First Detection of Carbapenem-Resistant Escherichia fergusonii Strains Harbouring Beta-Lactamase Genes from Clinical Samples
Abstract
:1. Introduction
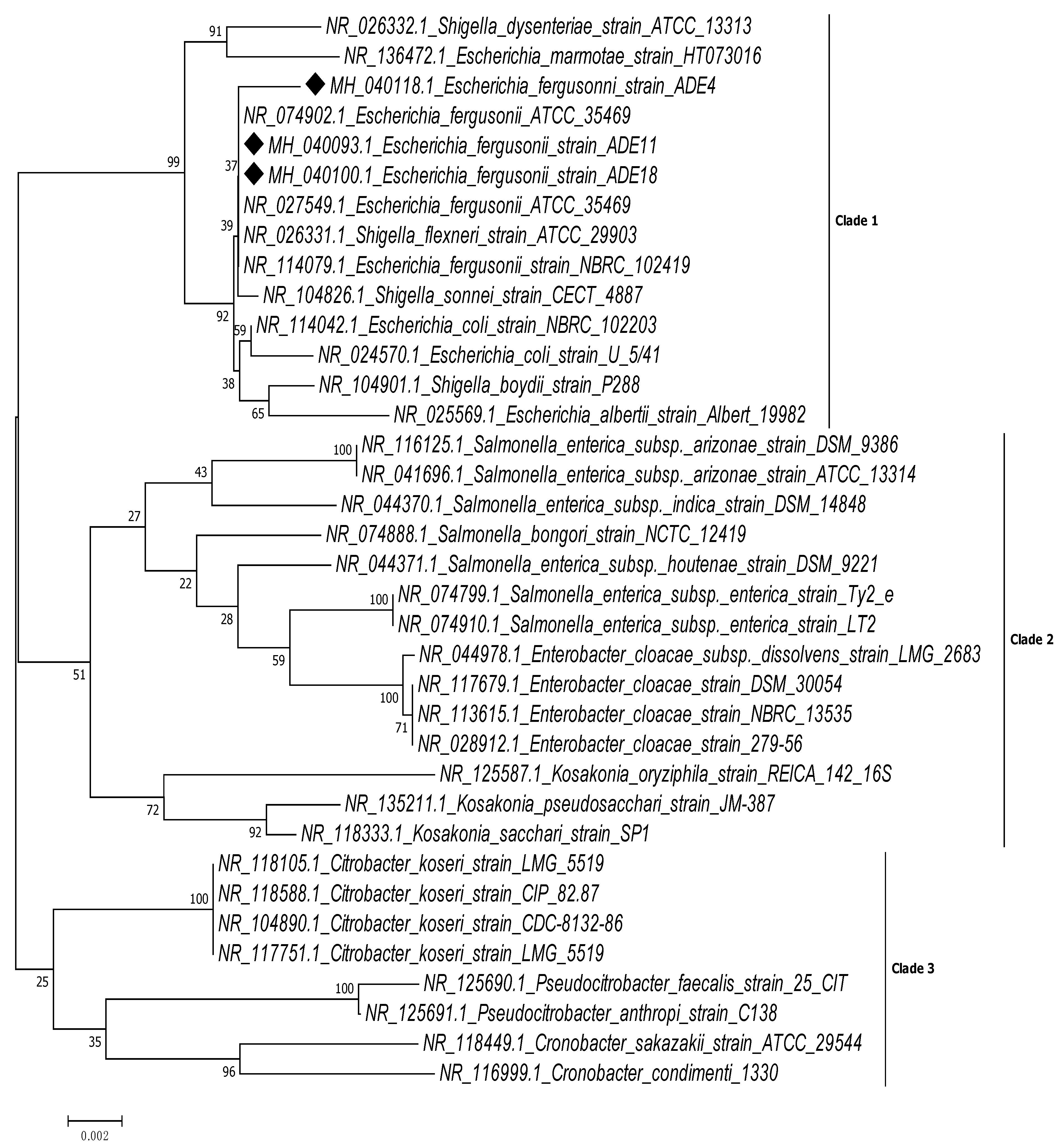
2. Results
3. Discussion
4. Materials and Methods
4.1. Culture and Identification of Clinical Isolates
4.1.1. Culture and Phenotypic Identification of Clinical Isolates
4.1.2. Genotypic Identification of Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. Detection of ESBL Production
4.4. DNA Extraction and Genotypic Detection of β-lactamase Genes
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manges, A.R.; Johnson, J.R. Reservoirs of Extraintestinal Pathogenic Escherichia coli. Microbiol. Spectr. 2015, 3, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S. Urinary tract infections attributed to diverse ExPEC strains in food animals: Evidence and data gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Gomi, R.; Matsuda, T.; Matsumura, Y.; Yamamoto, M.; Tanaka, M.; Ichiyama, S.; Yoneda, M. Whole-Genome Analysis of Antimicrobial-Resistant and Extraintestinal Pathogenic Escherichia coli in River Water. Appl. Environ. Microbiol. 2017, 83, e02703-16. [Google Scholar] [CrossRef] [PubMed]
- Poolman, J.T.; Wacker, M. Extraintestinal Pathogenic Escherichia coli, a Common Human Pathogen: Challenges for Vaccine Development and Progress in the Field. J. Infect. Dis. 2016, 213, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Glover, B.; Wentzel, J.; Jenkins, A.; Van Vuuren, M. The first report of Escherichia fergusonii isolated from non-human primates, in Africa. One Heal. 2017, 3, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.A.; Lee, K.; Chung, J.E. Risk factors and molecular features of sequence type (ST) 131 extended-Spectrum-β-lactamase-producing Escherichia coli in community-onset female genital tract infections. BMC Infect. Dis. 2018, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.; Lee, I.; Chu, S.; Lien, R.; Huang, H.; Chiang, M.; Fu, R.; Hsu, J.; Huang, Y. Clinical and Molecular Characteristics of Neonatal Extended-Spectrum β-Lactamase-Producing Gram-Negative Bacteremia: A 12-Year Case-Control-Control Study of a Referral Center in Taiwan. PLoS ONE 2016, 11, e0159744. [Google Scholar] [CrossRef]
- Matsumura, Y.; Pitout, J.D.D.; Gomi, R.; Matsuda, T.; Noguchi, T.; Yamamoto, M.; Peirano, G.; DeVinney, R.; Bradford, P.A.; Motyl, M.R.; et al. Global Escherichia coli Sequence Type 131 Clade with bla CTX-M-27 Gene. Emerg. Infect. Dis. 2016, 22, 1900–1907. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence Type ST131 and ST10 Complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria* *This study has been partially presented during the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) in Chicago, IL, September 2011. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [PubMed]
- Funke, G.; Hany, A.; Altwegg, M. Isolation of Escherichia fergusonii from four different sites in a patient with pancreatic carcinoma and cholangiosepsis. J. Clin. Microbiol. 1993, 31, 2201–2203. [Google Scholar] [PubMed]
- Gokhale, V.V.; Therese, K.L.; Bagyalakshmi, R.; Biswas, J. Detection of Escherichia fergusonii by PCR-based DNA sequencing in a case of delayed-onset chronic endophthalmitis after cataract surgery. J. Cataract Refract. Surg. 2014, 40, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Cheng, A.; Huang, Y.T.; Chung, K.P.; Lee, M.R.; Liao, C.H.; Hsueh, P.R. Escherichia fergusonii bacteremia in a diabetic patient with pancreatic cancer. J. Clin. Microbiol. 2011, 49, 4001–4002. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, A.; Mahapatra, S.; Mahapatra, A. Escherichia fergusonii: An emerging pathogen in South Orissa. Indian J. Med. Microbiol. 2005, 23, 204. [Google Scholar] [CrossRef] [PubMed]
- Maheux, A.F.; Boudreau, D.K.; Bergeron, M.G.; Rodriguez, M.J. Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 2014, 117, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Savini, V.; Catavitello, C.; Talia, M.; Manna, A.; Pompetti, F.; Favaro, M.; Fontana, C.; Febbo, F.; Balbinot, A.; Di Berardino, F.; et al. Multidrug-resistant Escherichia fergusonii: A case of acute cystitis. J. Clin. Microbiol. 2008, 46, 1551–1552. [Google Scholar] [CrossRef]
- Weiss, A.T.A.; Lübke-Becker, A.; Krenz, M.; van der Grinten, E. Enteritis and Septicemia in a Horse Associated With Infection by Escherichia fergusonii. J. Equine Vet. Sci. 2011, 31, 361–364. [Google Scholar] [CrossRef]
- Lagacé-Wiens, P.R.S.; Baudry, P.J.; Pang, P.; Hammond, G. First description of an extended-spectrum-beta-lactamase-producing multidrug-resistant Escherichia fergusonii strain in a patient with cystitis. J. Clin. Microbiol. 2010, 48, 2301–2302. [Google Scholar] [CrossRef]
- Kola, A.; Kohler, C.; Pfeifer, Y.; Schwab, F.; Kühn, K.; Schulz, K.; Balau, V.; Breitbach, K.; Bast, A.; Witte, W.; et al. High prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012, 67, 2631–2634. [Google Scholar] [CrossRef]
- Giwa, F.J.; Ige, O.T.; Haruna, D.M.; Yaqub, Y.; Lamido, T.Z.; Usman, S.Y. Extended - Spectrum Beta - lactamase Production and Antimicrobial Susceptibility Pattern of Uropathogens in a Tertiary Hospital in Northwestern Nigeria. Ann. Trop. Pathol. 2018, 9, 11–16. [Google Scholar] [CrossRef]
- Onyedibe, K.I.; Shobowale, E.O.; Okolo, M.O.; Iroezindu, M.O.; Afolaranmi, T.O.; Nwaokorie, F.O.; Opajobi, S.O.; Isa, S.E.; Egah, D.Z. Low Prevalence of Carbapenem Resistance in Clinical Isolates of Extended Spectrum Beta Lactamase (ESBL) Producing Escherichia coli in North Central, Nigeria. Adv. Infect. Dis. 2018, 8, 109–120. [Google Scholar] [CrossRef]
- Oli, A.N.; Eze, D.E.; Gugu, T.H.; Ezeobi, I.; Maduagwu, U.N.; Ihekwereme, C.P. Multi-antibiotic resistant extended-spectrum beta-lactamase producing bacteria pose a challenge to the effective treatment of wound and skin infections. Pan Afr. Med. J. 2017, 27. [Google Scholar] [CrossRef] [PubMed]
- Alaka, O.O.; Orimolade, E.A.; Ojo, O.O.; Onipede, A.O. The Phenotypic Detection of Carbapenem Resistant Organisms in Orthopaedic Wound nfections in Ile-Ife, Nigeria. Acta Sci. Microbiol. 2019, 2, 35–42. [Google Scholar]
- Yusuf, I.; Rabiu, A.T.; Haruna, M.; Abdullahi, S.A. Carbapenem-Resistant Enterobacteriaceae (CRE) in Intensive Care Units and Surgical Wards of hospitals with no history of carbapenem usage in Kano, North West Nigeria. Niger. J. Microbiol. 2015, 27, 2612–2618. [Google Scholar]
- Oduyebo, O.; Falayi, O.; Oshun, P.; Ettu, A. Phenotypic determination of carbapenemase producing enterobacteriaceae isolates from clinical specimens at a tertiary hospital in Lagos, Nigeria. Niger. Postgrad. Med. J. 2015, 22, 223. [Google Scholar] [PubMed]
- Igwe, J.C.; Olayinka, B.O.; Ehnimidu, J.O.; Onaolapo, J.A. Virulent Characteristics of Multidrug Resistant E. coli from Zaria, Nigeria. Clin. Microbiol. Open Access 2016, 5, 1–9. [Google Scholar]
- Olowe, B.; Oluyege, J.; Famurewa, O.; Ogunniran, A.; Adelegan, O. Molecular Identification of Escherichia coli and New Emerging Enteropathogen, Escherichia fergusonii, From Drinking Water Sources in Ado-Ekiti, Ekiti State, Nigeria. J. Microbiol. Res. 2017, 7, 45–54. [Google Scholar]
- Crawford-Miksza, L.K.; Himathongkham, S.; Dodd, M.L.; Badoiu, A.S.; Badoiu, O.M.; Guthertz, L.S. Misidentification of a variant biotype of Escherichia coli O157:H7 as Escherichia fergusonii by Vitek 2 Compact. J. Clin. Microbiol. 2009, 47, 872–873. [Google Scholar] [CrossRef]
- Rayamajhi, N.; Jung, B.Y.; Cha, S.B.; Shin, M.K.; Kim, A.; Kang, M.S.; Lee, K.M.; Yoo, H.S. Antibiotic resistance patterns and detection of blaDHA-1 in Salmonella species isolates from chicken farms in South Korea. Appl. Environ. Microbiol. 2010, 76, 4760–4764. [Google Scholar] [CrossRef]
- Farmer, J.J.; Fanning, G.R.; Davis, B.R.; O’Hara, C.M.; Riddle, C.; Hickman-Brenner, F.W.; Asbury, M.A.; Lowery, V.A., 3rd; Brenner, D.J. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 1985, 21, 77–81. [Google Scholar] [PubMed]
- Balqis, U.; Hambal, M.; Admi, M.; Meutia, N.; Abdullah, N.; Ferasyi, T.R.; Lubis, T.M.; Abrar, M.; Aceh, D.; Breeding, I.; et al. Escherichia fergusonii identified in preputial swabs from healthy Aceh cattle by phylogenetic 16S rRNA analysis. Malays. J. Microbiol. 2018, 14, 229–235. [Google Scholar]
- Nhung, P.H.; Ohkusu, K.; Mishima, N.; Noda, M.; Shah, M.M.; Sun, X.; Hayashi, M.; Ezaki, T. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn. Microbiol. Infect. Dis. 2007, 58, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Ohkusu, K.; Kusaba, K.; Ikeda, H.; Tanaka, T.; Matsuura, S.; Kawamura, Y.; Kobatake, S.; Nagasawa, Z.; Aoki, Y.; et al. Quantitative Microarray-Based DNA-DNA Hybridization Assay for Measuring Genetic Distances among Bacterial Species and Its Application to the Identification of Family Enterobacteriaceae. Microbiol. Immunol. 2005, 49, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Pettengill, E.A.; Pettengill, J.B.; Binet, R. Phylogenetic Analyses of Shigella and Enteroinvasive Escherichia coli for the Identification of Molecular Epidemiological Markers: Whole-Genome Comparative Analysis Does Not Support Distinct Genera Designation. Front. Microbiol. 2016, 6, 1573. [Google Scholar] [CrossRef]
- Ud-Din, A.; Wahid, S. Relationship among Shigella spp. and enteroinvasive Escherichia coli (EIEC) and their differentiation. Braz. J. Microbiol. 2014, 45, 1131–1138. [Google Scholar] [CrossRef]
- Doare, K.L.; Bielicki, J.; Heath, P.T.; Sharland, M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited Countries. J. Pediatric Infect. Dis. Soc. 2015, 4, 11–20. [Google Scholar] [CrossRef]
- Gaastra, W.; Kusters, J.G.; van Duijkeren, E.; Lipman, L.J.A. Escherichia fergusonii. Vet. Microbiol. 2014, 172, 7–12. [Google Scholar] [CrossRef]
- Sahl, J.W.; Morris, C.R.; Emberger, J.; Fraser, C.M.; Ochieng, J.B.; Juma, J.; Fields, B.; Breiman, R.F.; Gilmour, M.; Nataro, J.P.; et al. Defining the phylogenomics of Shigella species: A pathway to diagnostics. J. Clin. Microbiol. 2015, 53, 951–960. [Google Scholar] [CrossRef]
- Devanga Ragupathi, N.K.; Muthuirulandi Sethuvel, D.P.; Inbanathan, F.Y.; Veeraraghavan, B. Accurate differentiation of Escherichia coli and Shigella serogroups: Challenges and strategies. New Microbes New Infect. 2018, 21, 58–62. [Google Scholar] [CrossRef]
- Sims, G.E.; Kim, S.-H. Whole-genome phylogeny of Escherichia coli/Shigella group by feature frequency profiles (FFPs). Proc. Natl. Acad. Sci. USA 2011, 108, 8329–8334. [Google Scholar] [CrossRef] [PubMed]
- Maria Dannessa Delost Introduction to Diagnostic Microbiology for the Laboratory Sciences - Maria Dannessa Delost - Google Books; Jones & Bartlett Publishers: Burlington, MA, USA, 2015; ISBN 9781284032321.
- Morris, D.; O’Hare, C.; Glennon, M.; Maher, M.; Corbett-Feeney, G.; Cormican, M. Extended-spectrum β-lactamases in Ireland, including a novel enzyme, TEM-102. Antimicrob. Agents Chemother. 2003, 47, 2572–2578. [Google Scholar] [PubMed]
- Chaudhury, A.; Nath, G.; Tikoo, A.; Sanyal, S.C. Enteropathogenicity and antimicrobial susceptibility of new Escherichia spp. J. Diarrhoeal Dis. Res. 1999, 17, 85–87. [Google Scholar] [PubMed]
- Rayamajhi, N.; Cha, S.B.; Shin, S.W.; Jung, B.Y.; Lim, S.; Yoo, H.S. Plasmid Typing and Resistance Profiling of Escherichia fergusonii and Other Enterobacteriaceae Isolates from South Korean Farm Animals. Appl. Environ. Microbiol. 2011, 77, 3163–3166. [Google Scholar] [CrossRef] [PubMed]
- Willey, M.; Sherwood, L.; Woolverton, J. Prescott, Harley, and Klein’s Microbiology, 7th ed.; Colin Wheatley/Janice Roerig-Blong: New York, NY, USA, 2008; ISBN 978-0-07-299291-5. [Google Scholar]
- Attallah, A.G.; EL-Shaer, H.F.A.; Abd-El-Aal, S.K.H. Research Journal of Pharmaceutical, Biological and Chemical Sciences 16S rRNA Characterization of a Bacillus Isolates From Egyptian Soil and its Plasmid Profile. Res. J. Pharm. Biol. Chem. Sci. 2014, 5, 1590–1604. [Google Scholar]
- El Allaoui, A.; Rhazi, F.F.; Essahale, A.; Bouchrif, B.; Karraouan, B.; Ameur, N.; Aboulkacem, A. Characterization of antimicrobial susceptibility, virulence genes and identification by 16S ribosomal RNA gene sequencing of Salmonella serovars isolated from turkey meat in Meknes, Morocco. Int. J. Microbiol. Immunol. Res. 2013, 1, 68–79. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Jean, B.P.; Franklin, R.C.; George, M.E.; Stephen, G.J.; James, S.L.; Brandi, L. M100S Performance Standards for Antimicrobial; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; ISBN 1562389238. [Google Scholar]
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST. Available online: http://www.korld.edu.pl/pdf/eucast/EUCAST_QC_tables_5.0_routine_and_extended.pdf (accessed on 12 September 2017).
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Available online: http://www.eucast.org/resistance_mechanisms/ (accessed on 12 september 2017).
- Helmy, M.M.; Wasfi, R. Phenotypic and molecular characterization of plasmid mediated AmpC β-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. BioMed Res. Int. 2014, 2014. [Google Scholar]
- Greco, M.; Sáez, C.A.; Brown, M.T.; Bitonti, M.B. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model brown alga. PLoS ONE 2014, 9, e96470. [Google Scholar] [CrossRef]
- Daoud, Z.; Salem Sokhn, E.; Masri, K.; Cheaito, K.; Haidar-Ahmad, N.; Matar, G.M.; Doron, S. Corrigendum: Escherichia coli Isolated from Urinary Tract Infections of Lebanese Patients between 2005 and 2012: Epidemiology and Profiles of Resistance. Front. Med. 2015, 2, 1–11. [Google Scholar] [CrossRef]
- Rafiee, R.; Eftekhar, F.; Tabatabaei, S.A.; Tehrani, D.M. Prevalence of extended-spectrum and metallo β-lactamase production in AmpC β-lactamase producing Pseudomonas aeruginosa isolates from burns. Jundishapur J. Microbiol. 2014, 7, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.Y.; Li, Y.; Chan, E.W.C.; Chen, S. Functional categorization of carbapenemase- mediated resistance by a combined genotyping and two-tiered Modified Hodge Test approach. Front. Microbiol. 2015, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nwinyi, O.C.; Ajayi, O.O.; Amund, O.O. Degradation of polynuclear aromatic hydrocarbons by two strains of Pseudomonas. Braz. J. Microbiol. 2016, 47, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.H.; Song, X.Y.; Ma, X.B.; Zhang, S.Y.; Zhang, J.Q. Molecular characterization of multidrug-resistant klebsiella pneumoniae isolates. Braz. J. Microbiol. 2015, 46, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Lari, A.R.; Azimi, L.; Rahbar, M.; Alaghehbandan, R.; Sattarzadeh-Tabrizi, M. First report of Klebsiella pneumonia carbapenemase-producing Pseudomonas aeruginosa isolated from burn patients in Iran: Phenotypic and genotypic methods. GMS Hyg. Infect. Control 2014, 9, Doc06. [Google Scholar] [PubMed]
| Organism Name | Form | Margin | Elevation | Optical Property | Appearance | Consistency | Colour on NA | Colour on MacC | Gram Stain | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. fergusonii | circular | Entire | Flat | opaque | Glistening | Moist | White | Pink | Negative | ||||||
| Indole production | Methyl-red | Voges-Proskauer | Citrate | Urease | Hydrogen sulphide | Motility | Lactose | Maltose | Sucrose | Glucose (A/G) | Mannitol | Cellobiose | Adonitol | Sorbitol | Oxidase |
| + | + | − | − | − | − | + | − | + | − | +/+ | + | + | + | − | − |
| Isolate Name | Organism | Accession Number | Antibiotics Resistance Pattern | Beta-Lactamase Gene Encoded |
|---|---|---|---|---|
| CR11 | E. fergusoniiADE4 | MH040118 | AUG PEN G AMP CRO CRX CAZ LEV OFL CIP NIT GEN IMP MER | TEM CTX-M |
| CR35 | E. fergusoniiADE11 | MH040093 | AUG PEN G AMP CRO CRX CAZ LEV OFL CIP NIT GEN IMP MER | TEM CTX-M |
| CR49 | E. fergusoniiADE18 | MH040100 | AUG PEN G AMP CRO CRX CAZ IMP | SHV |
| Primer Name | Primer Sequence 5′ ⟶ 3′ | Annealing Temperature | Expected Amplicon Size | Reference |
|---|---|---|---|---|
| TEM | F:5′CGCCGCATACACTATTCTCAGAATGA3′ R:5′ACGCTCACCGGCTCCAGATTTAT3′ | 58 °C | 445 pb | [55] |
| SHV | F:5′CTTTATCGGCCCTCACTCAA3′ R:5′AGGTGCTCATCATGGGAAAG3′ | 58 °C | 273 bp | [55] |
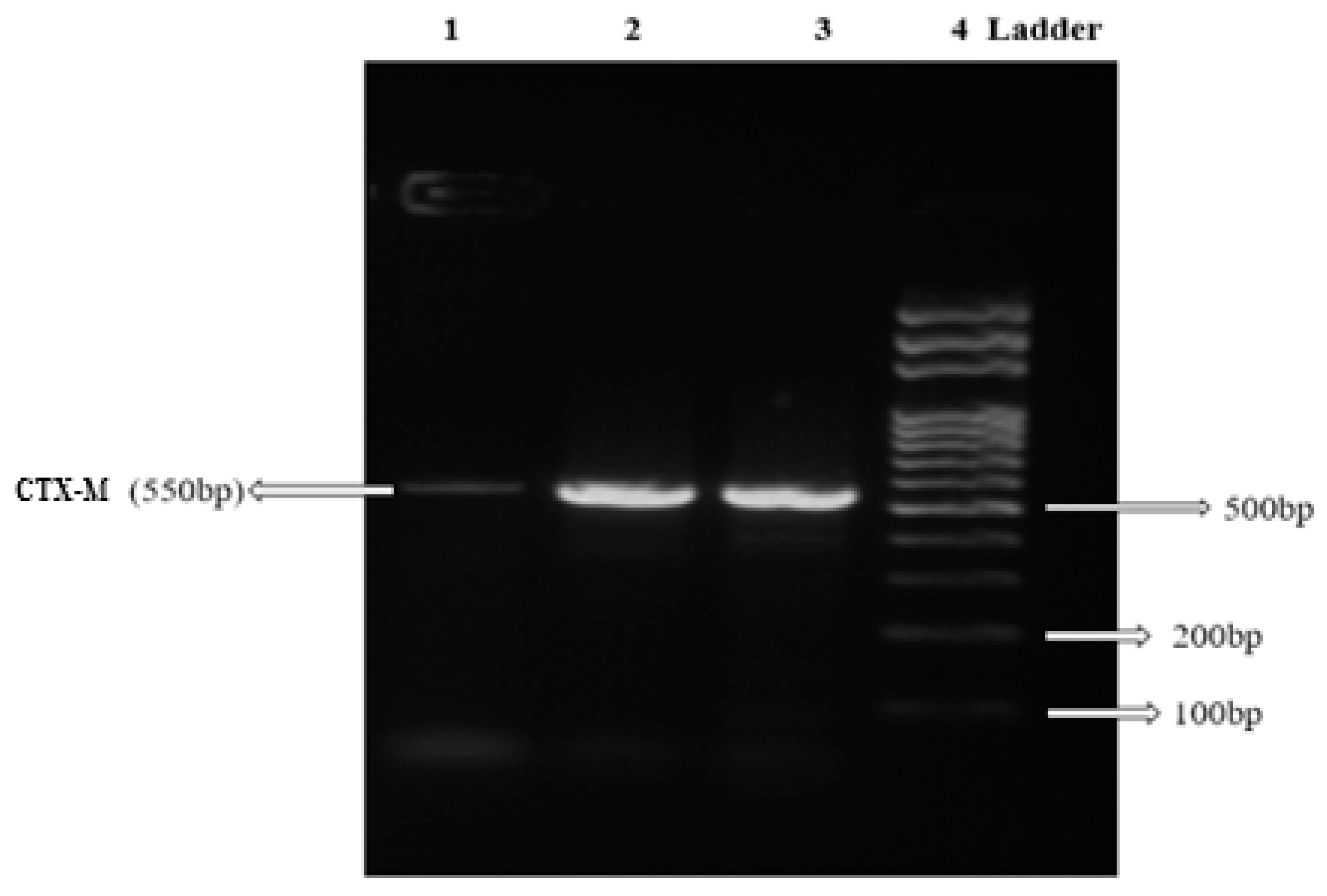
| CTX-M | F:5′CGCTTTGCGATGTGCAG3′ R:5′ACCGCGATATCGTTGGT3′ | 55 °C | 550 bp | [56] |
| IMP | F:5′-GGAATAGAGTGGCTTAAYTCTC-3′ R:5′-CCAAACYACTASGTTATCT-3′ | 53 °C | 188 bp | [59] |
| NDM | F:5′GGTTTGGCGATCTGGTTTTC R:5′CGGAATGGCTCATCACGATC | 62.8 °C | 621 bp | [57] |
| OXA | F:5′ACACAATACATATCAACTTCGC3′ R:5′AGTGTGTTTAGAATGGTGATC3′ | 62.8 °C | 813 bp | [55] |
| KPC | F:5′GTATCGCCGTCTAGTTCTGC3′ R:5′GGTCGTGTTTCCCTTTAGCCA3′ | 59.9 °C | 636 bp | [60] |
| VIM | F:5′-GATGGYGTTTGGTCGCATATCKCAAC3′ R:5′-CGAATGCGCAGCACCRGGATAGAA-3′ | 54.4 °C | 390 bp | [57] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adesina, T.; Nwinyi, O.; De, N.; Akinnola, O.; Omonigbehin, E. First Detection of Carbapenem-Resistant Escherichia fergusonii Strains Harbouring Beta-Lactamase Genes from Clinical Samples. Pathogens 2019, 8, 164. https://doi.org/10.3390/pathogens8040164
Adesina T, Nwinyi O, De N, Akinnola O, Omonigbehin E. First Detection of Carbapenem-Resistant Escherichia fergusonii Strains Harbouring Beta-Lactamase Genes from Clinical Samples. Pathogens. 2019; 8(4):164. https://doi.org/10.3390/pathogens8040164
Chicago/Turabian StyleAdesina, Tomilola, Obinna Nwinyi, Nandita De, Olayemi Akinnola, and Emmanuel Omonigbehin. 2019. "First Detection of Carbapenem-Resistant Escherichia fergusonii Strains Harbouring Beta-Lactamase Genes from Clinical Samples" Pathogens 8, no. 4: 164. https://doi.org/10.3390/pathogens8040164
APA StyleAdesina, T., Nwinyi, O., De, N., Akinnola, O., & Omonigbehin, E. (2019). First Detection of Carbapenem-Resistant Escherichia fergusonii Strains Harbouring Beta-Lactamase Genes from Clinical Samples. Pathogens, 8(4), 164. https://doi.org/10.3390/pathogens8040164





