; Xiao, S.Y.; Li, L.; Xu, B.J.; Fu, J.Y.; Lv, Y.J.; Amjad, N.; Tan, C.; Kim, K.S.; et al. Meningitic Escherichia coli-induced upregulation of PDGF-B and ICAM-1 aggravates blood-brain barrier disruption and neuroinflammatory response. J. Neuroinflamm. 2019, 16, 101. [Google Scholar] [CrossRef]
- Cushing, E.M.; Chi, X.; Sylvers, K.L.; Shetty, S.K.; Potthoff, M.J.; Davies, B.S.J. Angiopoietin-like 4 directs uptake of dietary fat away from adipose during fasting. Mol. Metab. 2017, 6, 809–818. [Google Scholar] [CrossRef]
- Qike, Y.; Akao, M.; Kubota, M.; Suda, T. Angiopoietin-like proteins: Potential new targets for metabolic syndrome therapy. Trends Mol. Med. 2005, 11, 473–479. [Google Scholar]
- Abu-Farha, M.; Cherian, P.; Qaddoumi, M.G.; AlKhairi, I.; Sriraman, D.; Alanbaei, M.; Abubaker, J. Increased plasma and adipose tissue levels of ANGPTL8/Betatrophin and ANGPTL4 in people with hypertension. Lipids Health Dis. 2018, 17, 35. [Google Scholar] [CrossRef]
- Bouleti, C.; Monnot, C.; Germain, S. ANGPTL4, a multifaceted protein at the cross-talk between metabolism and cardiovascular disorders. Int. J. Cardiol. 2018, 256, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, R.X.; Cao, X.L.; Huang, F.; Zhou, Y.J.; Chen, W.X. ANGPTL4 variants and their haplotypes are associated with serum lipid levels, the risk of coronary artery disease and ischemic stroke and atorvastatin cholesterol-lowering responses. Nutr. Metab. (Lond.) 2018, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Masuko, K. Angiopoietin-like 4: A molecular link between insulin resistance and rheumatoid arthritis. J. Orthop. Res. 2017, 35, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Phua, T.; Sng, M.K.; Tan, E.H.; Chee, D.S.; Li, Y.; Wee, J.W.; Teo, Z.; Chan, J.S.; Lim, M.M.; Tan, C.K.; et al. Angiopoietin-like 4 mediates colonic inflammation by regulating chemokine transcript stability via tristetraprolin. Sci. Rep. 2017, 7, 44351. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lu, P.; Chen, W.; Lu, L.; Zheng, Z. ANGPTL-4 induces diabetic retinal inflammation by activating Profilin-1. Exp. Eye Res. 2018, 166, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Huang, Y.; Xie, H.; Mei, X. Buyang Huanwu Tang improves denervation-dependent muscle atrophy by increasing ANGPTL4, and increases NF-kappaB and MURF1 levels. Mol. Med. Rep. 2018, 17, 3674–3680. [Google Scholar] [CrossRef] [PubMed]
- Conway, G.D.; Buzza, M.S.; Martin, E.W.; Duru, N.; Johnson, T.A.; Peroutka, R.J.; Pawar, N.R.; Antalis, T.M. Correction to: PRSS21/testisin inhibits ovarian tumor metastasis and antagonizes proangiogenic angiopoietins ANG2 and ANGPTL4. J. Mol. Med. 2019, 97, 1375. [Google Scholar] [CrossRef]
- Fekir, K.; Dubois-Pot-Schneider, H.; Desert, R.; Daniel, Y.; Glaise, D.; Rauch, C.; Morel, F.; Fromenty, B.; Musso, O.; Cabillic, F.; et al. Retrodifferentiation of human tumor hepatocytes to stem cells leads to metabolic reprogramming and chemoresistance. Cancer Res. 2019, 79, 1869–1883. [Google Scholar] [CrossRef]
- Hui, B.; Ji, H.; Xu, Y.; Wang, J.; Ma, Z.; Zhang, C.; Wang, K.; Zhou, Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019, 10, 207. [Google Scholar] [CrossRef]
- Yang, R.; Huang, F.; Fu, J.; Dou, B.; Xu, B.; Miao, L.; Liu, W.; Yang, X.; Tan, C.; Chen, H.; et al. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016, 6, 38903. [Google Scholar] [CrossRef]
- Heraud, C.; Pinault, M.; Lagree, V.; Moreau, V. p190RhoGAPs, the ARHGAP35- and ARHGAP5-encoded proteins, in health and disease. Cells 2019, 8, 351. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, S.Y.; Ji, F.Y.; Zhao, Y.F.; Zhong, Y.; Lv, X.J.; Wu, X.L.; Qian, G.S. Role of ANGPTL4 in vascular permeability and inflammation. Inflamm. Res. 2014, 63, 13–22. [Google Scholar] [CrossRef] [PubMed]
- La Paglia, L.; Listi, A.; Caruso, S.; Amodeo, V.; Passiglia, F.; Bazan, V.; Fanale, D. Potential role of ANGPTL4 in the cross talk between metabolism and cancer through PPAR signaling pathway. PPAR Res. 2017, 2017, 8187235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, X.; Chu, X.; Yu, X.; Zhao, Y.J.N.L. Protective effects of angiopoietin-like 4 on the blood-brain barrier in acute ischemic stroke treated with thrombolysis in mice. Neurosci. Lett. 2017, 645, 113–120. [Google Scholar] [CrossRef]
- Al-Obaidi, M.M.J.; Desa, M.N.M. Mechanisms of blood brain barrier disruption by different types of bacteria, and bacterial-host interactions facilitate the bacterial pathogen invading the brain. Cell. Mol. Neurobiol. 2018, 38, 1349–1368. [Google Scholar] [CrossRef]
- Kim, B.J.; Bee, O.B.; Mcdonagh, M.A.; Stebbins, M.J.; Palecek, S.P.; Doran, K.S.; Shusta, E.V.J.M. Modeling Group B Streptococcal and blood-brain barrier interaction by using induced pluripotent stem cell-derived brain endothelial cells. Msphere 2017, 2, 00398-17. [Google Scholar] [CrossRef]
- Kim, B.; Shusta, E.; Doran, K. Past and current perspectives in modeling bacteria and blood–brain barrier interactions. Front. Microbiol. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Deng, L.; Spencer, B.L.; Holmes, J.A.; Mu, R.; Rego, S.; Weston, T.A.; Hu, Y.; Sanches, G.F.; Yoon, S.; Park, N.J.P.P. The Group B Streptococcal surface antigen I/II protein, BspC, interacts with host vimentin to promote adherence to brain endothelium and inflammation during the pathogenesis of meningitis. PLoS Pathog. 2019, 15, e1007848. [Google Scholar] [CrossRef]
- Bonney, S.; Seitz, S.; Ryan, C.A.; Jones, K.L.; Clarke, P.; Tyler, K.L.; Siegenthaler, J.A. Gamma interferon alters junctional integrity via Rho kinase, resulting in blood-brain barrier leakage in experimental viral encephalitis. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- England, J.; Loughna, S. Heavy and light roles: Myosin in the morphogenesis of the heart. Cell. Mol. Life Sci. 2013, 70, 1221–1239. [Google Scholar] [CrossRef]
- Wang, S.; Chen, C.; Su, K.; Zha, D.; Liang, W.; Hillebrands, J.L.; Goor, H.; Ding, G. Angiotensin II induces reorganization of the actin cytoskeleton and myosin light-chain pshosphorylation in podocytes through rho/ROCK-signaling pathway. Ren. Fail. 2016, 38, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, S.T.; Feng, Y.L.; Wan, T.; Gu, H.F.; Xu, J.; Yuan, L.J.; Zhou, Y.; Yu, X.J.; Huang, L.; et al. The bidirectional regulation between MYL5 and HIF-1alpha promotes cervical carcinoma metastasis. Theranostics 2017, 7, 3768–3780. [Google Scholar] [CrossRef] [PubMed]
- Lum, H.; Malik, A.B. Regulation of vascular endothelial barrier function. Am. J. Physiol. 1994, 267, L223–L241. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.H.; Ditzler, C.E.; Germain, A.; Irwin, P.M.; Vogt, E.T.; Yang, S.; Wu, X.; Shuster, C.B. The ultrastructural organization of actin and myosin II filaments in the contractile ring: New support for an old model of cytokinesis. Mol. Biol. Cell 2017, 28, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.; Cho, E.H.; Park, J.G.; Kim, J.Y.; Alfajaro, M.M.; Baek, Y.B.; Kim, D.S.; Kang, M.I.; Park, S.I.; Cho, K.O. Rotavirus-induced early activation of the RhoA/ROCK/MLC signaling pathway mediates the disruption of tight junctions in polarized MDCK cells. Sci. Rep. 2018, 8, 13931. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Li, L.; Yang, X.; Yang, R.; Amjad, N.; Liu, L.; Tan, C.; Chen, H.; Wang, X. Transactivated epidermal growth factor receptor recruitment of alpha-actinin-4 from F-actin contributes to invasion of brain microvascular endothelial cells by meningitic Escherichia coli. Front. Cell Infect. Microbiol. 2018, 8, 448. [Google Scholar] [CrossRef]
- Tan, C.; Xu, Z.; Zheng, H.; Liu, W.; Tang, X.; Shou, J.; Wu, B.; Wang, S.; Zhao, G.P.; Chen, H. Genome sequence of a porcine extraintestinal pathogenic Escherichia coli strain. J. Bacteriol. 2011, 193, 5038. [Google Scholar] [CrossRef]
- Liu, C.; Zheng, H.; Yang, M.; Xu, Z.; Wang, X.; Wei, L.; Tang, B.; Liu, F.; Zhang, Y.; Ding, Y.; et al. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genom. 2015, 16, 717. [Google Scholar] [CrossRef]
- Stins, M.F.; Gilles, F.; Kim, K.S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J. Neuroimmunol. 1997, 76, 81–90. [Google Scholar] [CrossRef]
- Szulcek, R.; Bogaard, H.J.; van Nieuw Amerongen, G.P. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J. Vis. Exp. 2014. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Li, J.; Huo, D.; Peng, Z.; Yang, R.; Fu, J.; Xu, B.; Yang, B.; Chen, H.; Wang, X. Meningitic Escherichia coli Induction of ANGPTL4 in Brain Microvascular Endothelial Cells Contributes to Blood–Brain Barrier Disruption via ARHGAP5/RhoA/MYL5 Signaling Cascade. Pathogens 2019, 8, 254. https://doi.org/10.3390/pathogens8040254
Liu L, Li J, Huo D, Peng Z, Yang R, Fu J, Xu B, Yang B, Chen H, Wang X. Meningitic Escherichia coli Induction of ANGPTL4 in Brain Microvascular Endothelial Cells Contributes to Blood–Brain Barrier Disruption via ARHGAP5/RhoA/MYL5 Signaling Cascade. Pathogens. 2019; 8(4):254. https://doi.org/10.3390/pathogens8040254
Chicago/Turabian StyleLiu, Lu, Jixuan Li, Dong Huo, Zhong Peng, Ruicheng Yang, Jiyang Fu, Bojie Xu, Bo Yang, Huanchun Chen, and Xiangru Wang. 2019. "Meningitic Escherichia coli Induction of ANGPTL4 in Brain Microvascular Endothelial Cells Contributes to Blood–Brain Barrier Disruption via ARHGAP5/RhoA/MYL5 Signaling Cascade" Pathogens 8, no. 4: 254. https://doi.org/10.3390/pathogens8040254
APA StyleLiu, L., Li, J., Huo, D., Peng, Z., Yang, R., Fu, J., Xu, B., Yang, B., Chen, H., & Wang, X. (2019). Meningitic Escherichia coli Induction of ANGPTL4 in Brain Microvascular Endothelial Cells Contributes to Blood–Brain Barrier Disruption via ARHGAP5/RhoA/MYL5 Signaling Cascade. Pathogens, 8(4), 254. https://doi.org/10.3390/pathogens8040254





