Differential Cell Lysis Among Periodontal Strains of JP2 and Non-JP2 Genotype of Aggregatibacter actinomycetemcomitans Serotype B Is Not Reflected in Dissimilar Expression and Production of Leukotoxin
Abstract
1. Introduction
2. Results
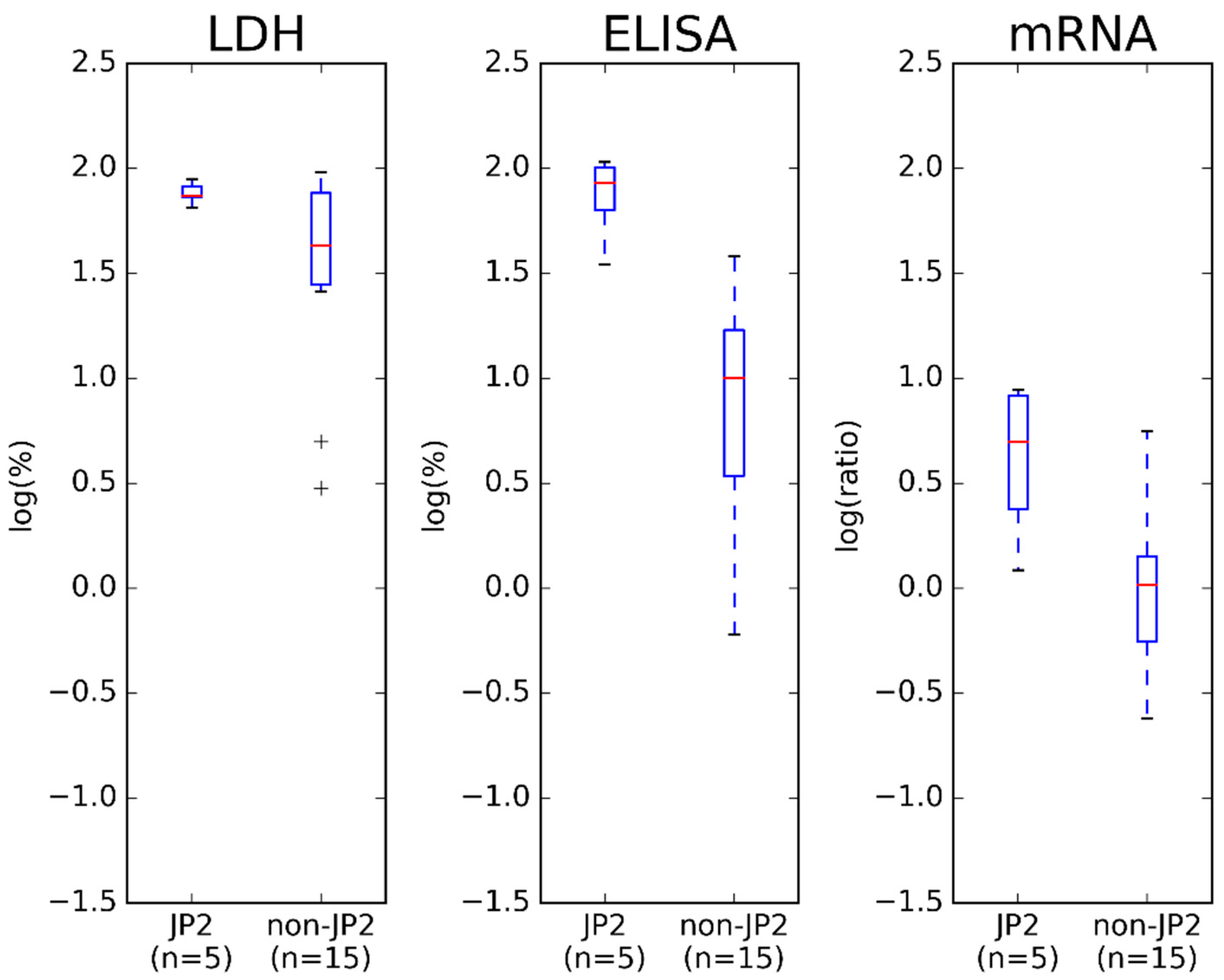
2.1. LtxA Expression and Production of the 20 Ghanaian Strains by Western Blotting, ELISA and mRNA Expression Assay
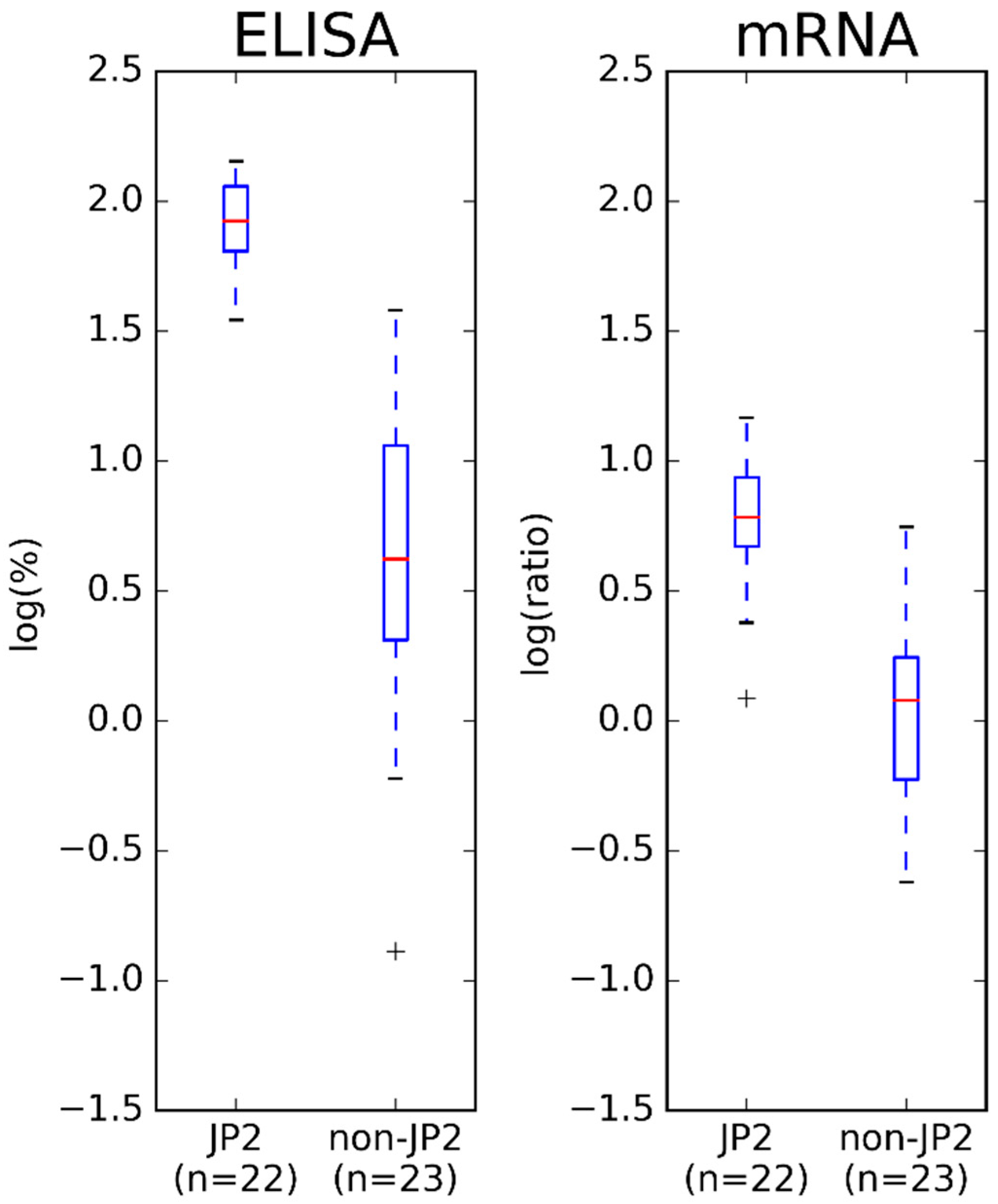
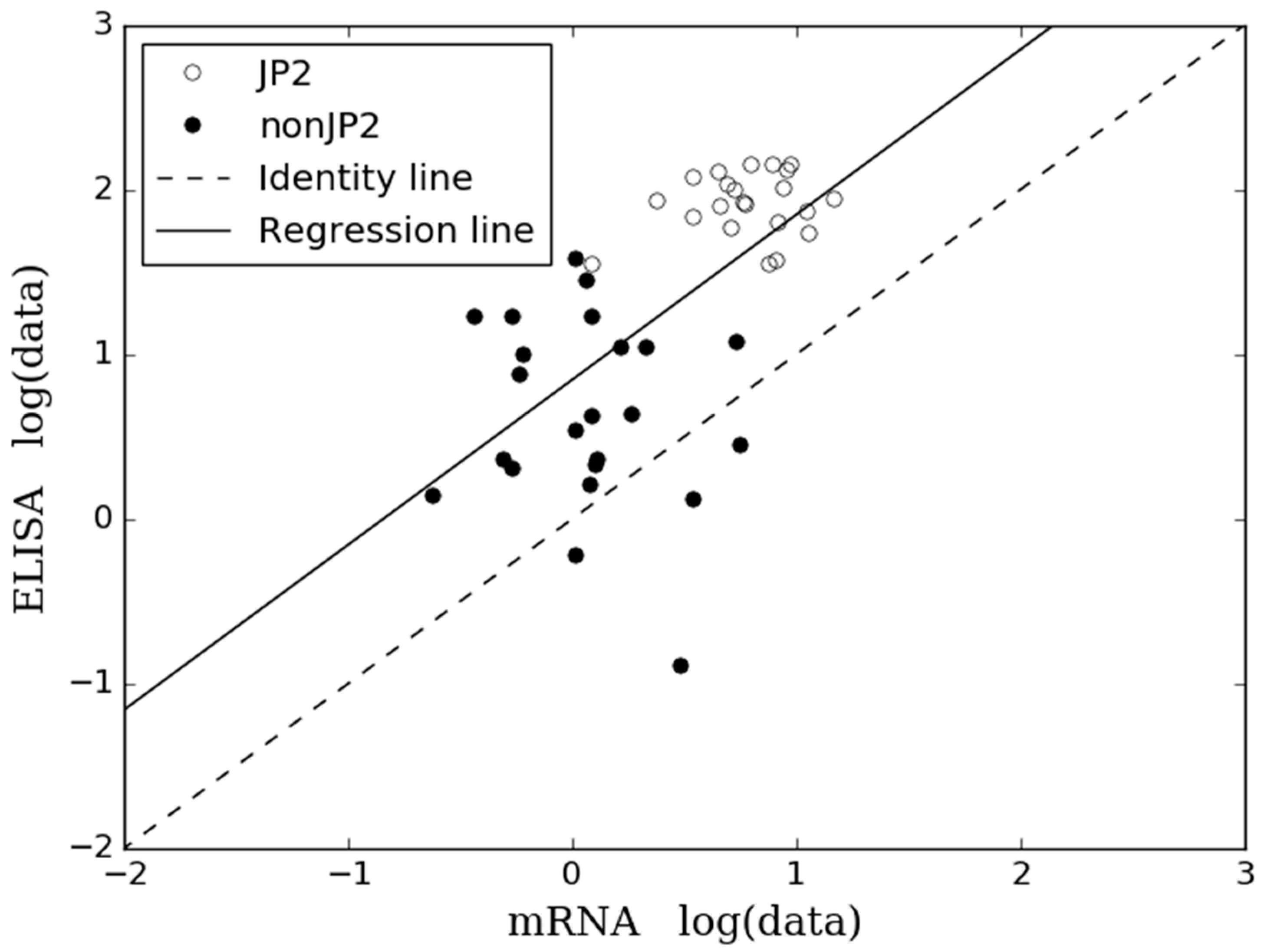
2.2. Comparison of Results Obtained by ELISA and mRNA Expression Assay
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cell Lysis Assay
4.2. LtxA Isolation from the Cell Pellet and the Growth Supernatant
4.3. Western Blotting
4.4. ELISA for Quantification of LtxA in Growth Supernatants
4.5. mRNA Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Slots, J.; Ting, M. Actinobacillus actinomycetemcomitans and Porphyromonas Gingivalis in Human Periodontal Disease: Occurrence and Treatment. Periodontology 2000 1999, 20, 82–121. [Google Scholar] [CrossRef] [PubMed]
- Zambon, J.J. Actinobacillus actinomycetemcomitans in Human Periodontal Disease. J. Clin. Periodontol. 1985, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Fives-Taylor, P.M.; Meyer, D.H.; Mintz, K.P.; Brissette, C. Virulence Factors of Actinobacillus actinomycetemcomitans. Periodontology 2000 1999, 20, 136–167. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Ward, J.M.; Ready, D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: A Triple A* Periodontopathogen? Periodontology 2000 2010, 54, 78–105. [Google Scholar] [CrossRef]
- Norskov-Lauritsen, N. Classification, Identification, and Clinical Significance of Haemophilus and Aggregatibacter Species with Host Specificity for Humans. Clin. Microbiol. Rev. 2014, 27, 214–240. [Google Scholar] [CrossRef]
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal Diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef]
- Lally, E.T.; Kieba, I.R.; Demuth, D.R.; Rosenbloom, J.; Golub, E.E.; Taichman, N.S.; Gibson, C.W. Identification and Expression of the Actinobacillus actinomycetemcomitans Leukotoxin Gene. Biochem. Biophys. Res. Commun. 1989, 159, 256–262. [Google Scholar] [CrossRef]
- Korostoff, J.; Wang, J.F.; Kieba, I.; Miller, M.; Shenker, B.J.; Lally, E.T. Actinobacillus actinomycetemcomitans Leukotoxin Induces Apoptosis in HL-60 Cells. Infect. Immun. 1998, 66, 4474–4483. [Google Scholar]
- Kachlany, S.C.; Fine, D.H.; Figurski, D.H. Purification of Secreted Leukotoxin (LtxA) from Actinobacillus actinomycetemcomitans. Protein Expr. Purif. 2002, 25, 465–471. [Google Scholar] [CrossRef]
- Johansson, A.; Claesson, R.; Hänström, L.; Kalfas, S. Serum-Mediated Release of Leukotoxin from the Cell Surface of the Periodontal Pathogen Actinobacillus actinomycetemcomitans. Eur. J. Oral Sci. 2003, 111, 209–215. [Google Scholar] [CrossRef]
- Balashova, N.V.; Shah, C.; Patel, J.K.; Megalla, S.; Kachlany, S.C. Aggregatibacter actinomycetemcomitans LtxC Is Required for Leukotoxin Activity and Initial Interaction between Toxin and Host Cells. Gene 2009, 443, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Balashova, N.; Dhingra, A.; Boesze-Battaglia, K.; Lally, E.T. Aggregatibacter actinomycetemcomitans Leukotoxin Induces Cytosol Acidification in LFA-1 Expressing Immune Cells. Mol. Oral Microbiol. 2016, 31, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A. Aggregatibacter actinomycetemcomitans Leukotoxin: A Powerful Tool with Capacity to Cause Imbalance in the Host Inflammatory Response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef] [PubMed]
- Reinholdt, J.; Poulsen, K.; Brinkmann, C.R.; Hoffmann, S.V.; Stapulionis, R.; Enghild, J.J.; Jensen, U.B.; Boesen, T.; Vorup-Jensen, T. Monodisperse and LPS-Free Aggregatibacter actinomycetemcomitans Leukotoxin: Interactions with Human Β2 Integrins and Erythrocytes. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 546–558. [Google Scholar] [CrossRef]
- Brogan, J.M.; Lally, E.T.; Poulsen, K.; Kilian, M.; Demuth, D.R. Regulation of Actinobacillus actinomycetemcomitans Leukotoxin Expression: Analysis of the Promoter Regions of Leukotoxic and Minimally Leukotoxic Strains. Infect. Immun. 1994, 62, 501–508. [Google Scholar]
- Haubek, D.; Ennibi, O.-K.; Poulsen, K.; Væth, M.; Poulsen, S.; Kilian, M. Risk of Aggressive Periodontitis in Adolescent Carriers of the JP2 Clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A Prospective Longitudinal Cohort Study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef]
- Åberg, C.H.; Kwamin, F.; Claesson, R.; Johansson, A.; Haubek, D. Presence of JP2 and Non-JP2 Genotypes of Aggregatibacter actinomycetemcomitans and Attachment Loss in Adolescents in Ghana. J. Periodontol. 2012, 83, 1520–1528. [Google Scholar] [CrossRef]
- Spitznagel, J.; Kraig, E.; Kolodrubetz, D. Regulation of Leukotoxin in Leukotoxic and Nonleukotoxic Strains of Actinobacillus actinomycetemcomitans. Infect. Immun. 1991, 59, 1394–1401. [Google Scholar]
- Diaz, R.; Al Ghofaily, L.; Patel, J.; Balashova, N.V.; Freitas, A.C.; Labib, I.; Kachlany, S.C. Characterization of Leukotoxin from a Clinical Strain of Actinobacillus actinomycetemcomitans. Microb. Pathog. 2006, 40, 48–55. [Google Scholar] [CrossRef]
- Höglund Åberg, C.; Haubek, D.; Kwamin, F.; Johansson, A.; Claesson, R. Leukotoxic Activity of Aggregatibacter actinomycetemcomitans and Periodontal Attachment Loss. PLoS ONE 2014, 9, e104095. [Google Scholar] [CrossRef]
- Sampathkumar, V.; Velusamy, S.K.; Godboley, D.; Fine, D.H. Increased Leukotoxin Production: Characterization of 100 Base Pairs within the 530 Base Pair Leukotoxin Promoter Region of Aggregatibacter actinomycetemcomitans. Sci. Rep. 2017, 7, 1887. [Google Scholar] [CrossRef] [PubMed]
- Balashova, N.V.; Diaz, R.; Balashov, S.V.; Crosby, J.A.; Kachlany, S.C. Regulation of Aggregatibacter (Actinobacillus) actinomycetemcomitans Leukotoxin Secretion by Iron. J. Bacteriol. 2006, 188, 8658–8661. [Google Scholar] [CrossRef] [PubMed]
- Kachlany, S.C.; Fine, D.H.; Figurski, D.H. Secretion of RTX Leukotoxin by Actinobacillus actinomycetemcomitans. Infect. Immun. 2000, 68, 6094–6100. [Google Scholar] [CrossRef] [PubMed]
- Fukui, K.; Tada, T.; Tanimoto, I.; Inoue, T.; Ohta, H.; Ohashi, T. Fermentable-Sugar-Level-Dependent Regulation of Leukotoxin Synthesis in a Variably Toxic Strain of Actinobacillus actinomycetemcomitans. Microbiology 2001, 147, 2749–2756. [Google Scholar] [CrossRef][Green Version]
- Longo, P.L.; Nunes, A.C.R.; Umeda, J.E.; Mayer, M.P.A. Gene Expression and Phenotypic Traits of Aggregatibacter actinomycetemcomitans in Response to Environmental Changes. J. Periodontal Res. 2013, 48, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Nowotny, A.; Behling, U.H.; Hammond, B.; Lai, C.H.; Listgarten, M.; Pham, P.H.; Sanavi, F. Release of Toxic Microvesicles by Actinobacillus actinomycetemcomitans. Infect. Immun. 1982, 37, 151–154. [Google Scholar] [PubMed]
- Berthold, P.; Forti, D.; Kieba, I.R.; Rosenbloom, J.; Taichman, N.S.; Lally, E.T. Electron Immunocytochemical Localization of Actinobacillus actinomycetemcomitans Leukotoxin. Oral Microbiol. Immunol. 1992, 7, 24–27. [Google Scholar] [CrossRef]
- Nice, J.; Balashova, N.; Kachlany, S.; Koufos, E.; Krueger, E.; Lally, E.; Brown, A. Aggregatibacter actinomycetemcomitans Leukotoxin Is Delivered to Host Cells in an LFA-1-Indepdendent Manner When Associated with Outer Membrane Vesicles. Toxins 2018, 10, 414. [Google Scholar] [CrossRef]
- Kato, S.; Kowashi, Y.; Demuth, D.R. Outer Membrane-like Vesicles Secreted by Actinobacillus actinomycetemcomitans Are Enriched in Leukotoxin. Microb. Pathog. 2002, 32, 1–13. [Google Scholar] [CrossRef]
- Johansson, A.; Claesson, R.; Höglund Åberg, C.; Haubek, D.; Oscarsson, J. The CagE Gene Sequence as a Diagnostic Marker to Identify JP2 and Non-JP2 Highly Leukotoxic Aggregatibacter actinomycetemcomitans Serotype b Strains. J. Periodontal Res. 2017, 52, 903–912. [Google Scholar] [CrossRef]
- Johansson, A.; Hänström, L.; Kalfas, S. Inhibition of Actinobacillus actinomycetemcomitans Leukotoxicity by Bacteria from the Subgingival Flora. Oral Microbiol. Immunol. 2000, 15, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, S.K.; Sampathkumar, V.; Godboley, D.; Fine, D.H. Survival of an Aggregatibacter actinomycetemcomitans Quorum Sensing Lux S Mutant in the Mouths of Rhesus Monkeys: Insights into Ecological Adaptation. Mol. Oral Microbiol. 2017, 32, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Ludbrook, J. Statistical Techniques for Comparing Measurers and Methods of Measurement: A Critical Review. Clin. Exp. Pharmacol. Physiol. 2002, 29, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Measurement in Medicine: The Analysis of Method Comparison Studies. J. R. Stat. Soc. Ser. D (Stat.) 1983, 32, 307–317. [Google Scholar] [CrossRef]
- Haubek, D.; Poulsen, K.; Westergaard, J.; Dahlèn, G.; Kilian, M. Highly Toxic Clone of Actinobacillus actinomycetemcomitans in Geographically Widespread Cases of Juvenile Periodontitis in Adolescents of African Origin. J. Clin. Microbiol. 1996, 34, 1576–1578. [Google Scholar] [PubMed]
- Haubek, D.; Poulsen, K.; Kilian, M. Microevolution and Patterns of Dissemination of the JP2 Clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 2007, 75, 3080–3088. [Google Scholar] [CrossRef]
- Nørskov-Lauritsen, N.; Søndergaard, A.; Lund, M. TEM-1-Encoding Small Plasmids Impose Dissimilar Fitness Costs on Haemophilus influenzae and Haemophilus parainfluenzae. Microbiology 2015, 161, 2310–2315. [Google Scholar] [CrossRef]
- SciPy—SciPy v1.3.0 Reference Guide. Available online: https://docs.scipy.org/doc/scipy-1.3.0/reference/ (accessed on 8 July 2019).


| Bacterial Strains | Genotype | Origin | Leukotoxicity * |
|---|---|---|---|
| 1 G | non-JP2 | Ghana | Average |
| 153 G | non-JP2 | Ghana | Average |
| 212 G | non-JP2 | Ghana | Average |
| 217 G | non-JP2 | Ghana | Average |
| 640 G | non-JP2 | Ghana | Average |
| 443 G | non-JP2 | Ghana | High |
| 486 G | non-JP2 | Ghana | High |
| 575 G | non-JP2 | Ghana | High |
| 605 G | non-JP2 | Ghana | High |
| 638 G | non-JP2 | Ghana | High |
| 369 G | non-JP2 | Ghana | Low |
| 467 G | non-JP2 | Ghana | Low |
| 493 G | non-JP2 | Ghana | Low |
| 708 G | non-JP2 | Ghana | Low |
| 744 G | non-JP2 | Ghana | Low |
| 331 M | non-JP2 | Morocco | |
| 394 M | non-JP2 | Morocco | |
| 416 M | non-JP2 | Morocco | |
| HK1605 | non-JP2 | USA | |
| HK908 | non-JP2 | Porto-Brazil 1 | |
| HK911 | non-JP2 | Holland | |
| HK912 | non-JP2 | Holland | |
| HK975 (Y4) | non-JP2 | USA | |
| 437 G | JP2 | Ghana | High |
| 488 G | JP2 | Ghana | High |
| 524 G | JP2 | Ghana | High |
| 654 G | JP2 | Ghana | High |
| 666 G | JP2 | Ghana | High |
| HK1199 | JP2 | USA | |
| HK1507 | JP2 | Cape Verde Isands-Sweden 2 | |
| HK1519 | JP2 | Cape Verde Islands-Sweden 2 | |
| HK1547 | JP2 | USA | |
| HK1609 | JP2 | USA | |
| HK1615 | JP2 | USA | |
| HK1626 | JP2 | Tel Aviv-Switzerland 3 | |
| HK1630 | JP2 | Algeria-Denmark 4 | |
| HK1631 | JP2 | Morocco-Denmark 5 | |
| HK1651 | JP2 | Ghana-Denmark 6 | |
| HK1659 | JP2 | Morocco-Denmark 5 | |
| HK1702 | JP2 | Brazil | |
| HK1707 | JP2 | Brazil | |
| HK1990 | JP2 | Portugal-Holland | |
| HK2000 | JP2 | Morocco | |
| HK2017 | JP2 | Tel Aviv | |
| HK921 (JP2) | JP2 | USA |
| Gene | Primer F (5’ → 3’) | Primer R (3’ → 5’) | Probe (5’ → 3’) |
|---|---|---|---|
| pgi | TAACCATGCACTTCGTGTCTAAC | CACAAGGGTGGTTTCCGGATT | TGGACGGCACGCACATTGCGGAAAC |
| adk | GATATGTTACGTTCCGCGATCA | GCACTAATTGACCGGCATCCA | AGCTGGCACCGAGTTAGGCAAACAAGC |
| ltxA | CTGTCGCAGGGTTAATTGCCT | GATCAAATTGTTTCGCAATACCTAG | TGTGGTCAGCTTGGCAATCAGCCCTTTG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jensen, A.B.; Lund, M.; Nørskov-Lauritsen, N.; Johansson, A.; Claesson, R.; Reinholdt, J.; Haubek, D. Differential Cell Lysis Among Periodontal Strains of JP2 and Non-JP2 Genotype of Aggregatibacter actinomycetemcomitans Serotype B Is Not Reflected in Dissimilar Expression and Production of Leukotoxin. Pathogens 2019, 8, 211. https://doi.org/10.3390/pathogens8040211
Jensen AB, Lund M, Nørskov-Lauritsen N, Johansson A, Claesson R, Reinholdt J, Haubek D. Differential Cell Lysis Among Periodontal Strains of JP2 and Non-JP2 Genotype of Aggregatibacter actinomycetemcomitans Serotype B Is Not Reflected in Dissimilar Expression and Production of Leukotoxin. Pathogens. 2019; 8(4):211. https://doi.org/10.3390/pathogens8040211
Chicago/Turabian StyleJensen, Anne Birkeholm, Marianne Lund, Niels Nørskov-Lauritsen, Anders Johansson, Rolf Claesson, Jesper Reinholdt, and Dorte Haubek. 2019. "Differential Cell Lysis Among Periodontal Strains of JP2 and Non-JP2 Genotype of Aggregatibacter actinomycetemcomitans Serotype B Is Not Reflected in Dissimilar Expression and Production of Leukotoxin" Pathogens 8, no. 4: 211. https://doi.org/10.3390/pathogens8040211
APA StyleJensen, A. B., Lund, M., Nørskov-Lauritsen, N., Johansson, A., Claesson, R., Reinholdt, J., & Haubek, D. (2019). Differential Cell Lysis Among Periodontal Strains of JP2 and Non-JP2 Genotype of Aggregatibacter actinomycetemcomitans Serotype B Is Not Reflected in Dissimilar Expression and Production of Leukotoxin. Pathogens, 8(4), 211. https://doi.org/10.3390/pathogens8040211






