Abstract
Control of Salmonella spp. in food production chains is very important to ensure safe foods and minimize the risks of foodborne disease occurrence. This study aimed to identify the prevalence and main contamination sources of Salmonella spp. in a pig production chain in southern Brazil. Six lots of piglets produced at different farms were tracked until their slaughter, and samples were subjected to Salmonella spp. detection. The obtained isolates were serotyped, subjected to antimicrobial resistance testing, and pulsed field gel electrophoresis (PFGE). Salmonella spp. was detected in 160 (10.2%) samples, and not detected in pig carcasses after final washing or chilling. Among the 210 Salmonella spp. isolates, S. Typhimurium was the most prevalent (n = 101) and resistant to at least one antimicrobial. High resistance rates were detected against tetracycline (83.8%), chloramphenicol (54.3%), and trimethoprim-sulfamethoxazole (33.3%). The isolates that were non-susceptible to three or more classes of antimicrobials (n = 60) were considered multidrug-resistant (MDR), and isolates resistant to up to six of the tested antimicrobials were found. PFGE allowed the identification of genetic diversity and demonstrated that farm environment and feed supply may be sources for the dissemination of Salmonella spp. along the production chain. The results revealed the sources of Salmonella contamination in the pig production chain and highlighted the risks of antimicrobial resistance spread.
1. Introduction
Pork is the second most widely animal protein consumed worldwide, and Brazil is the fourth world producer and exporter [1]. The southern Brazilian region has a concentration of more than 60% of pig production, according to data from the Brazilian Association of Animal Protein [2]. Pork meat exportation has increased in Brazil since 2000, leading to modifications across the industries to fit the international requirements from the World Trade Organization regarding safety and quality [3]. Based on safety aspects, the control of pathogens such as Salmonella spp. is mandatory to ensure safe foods to consumers and minimize the risks of the occurrence of foodborne disease. Ensuring food quality and safety is very important mainly due to the changing food habits of the population, the popularization of mass feeding establishments, and the globalization of food supply [4,5,6,7].
Salmonella spp. can be considered as a component of pig intestinal microbiota [5]. They are usually asymptomatic carriers, excreting the pathogen intermittently or when stressed [8]. Nowadays, pig farms usually breed animals in high density barns, facilitating the spread and persistence of Salmonella spp. in the environment [6,9]. Many countries are concerned with regard to the spread and persistence of multidrug-resistant (MDR) Salmonella, and these strains have been reported in pigs, slaughterhouses, and the final products as well as in clinical samples [10,11,12,13,14]. The use of antibiotics in different steps of pig production can contribute to the spread of Salmonella resistant strains [10]. Genes related to resistance against antimicrobials are often transferred through mobile elements (particularly plasmids) amongst bacteria including Salmonella, which makes the fight against antimicrobial resistance a challenge [15,16].
Salmonella spp. is an important foodborne pathogen in Brazil, and according to data presented by the Brazilian Ministry of Health, this pathogen was responsible for more than 11% of the recorded and investigated foodborne diseases in the country between 2009 and 2018. Pork products were considered relevant foods associated with these cases/outbreaks [17], but these data are considered to be underestimated in Brazil. Data from the United States and European Union also indicate Salmonella spp. as the main pathogen related to foodborne diseases caused by the ingestion of contaminated meat products including pork [18,19].
The identification of Salmonella spp. contamination sources along the pig production chain is a key point for taking preventive and corrective measures. The proper characterization of Salmonella spp. isolates and the identification of the main contamination routes as well as the spread of multidrug-resistant strains can subsidize the development of reliable programs to control and limit the spread of antimicrobial resistance. Considering these aspects, the present study aimed to determine the prevalence of Salmonella spp. along the pig production chain in Paraná State (southern Brazil), determine the antimicrobial resistance profiles, and possible contamination routes within this meat production chain.
2. Results and Discussion
Pig production is one of the most important husbandry activities in Brazil, therefore studies focusing on the detection of pathogens are important to ensure sanitary control during the production and safety of the meat distributed for sale. The occurrence of Salmonella spp. in the pig production chain evaluated in the present study is presented according to the site and type of sample in Table 1. The results showed that in the nursery (maternity and piglets barns), only 4.8% (32/662) of samples were positive for Salmonella, while in the pig finishing farms, the prevalence increased to 13.9% (88/636). The overall prevalence of Salmonella at the slaughterhouse was 14.8% (40/270), which was detected mainly from feces (21/25); all carcasses after end washing and chilling tested negative (Table 1). The presence of the Salmonella in the feed supply certainly contributes to pig contamination and consequently persistence in the barn environment. High discharge of Salmonella in feces collected at the slaughterhouse confirm the constant re-introduction of the pathogen in this environment.

Table 1.
Detection of Salmonella spp. along the pig production chain in Paraná State, southern Brazil.
The higher frequencies of Salmonella spp. in the pig finishing farms when compared to the nursery can be explained by two key factors: (1) maternal immunity is transferred from sows to their piglets via colostrum and milk, which contributes to the development of a protecting microbiota against pathogens during this period [20]; and (2) in the finishing farms, the longer period of stay (110 days in average) [21] and close contact among animals from different production farms facilitates the contamination and leads to the high excretion of Salmonella spp due to stress conditions. In agreement with our findings, a study conducted in Brazil by Kich et al. [22] demonstrated higher frequencies of Salmonella spp. carrier animals in finishing farms when compared to the previous production steps. Stress conditions (observed during transport and before slaughter) may contribute to the major discharge of the pathogen at the slaughterhouse [8,23].
The presence of Salmonella spp. in pig jowls may be explained by the high pathogen excretion in feces, associated with the habit of coprophagy [21,24]. Salmonella spp. in the pig oral cavity can often lead to contamination of the jowls as well as its presence in the animal’s intestine leads to contamination in the mesenteric lymph nodes. Several studies have demonstrated that the incision of lymphatic tissues can significantly affect bacterial contamination on the pig carcass [25,26]. In the present study, Salmonella spp. was not identified among the 50 carcasses sampled after final washing and chilling, demonstrating that procedures adopted during slaughter ensured the microbiological safety of the carcasses (Table 1). Focusing on microbial hazards such as Salmonella, the official guidelines for swine slaughtering in Brazil have changed, being more based in a risk analysis approach [27].
A total of 210 Salmonella spp. isolates obtained from different sources along the pig production chain were subjected to serological identification (Table 2). Serotypes Typhimurium, Mbandaka, Panama, Derby, and Agona were the most prevalent, similar to those observed in another study conducted in the southern region of Brazil (Kich, Coldebella, Morés, Nogueira, Cardoso, Fratamico, Call, Fedorka-Cray, and Luchansky [22]. S. Typhimurium was the most common serotype isolated in both farms and slaughterhouses (Table 2); this serotype has already been described as the most frequent in pig production worldwide [28,29,30,31], and is most associated with human salmonellosis outbreaks in the European Union [18,19].

Table 2.
Salmonella serotypes identified in 210 isolates obtained from the pig production chain in Paraná State, southern Brazil.
The isolates obtained from feces showed the highest serotype diversity, mainly those from piglet production and pig finishing farms (data not shown). Interestingly, various serotypes were isolates from both farms and slaughterhouses, while S. Give, S. Meleagridis, and S. Worthington were isolated only from feces samples from animals in the slaughterhouse (Table 2), suggesting that contamination occurred during transportation, since animal transport is relevant for the spread of Salmonella spp. [32,33,34].
Overall, only 17 (8.1%) of the 210 isolates were sensitive to all tested antimicrobials: resistance to tetracycline was the most common (83.8%), followed by chloramphenicol (54.3%), and trimethoprim-sulfamethoxazole (33.3%). The antimicrobial resistance profiles of the isolates, according to the number of antibiotics to which the strain was resistant, are presented in Table 3. From the 193 isolates resistant at least to one tested antibiotic, it was identified that many isolates exhibited resistance to at least three (n = 43), four (n = 12), five (n = 4), and six (n = 1) antimicrobials (Table 3). All S. Typhimurium (n = 101) isolates obtained in the present study were resistant to at least one tested antibiotic and 30 isolates were resistant to at least three antimicrobial agents. A total of 111 isolates from different serotypes exhibited simultaneous resistance to chloramphenicol and tetracycline. The emergence of MDR Salmonella is a global health concern [10,13]. Herein, simultaneous resistance to chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole was recorded in 57 isolates, representative of different serotypes, while 14 isolates were resistant to these antimicrobials plus ampicillin (Table 3). In addition, one isolate from serotype Panama was resistant to six antibiotics. Despite the efforts of the Brazilian Ministry of Agriculture to regulate the use of antimicrobials in animal production since the 1990s as a growth promoter and clinical treatment, the obtained data might be associated with the previously extensive use of these drugs in pig production [10,35]. Besides data regarding the emergence of antibiotic resistance in food production, a National Prevention and Control Program was recently established by the Brazilian Ministry of Agriculture. This program aims to promote strategic actions such as epidemiological studies, the implementation of infection prevention and other control measures to promote rational use to prevent antibiotic resistance spread [36]. Our results revealed that the situation is worrying, and further studies will allow the proper identification of molecular mechanisms associated with antimicrobial resistance exhibited by the MDR isolates recovered in the present study.

Table 3.
Antimicrobial resistance profile of Salmonella isolates recovered along the pig production chain in southern Brazil. All isolates that exhibited resistance against antimicrobials from three or more classes were considered as multidrug-resistant.
Based on previous results, 59 Salmonella spp. isolates representing positive samples sources were selected for PFGE. Considering the adopted UPGMA parameters, the isolates were grouped in five main clusters with similarities ranging from 77 to 94% (Figure 1). It was possible to identify in each cluster the isolates from different sources and pig production steps, from the nursery to the slaughter. Identical Salmonella profiles were also obtained from samples recovered from different lots, indicating that genetically close isolates are spread across Paraná State (Figure 1). We identified isolates belonging to serotype Typhimurium and its monophasic variant 4,5,12:i:-, which presented identical genetic profiles by PFGE, and can be explained by the fact that they are from the same serogroup and share a high genetic similarity [14].
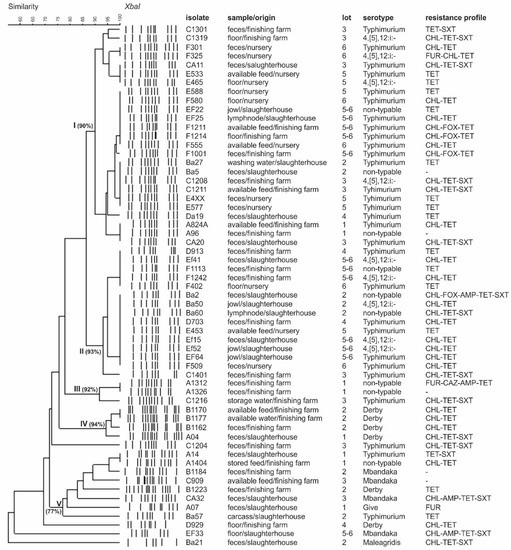
Figure 1.
Schematic representation (genetic profile, isolate identification, isolation source, lot, serotype, and antibiotic resistance profile) of 59 Salmonella isolates obtained from the pork production chain in southern Brazil. Macro-restriction was conducted with XbaI. Identity was estimated using the Dice coefficient (5% tolerance). Lot “5-6” was composed of animals from lots 5 and 6, mixed after nursery. Antibiotics: FUR: furazolidone (15 µg), CHL: chloranphenicol (30 µg), CAZ: ceftazidime (30 µg), FOX: cefoxitin (30 µg), AMP: ampicillin (10 µg), TET: tetracycline (30 µg), SXT: trimethoprim-sulfamethoxazole (25 µg).
The PFGE has been useful and accurate for tracking contamination sources, allowing the identification of Salmonella persistence, cross contamination, and distribution in swine production and pork processing [22,28,29,34,37]. Herein, the PFGE results demonstrate the possible role of the farm environment and feed supply in the dissemination of Salmonella spp. along the production chain (Figure 1), as revealed by previous studies [9,38]. Interestingly, isolates obtained from different nurseries and different pig finishing farms (feces and environment) and isolates from the slaughterhouse (environment, jowls, mesenteric lymph nodes) shared identical genetic profiles, as observed in clusters I and II (Figure 1). This might indicate cross-contamination or possible contamination from the same source in the nurseries, farms, or slaughterhouse.
The present study identified critical contamination points in the pig production chain in southern Brazil including feed supply, which may have contributed to the Salmonella introduction or maintenance in the pig barn environment. The isolates exhibiting an identical genetic profile isolated from different sources and farms revealed the spread of Salmonella in Parana State. In addition, Salmonella isolated from the same lots exhibiting different PFGE profiles demonstrated that the animals were exposed to a diversity of Salmonella throughout their life (Figure 1).
3. Materials and Methods
3.1. Sampling
Six pig lots were selected in three different farms located in Paraná State, southern Brazil. The lot selection criterion enables sampling during breeding (piglet production and pig finishing) and slaughtering steps, at 10 day intervals (day 0 to day 150). The piglet lots were selected on the birth date and each lot was sampled throughout the nurseries (maternity and piglet barns), pig finishing farms (pig barns), and slaughterhouses (environment and carcasses), resulting in a total of 1568 samples.
At pig farms, the following samples were collected: feed stored (250 g), feed available for animal consumption (250 g), water from nipple (250 mL), water from storage (250 mL), feces collected directly from the floor (80 g), and barn floor (100 cm2). All animals sampled in the pig finishing farms were sent to the same commercial slaughterhouse inspected by the Brazilian Ministry of Agriculture. At the slaughterhouse, the following samples were collected: clean lairage floor (100 cm2); feces (80 g); scalding water (250 mL); pig wash water (250 mL); evisceration table (100 cm2); carcass saw (100 cm2); carcass wash water (250 mL); carcasses after evisceration, after final washing, and after chilling, (100 cm2 each); pig jowls (300 g); and mesenteric lymph nodes (50 g). Surface sampling was conducted by swabbing one sterile sponge for each point (3M Microbiology, St. Paul, MN, USA), previously moistened with 10 mL of buffered peptone (1% w/v) saline (0.85% w/v) solution (BPS, Oxoid Ltd., Basingstoke, UK).
3.2. Salmonella spp. Detection and Serotyping
Pre-enrichment of the samples was conducted in buffered peptone water at 1% (w/v) (BPW, Oxoid Ltd., Basingstoke, UK), which was 25 g of solid sample added to 225 mL of BPW, sponges were added to 90 mL of BPW, and 200 mL of the water samples were centrifuged at 3500× g for 15 min and the pellet was added to 100 mL of BPW incubated at 35 °C for 18 to 24 h. Salmonella isolation was conducted according to the protocol described by USDA [39]. Colonies with typical Salmonella characteristics on triple sugar agar and lysine iron agar (Oxoid) were subjected to a serological test by using polyvalent somatic and flagellar antisera (Probac do Brasil, São Paulo, SP, Brazil), and further biochemical characterization by the following tests: urease, indole, methyl red, Voges-Proskauer, citrate, motility, and malonate [39]. Isolates identified as Salmonella spp. (n = 210) were serotyped by slide agglutination (Kauffmann–White–Le Minor scheme) using diverse somatic and flagellar antisera in the Bacteriology Section of the Instituto Adolfo Lutz (São Paulo, SP, Brazil) and Enterobacteriaceae Laboratory from the Fundação Oswaldo Cruz (Rio de Janeiro, RJ, Brazil). Isolates were kept stored in trypticase soya broth (TSB, Oxoid) added to glycerol at 10% at −20 °C.
3.3. Antimicrobial Resistance Testing
Salmonella isolates (n = 210) were subjected to the disk-diffusion assay, as described by the Clinical & Laboratory Standards Institute [40]. Isolates were grown in BHI (Oxoid) at 37 °C for 24 h, the inoculum was adjusted by the McFarland standard and a swab was dipped into the adjusted suspension, then spread onto the surface of plates containing Mueller Hinton agar (Oxoid), where disks of the following antimicrobials were added: ampicillin (10 μg), cefoxitin (30 μg), chloramphenicol (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), tetracycline (30 μg), imipenem (10 μg), furazolidone (15 μg), amikacin (30 μg), and trimethoprim-sulfamethoxazole (25 μg). The plates were incubated at 35 °C for 18 h. All antimicrobials were purchased from Oxoid and Escherichia coli ATCC 25922 was used as a pan-susceptible quality control. The diameters of the zones of inhibition were measured and the Salmonella isolates were characterized as resistant or susceptible according the Clinical & Laboratory Standards Institute [41]; in this study, Salmonella isolates that exhibited an intermediate resistance were considered as resistant.
3.4. Pulsed Field Gel Electrophoresis (PFGE)
Based on Salmonella spp. positive results for each step of pig production, 59 isolates were selected for PFGE. Aliquots of the selected isolates were transferred to TSB (Oxoid), incubated at 37 °C overnight, and added to agarose (Bio-Rad Laboratories, Hercules, CA, USA) for plug preparation. Plugs were subjected to lysis and washed, and the obtained DNA was subjected to macrorestriction with 50 IU of XbaI (Promega, Madison, WI, USA) at 37 °C for 2 h, as recommended by the Centers for Disease Control and Prevention (CDC) and described by Ribot et al. [42]. Macrorestriction products were separated in agarose gels (Agarose Seakem Gold 1%, w/v, buffer TE 0.5X) in a CHEF-DR II system (Bio-Rad) following the parameters recommended by the CDC. The gels were stained with GelRed (Biotium Inc., Hayward, CA, USA) and the band patterns were analyzed by using BioNumeric 6.6 software (Applied Maths NV, Sint-Martens-Latem, Belgium). Pulse Marker 50 (1000 kb, Sigma-Aldrich Corp., St. Louis, MO, USA) was added to each gel for band normalization. Isolate similarities were calculated considering the Dice coefficient, 5% of tolerance, and the dendrogram with the genetic profiles of selected isolates was constructed considering the unweighted pairwise grouping with mathematical averaging (UPGMA) algorithm.
4. Conclusions
Taken together, our results revealed that serotype Typhimurium is spread along the pig production chain in Paraná State, southern Brazil, which is introduced continuously into the slaughterhouse through animal carriers. Our results showed that farms must adopt measures to control Salmonella spp. in the feed supply to prevent pig contamination through this foodborne pathogen. Even through Salmonella spp. was identified at different sampling sites including mesenteric lymph nodes and jowls, all 50 carcasses sampled after final washing and chilling tested negative for this pathogen, indicating the good manufacturing practices were properly applied in the slaughterhouse. However, the high rate of MDR strains isolated in the pig production chain highlight that implementation measures are needed to prevent the spread of antibiotic resistance in the environment. Monitoring antimicrobial resistance in the pork production chain is very relevant as resistant strains can be transferred to humans. Our study emphasizes the high genotypic diversity of Salmonella and the importance of the swine environment as a reservoir of MDR Salmonella.
Author Contributions
Conceptualization, L.d.S.B., J.P.d.A.N.P., L.A.N. and M.T.D.; Funding acquisition, M.T.D.; Investigation, L.d.S.B., V.Q.C., C.V., R.C.K.B. and A.C.C.; Methodology, L.d.S.B., J.P.d.A.N.P., L.A.N. and M.T.D; Project administration, M.T.D; Supervision, J.P.d.A.N.P. and M.T.D.; Writing—Original draft, L.d.S.B., V.Q.C., C.V., A.C.C., L.A.N., and M.T.D; Writing—Review & editing, L.d.S.B., A.C.C., C.V., and L.A.N.
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil) Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil-001) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guimarães, D.D.; Amaral, G.F.; Maia, G.B.d.S.; Lemos, M.L.F.; Ito, M.; Custodio, S. Pig farming: Productive chain structure, panorama of the sector in Brazil and in the world and BNDES’ support. BNDES Setorial 2017, 45, 85–136. Available online: https://web.bndes.gov.br/bib/jspui/bitstream/1408/11794/1/BS%2045%20Suinocultura%20-%20estrutura%20da%20cadeia%20produtiva%2C%20panorama%20do%20setor%20no%20Brasil%5B...%5D_P.pdfpage (accessed on 19 May 2019).
- ABPA. Suinocultura. Available online: http://abpa-br.com.br/setores/suinocultura (accessed on 27 June 2017).
- Maertens, M.; Swinnen, J. Agricultural Trade and Development: A Value Chain Perspective; WTO Working Paper ERSD-2015-04; WTO—Economic Research and Statistics Division: Geneva, Switzerland, 2015; p. 38. [Google Scholar]
- Lee, K.-M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Baer, A.A.; Miller, M.J.; Dilger, A.C. Pathogens of interest to the pork industry: A review of research on interventions to assure food safety. Compr. Rev. Food Sci. Food Saf. 2013, 12, 183–217. [Google Scholar] [CrossRef]
- Motarjemi, Y.; Käferstein, F. Food safety, Hazard Analysis and Critical Control Point and the increase in foodborne diseases: A paradox? Food Control 1999, 10, 325–333. [Google Scholar] [CrossRef]
- Santini, A.; Novellino, E.; Armini, V.; Ritieni, A. State of the art of Ready-to-Use Therapeutic Food: A tool for nutraceuticals addition to foodstuff. Food Chem. 2013, 140, 843–849. [Google Scholar] [CrossRef]
- Simons, R.R.L.; Hill, A.A.; Swart, A.; Kelly, L.; Snary, E.L. A Transport and lairage model for Salmonella transmission between pigs applicable to EU member States. Risk Anal. 2016, 36, 482–497. [Google Scholar] [CrossRef]
- Pellegrini, D.C.P.; Paim, D.S.; Lima, G.J.M.M.; Pissetti, C.; Kich, J.D.; Cardoso, M.R.I. Distribution of Salmonella clonal groups in four Brazilian feed mills. Food Control 2015, 47, 672–678. [Google Scholar] [CrossRef]
- Lopes, V.; Pissetti, C.; Pellegrini, D.D.C.P.; Silva, L.E.d.; Cardoso, M. Resistance Phenotypes and Genotypes of Salmonella enterica subsp. enterica Isolates from Feed, Pigs, and Carcasses in Brazil. J. Food Prot. 2015, 78, 407–413. [Google Scholar] [CrossRef]
- Yan-Bing, Z.; Li-Gen, X.; Mei-Fang, T.; Hai-Qin, L.; Han, Y.; Li, Z.; De-Feng, Y.; Zhao-Feng, K.; Qi-Peng, W.; Lin-Guang, L. Prevalence and Antimicrobial Resistance of Salmonella in Pork, Chicken, and Duck from Retail Markets of China. Foodborne Pathog. Dis. 2019, 16, 339–345. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Singh, A.; Zhao, S.; Bartholomew, M.; Womack, N.; Ayers, S.; Fields, P.I.; McDermott, P.F. Antimicrobial Resistance in Salmonella in the United States from 1948 to 1995. Antimicrob. Agents Chemother. 2016, 60, 2567–2571. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-H.; Zhu, Y.-H.; Ren, T.-Y.; Guo, L.; Yang, G.-Y.; Jiao, L.-G.; Wang, J.-F. Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China. Microorganisms 2018, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.; Lincopan, N.; Fedorka-Cray, P.J.; Landgraf, M. Current insights on high priority antibiotic-resistant Salmonella enterica in food and foodstuffs: A review. Curr. Opin. Food Sci. 2019, 26, 35–46. [Google Scholar] [CrossRef]
- Argüello, H.; Guerra, B.; Rodríguez, I.; Rubio, P.; Carvajal, A. Characterization of Antimicrobial Resistance Determinants and Class 1 and Class 2 Integrons in Salmonella enterica spp., Multidrug-Resistant Isolates from Pigs. Genes 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- ANVISA. Ministério da Saúde. Surtos de Doenças Transmitidas por Alimentos no Brasil. Available online: http://portalarquivos2.saude.gov.br/images/pdf/2019/fevereiro/15/Apresenta----o-Surtos-DTA---Fevereiro-2019.pdf (accessed on 24 July 2019).
- CDC. Salmonella. Available online: https://www.cdc.gov/salmonella/ (accessed on 28 June 2019).
- EFSA. Salmonella. Available online: https://www.efsa.europa.eu/en/topics/topic/salmonella (accessed on 28 June 2019).
- Poonsuk, K.; Zimmerman, J. Historical and contemporary aspects of maternal immunity in swine. Anim. Health Res. Rev. 2018, 19, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Proux, K.; Cariolet, R.; Fravalo, P.; Houdayer, C.; Keranflech, A.; Madec, F. Contamination of pigs by nose-to-nose contact or airborne transmission of Salmonella Typhimurium. Vet. Res. 2001, 32, 591–600. [Google Scholar] [CrossRef]
- Kich, J.D.; Coldebella, A.; Morés, N.; Nogueira, M.G.; Cardoso, M.; Fratamico, P.M.; Call, J.E.; Fedorka-Cray, P.; Luchansky, J.B. Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina, Brazil. Int. J. Food Microbiol. 2011, 151, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.K.; Kim, H.B.; Borewicz, K.; Isaacson, R.E. Towards an understanding of Salmonella enterica serovar Typhimurium persistence in swine. Anim. Health Res. Rev. 2016, 17, 159–168. [Google Scholar] [CrossRef]
- Berriman, A.D.C.; Clancy, D.; Clough, H.E.; Armstrong, D.; Christley, R.M. Effectiveness of simulated interventions in reducing the estimated prevalence of Salmonella in UK pig herds. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Van Damme, I.; Berkvens, D.; Vanantwerpen, G.; Baré, J.; Houf, K.; Wauters, G.; De Zutter, L. Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: Distribution, quantification and identification of risk factors. Int. J. Food Microbiol. 2015, 204, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Biasino, W.; De Zutter, L.; Mattheus, W.; Bertrand, S.; Uyttendaele, M.; Van Damme, I. Correlation between slaughter practices and the distribution of Salmonella and hygiene indicator bacteria on pig carcasses during slaughter. Food Microbiol. 2018, 70, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa 79, de 14 de dezembro de 2018. Inspeção ante e post mortem de suínos com base em risco. Diário Oficial da União. Diário Oficial da União, 17 de dezembro de 2018. Brasília; 2018. Available online: http://www.in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/55444279/do1-2018-12-17-instrucao-normativa-n-79-de-14-de-dezembro-de-2018-55444116 (accessed on 10 June 2019).
- De Busser, E.V.; Maes, D.; Houf, K.; Dewulf, J.; Imberechts, H.; Bertrand, S.; De Zutter, L. Detection and characterization of Salmonella in lairage, on pig carcasses and intestines in five slaughterhouses. Int. J. Food Microbiol. 2011, 145, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Neves, E.; Antunes, P.; Tavares, A.; Themudo, P.; Cardoso, M.F.; Gärtner, F.; Costa, J.M.; Peixe, L. Salmonella cross-contamination in swine abattoirs in Portugal: Carcasses, meat and meat handlers. Int. J. Food Microbiol. 2012, 157, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Marier, E.A.; Snow, L.C.; Floyd, T.; McLaren, I.M.; Bianchini, J.; Cook, A.J.C.; Davies, R.H. Abattoir based survey of Salmonella in finishing pigs in the United Kingdom 2006–2007. Prev. Vet. Med. 2014, 117, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Swanenburg, M.; Van der Wolf, P.J.; Urlings, H.A.P.; Snijders, J.M.A.; van Knapen, F. Salmonella in slaughter pigs: The effect of logistic slaughter procedures of pigs on the prevalence of Salmonella in pork. Int. J. Food Microbiol. 2001, 70, 231–242. [Google Scholar] [CrossRef]
- Fernandes, L.; Centeno, M.M.; Couto, N.; Nunes, T.; Almeida, V.; Alban, L.; Pomba, C. Longitudinal characterization of monophasic Salmonella Typhimurium throughout the pig’s life cycle. Vet. Microbiol. 2016, 192, 231–237. [Google Scholar] [CrossRef]
- Gebreyes, W.A.; Davies, P.R.; Turkson, P.-K.; Morrow, W.E.M.; Funk, J.A.; Altier, C.; Thakur, S. Characterization of antimicrobial-resistant phenotypes and genotypes among Salmonella enterica recovered from pigs on farms, from transport trucks, and from pigs after slaughter. J. Food Prot. 2004, 67, 698–705. [Google Scholar] [CrossRef]
- Magistrali, C.; Dionisi, A.M.; De Curtis, P.; Cucco, L.; Vischi, O.; Scuota, S.; Zicavo, A.; Pezzotti, G. Contamination of Salmonella spp. in a pig finishing herd, from the arrival of the animals to the slaughterhouse. Res. Vet. Sci. 2008, 85, 204–207. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa n 41, de 23 de outubbro de 2017. Programa Nacional de Prevenção e Controle da Resistência aos Antimicrobianos na Agropecuária. Diário Oficial da União, 09 de novembro de 2017. Brasília; 2017. Available online: http://www.in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/19401380/do1-2017-11-09-instrucao-normativa-n-41-de-23-de-outubro-de-2017-19401312 (accessed on 10 June 2019).
- Zhou, Z.; Jin, X.; Zheng, H.; Li, J.; Meng, C.; Yin, K.; Xie, X.; Huang, C.; Lei, T.; Sun, X.; et al. The prevalence and load of Salmonella, and key risk points of Salmonella contamination in a swine slaughterhouse in Jiangsu province, China. Food Control 2018, 87, 153–160. [Google Scholar] [CrossRef]
- Molla, B.; Sterman, A.; Mathews, J.; Artuso-Ponte, V.; Abley, M.; Farmer, W.; Rajala-Schultz, P.; Morrow, W.E.M.; Gebreyes, W.A. Salmonella enterica in commercial swine feed and subsequent isolation of phenotypically and genotypically related strains from fecal samples. Appl. Environ. Microbiol. 2010, 76, 7188–7193. [Google Scholar] [CrossRef] [PubMed]
- USDA. Isolation and Identification of Salmonella from Meat, Poultry and Egg Products. In Microbiology Laboratory Guidebook; USDA: Washington, DC, USA, 2002; Volume MLG 4.02. [Google Scholar]
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; M02; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Ribot, E.M.; Fair, M.A.; Gautom, R.; Cameron, D.N.; Hunter, S.B.; Swaminathan, B.; Barrett, T.J. Standardization of Pulsed-Field Gel Electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 2006, 3, 59–67. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).