Abstract
Pneumococcal proteins unrelated to serotypes are considered to be candidates of antigens in next-generation vaccines. In the present study, the prevalence of vaccine candidate protein genes, along with serotypes and antimicrobial resistance determinants, was investigated in a total of 57 isolates obtained from a tertiary care hospital in Japan. All of the pediatric isolates and 76.6% of the adult isolates did not belong to PCV13 (a 13-valent pneumococcal conjugate vaccine) serotypes, and 70.2% of all isolates showed multidrug resistance. All of the isolates had ply, pavA, nanA, and nanB, and high prevalence was noted for the pspA and pspC genes (96.5% and 78.9%, respectively). Detection rates for the pneumococcal histidine triad protein (Pht) genes phtA, phtB, phtD, and phtE were 49.1%, 26.3%, 61.4%, and 100%, respectively. Two fusion-type genes, phtA/B and phtA/D, were identified, with a prevalence of 36.9% and 14.0%, respectively. These fusion types showed 78.1–90.0% nucleotide sequence identity with phtA, phtB, and phtD. The most prevalent pht profile was phtA + phtD + phtE (26.3%), followed by phtA/B + phtE (19.3%) and phtA/B + phtD + phtE (17.5%), while pht profiles including phtD and/or phtA/phtD were found in 71.9% of isolates. The present study revealed the presence of two fusion types of Pht and their unexpectedly high prevalence. These fusion types, as well as PhtA and PhtB, contained sequences similar to the B cell epitopes that have been previously reported for PhtD.
1. Introduction
Streptococcus pneumoniae (pneumococcus) occasionally causes both invasive and noninvasive pneumococcal disease (IPD and non-IPD, respectively), such as sepsis, meningitis, and community-acquired pneumonia [1], while this bacterium colonizes and persists on the human nasopharynx [2]. Pneumococcal diseases are considered preventable by vaccine. The capsular polysaccharide (CPS) is a principal virulence factor of S. pneumoniae and an essential component of commercially available vaccines against pneumococcal infections [3]. Currently, two classes of pneumococcal vaccines, a 23-valent pneumococcal polysaccharide vaccine (PPSV23) and 7-, 10-, and 13-valent pneumococcal conjugate vaccines (PCVs), both of which contain CPS as an immunogen, have been introduced in many countries. The unconjugated PPSV23 is widely offered to only adults because of poorer immunogenicity in children under 2 years of age [4,5], whereas the recently approved PCVs have been used in routine childhood vaccination programs across the world since 2000 [6].
Routine immunization with PCVs in children has greatly reduced the infections caused by pneumococci through the serotypes included in the vaccine (vaccine serotypes). However, following the introduction of PCV7 and PCV13 in children, the isolation rate of pneumococci with nonvaccine types increased globally as a result of the vaccine selection pressure, and recent studies in different countries have reported the occurrence of immediate chronological changes in serotypes [7,8,9]. In Japan, PCV7 was introduced in 2010 and was replaced with PCV13 in 2013. Our previous surveillance studies demonstrated that the rate of non-PCV13 serotypes in pediatric isolates increased from 39.7% in 2011 to 87.9% in 2016 [10,11]. The high prevalence rate of non-PCV13 serotypes was also documented in pediatric carriage isolates in recent studies [12,13]. Moreover, the widespread implementation of PCVs in children has been associated with the emergence and spread of drug-resistant clones with nonvaccine serotypes [14,15,16], posing a major public health concern. The currently available pneumococcal vaccines show protection against infections solely due to serotypes included in the vaccine, i.e., vaccine serotype-specific immunity, which is considered to be a limitation of the vaccines. For these reasons, the development of new pneumococcal vaccines in which the protective effect does not depend on serotypes has been anticipated.
Multiple pneumococcal proteins have been comparatively well investigated as promising targets for future non-serotype-specific protein-based pneumococcal vaccines, such as pneumococcal surface protein A (PspA) [17,18], pneumococcal surface protein C (PspC) [19,20], pneumococcal choline-binding protein (PcpA) [21], three neuraminidases (NanA, NanB, and NanC) [22,23], pneumolysin (Ply) [24], and four pneumococcal histidine triad proteins (PhtA, PhtB, PhtD, and PhtE) [25,26,27,28]. At present, an investigational pneumococcal vaccine is undergoing Phase I/II clinical trials in infants [29,30], children [31], and adults [32]. In these studies, two pneumococcal proteins, Ply toxoid (dPly) and PhtD, are included in the vaccines. A protein-based pneumococcal vaccine containing PhtD–dPly was shown to induce protection against pneumococcal pneumonia in a rhesus macaque study [33]. All four Pht proteins were exposed on the cell surface of pneumococcus and were characterized by histidine triad motifs in their amino acid sequences [34]. Among them, PhtD was described to be the least genetically variable [35] and was shown to induce an immune response in adults and infants [36], with immunodominant B cell epitopes being identified [27]. In addition to the PhtD protein, previous studies have indicated the presence of hybrid types of Pht in pneumococcus between PhtA and PhtB and between PhtA and PhtD [34,35]. However, genetic organization and sequences of the Pht hybrid/fusion types in S. pneumoniae have not yet been clearly described, and the prevalence of the hybrid types among clinical isolates remains to be determined.
The purpose of the present study was to investigate the prevalence of vaccine candidate protein genes in clinical isolates of S. pneumoniae collected from IPD and non-IPD patients, along with serotypes and antimicrobial resistance. Particularly, we focused on an identification and genetic analysis of the Pht fusion types that have not been well characterized, as well as the known Pht types. Consequently, we reveal the existence of two unique fusion types (PhtA/B and PhtA/D) in various serotypes of S. pneumoniae. The prevalence and profiles of authentic Pht and Pht fusion types in the clinical isolates are described in parallel.
2. Results
2.1. Serotypes and Sequence Types (STs) of Pneumococcal Isolates
For the 57 isolates analyzed, 21 different serotypes and 33 different STs, including two new STs, were identified. Serotypes and STs of all the isolates are listed in Table 1 with the designation of Pneumococcal Molecular Epidemiology Network (PMEN) international clones. All of the pediatric isolates belonged to non-PCV13 serotypes, i.e., 15B (ST199; Netherlands15B-37 PMEN clone), 23A (ST338; Colmbia23F-26 PMEN clone or its single-locus variant (SLV)), 24F (ST2572 or its SLV), or 35B (ST558, ST2755). Among isolates from adults, the three most prevalent serotypes were 15A (mostly ST63; Sweden15A-25 PMEN clone or its SLV, ST292 SLV), 3 (mostly ST180; Netherlands3-31 PMEN clone), and 35B (ST558, ST2755). Serotypes covered by PCV13 and PPSV23 in all adults were 23.4% (n = 11/47) and 44.7% (n = 21/47), respectively.

Table 1.
Serotypes and multilocus sequence types of all the isolates analyzed in this study.
2.2. Detection of Drug Resistance Genes and Antimicrobial Susceptibility
The antimicrobial susceptibility of all the isolates with individual serotypes is shown in the Supplementary Materials, Table S1. The highest nonsusceptibility rates were observed for erythromycin (91.2%) and tetracycline (86.0), which corresponded to the high prevalence of resistance genes to macrolides (erm(B) and/or mef(A/E)) and tetracycline (tetM) (94.7% and 93.0%, respectively). Nonsusceptibility to penicillin was detected in 38.9% of isolates, among which two isolates (serotypes 6C and 15A) were resistant to penicillin. All of the isolates showing nonsusceptibility to penicillin had the penicillin-binding protein (PBP) genotype penicillin-resistant S. pneumoniae (gPRSP) (alterations in three genes: pbp1a, pbp2x, and pbp2b). The prevalence of the multidrug resistance (MDR) phenotype (defined as resistance to three or more different classes of antibiotics) was 70.2%. Three MDR isolates (5.3%, 3/57) belonging to serotypes 14 (n = 1) and 15A (n = 2) were resistant to levofloxacin, which is associated with mutations in the quinolone resistance-determining region (QRDR) (positions of mutations are shown in the Supplementary Materials, Table S1, footnote).
2.3. Detection of the Vaccine Candidate Pneumococcal Protein Genes and the Novel Pht Fusion Proteins
The prevalence of various pneumococcal proteins among individual serotypes is summarized in Table 2. All of the isolates carried the ply and pavA genes. The prevalence of the pspA and pspC genes was 96.5% and 78.9%, respectively. Among the three nan genes, nanA and nanB were detected in all of the isolates, while nanC was in 52.6% of the isolates. Detection rates for phtA, phtB, phtD, and phtE were 49.1%, 26.3%, 61.4%, and 100%, respectively. In addition to the four pht genes, two fusion-type genes, phtA/B and phtA/D, were identified, with the prevalence at 36.9% and 14.0%, respectively. Two serotype 15B isolates had both phtD and phtA/D.

Table 2.
Prevalence of pneumococcal protein genes in all isolates among individual serotypes.
2.4. Prevalence of the Pht Pattern and Sequence Analysis of the Pht Fusion Types
Pht profiles are summarized in Table 3. The most prevalent pht pattern was phtA + phtD + phtE (26.3%), followed by phtA/B + phtE (19.3%), phtA/B + phtD + phtE (17.5%), and phtA + phtB + phtD + phtE (14.0%). Pht gene profiles, including pht fusion type, were detected in 50.9% of all isolates. Structural organizations of the fusion types PhtA/B and PhtA/D (compared to PhtA, PhtB, or PhtD) are schematically represented in Figure 1 and Figure 2, respectively. The sequence alignment of these genes is shown in the Supplementary Materials, Figures S1 and S2. These fusion-type Pht proteins were constituted by the N-terminal half of the phtA-like region and the phtB- or phtD-like region in the C-terminal side. The phtA-like region of phtA/B from the four isolates (aa.1–387) had 94.5–97.1% sequence identity with phtA in a reference strain (AF291695) and 76.7–77.9% identity with phtB in a reference strain (AF318954). In contrast, the phtB-like region (aa.387–) of phtA/B showed 99.3–99.6% identity with the phtB sequence. Similarly, phtA/D of the four representative isolates showed 93.3–97.0% identity with phtA (AF291695) in the phtA-like region and 97.9–99.7% identity with phtD (AF318955 and KP127692) in the phtD-like region. The nucleotide sequence identity within phtA/B was 96.6–98.4%, and similarly, 97.6–97.9% identity was found within phtA/D (Supplementary Materials, Table S2). In the present study, nucleotide sequences of phtB were determined for five isolates (Supplementary Materials, Table S3), which showed 97.6–99.4% identity with the phtB gene reference strain AF318954. In addition, phtA had 69.2–70.5% identity with phtB and phtD; 92.6–93.0% identity was found between phtB and phtD; and phtA/B and phtA/D had 78.1–90.0% identity with phtA, phtB, and phtD.

Table 3.
Pht profiles of all the Streptococcus pneumoniae isolates.
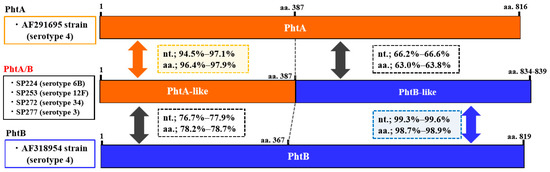
Figure 1.
Genetic structure of the phtA/B (fusion type of phtA and phtB) gene identified in the present study and phtA and phtB in reference strains (GenBank Accession Nos. AF291695 and AF318954, respectively). The serotype of each isolate is indicated in parentheses. The phtA/phtA-like and phtB/phtB-like sequences are shown in orange and blue, respectively. Nucleotide (nt.) and amino acid (aa.) sequence identities of phtA-like and phtB-like regions of phtA/B gene with those of phtA and phtB genes are shown in squares with dotted lines.
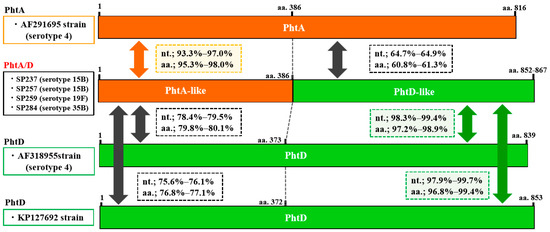
Figure 2.
Genetic structure of the phtA/D (fusion type of phtA and phtD) gene identified in the present study and phtA and phtD in reference strains (GenBank Accession Nos. AF291695, AF318954, and KP127692, respectively). The serotype of each isolate is indicated in parentheses. The phtA/phtA-like and phtD/phtD-like sequences are shown in orange and green, respectively. Nucleotide (nt.) and amino acid (aa.) sequence identities of phtA-like and phtD-like regions of phtA/D gene with those of phtA and phtD genes are shown in squares with dotted lines.
2.5. Sequences of B Cell Epitopes in PhtA, PhtB, PhtD, and Pht Fusion Types
Immunodominant B cell epitopes in PhtD have been identified by Lagousi and coworkers [27] and have been mapped into three regions, i.e., amino acids 88–107 (epitope I, pep11), 172–191 (epitope II, pep17), and 200–219 (epitope III, pep19). The alignment of Pht amino acid sequences indicated that the three epitopes were highly conserved in different Pht proteins, including the fusion types (Supplementary Materials, Figures S1–S3). In particular, epitopes I and III showed almost identical sequences, with only a few substitutions with similar amino acids.
3. Discussion
In the present study, we investigated the prevalence of 15 genes in promising vaccine candidate proteins and revealed the existence of two Pht fusion types besides the four known Pht proteins in pneumococcus. To our knowledge, this is the first report to show the prevalence of the fusion types PhtA/B and PhtA/D in various serotypes of pneumococcal isolates.
At present, for protein-based vaccines, pneumococcal proteins such as PhtD [37], dPly [38], PspA [39], and PcpA [40] have been proven to show effective protection against pneumococcal diseases in studies on human subjects. Further, a pneumococcal protein-based vaccine containing the PhtD protein and/or dPly has been investigated in a Phase I/II randomized clinical study [29,30,31,32]. In our present study, while the prevalence of ply was high (100%), phtD was detected in 61.4% of all isolates. A similar prevalence of phtD (61.0%) was reported in pneumococcal isolates from meningitis in France [41]. In contrast, Rioux et al. reported a high prevalence of phtD (100%) in the pneumococcal strains analyzed [35]. In the present study, we identified the Pht fusion types in clinical isolates of pneumococcus, which were classified into two distinct types, PhtA/B and PhtA/D. The prevalence of phtA/B and phtA/D was 36.9% and 14.0%, respectively, and phtD and/or phtA/D were found in 71.9% of all the isolates. Although the number of isolates analyzed in the present study was not sufficient, the prevalence of phtD and/or phtA/D was considered to be high (>70%). The prevalence of phtD (phtA/D) may vary depending on study subjects (e.g., serotypes/genotypes of the clinical isolates, country, infection types) and will be further clarified if a detection method for the fusion type phtA/D is established.
Although only two studies have described the hybrid/fusion types of Pht in pneumococcus to date [34,35], their sequence data have not yet been published. In our present study, sequences and genetic organizations of the hybrid types of pht genes were revealed for the first time, and these were found to be distributed to half of the isolates, including 13 different serotypes. Furthermore, in a BLAST search, we found that the sequences of phtA/B (strain SP224, serotype 6B) and phtA/D (strain SP284, serotype 35B) in the present study were similar to those in the S. pneumoniae complete genome of strain G54 (serotype 19F, GenBank Accession No. CP001015) (99.2% identity) [42] and strain Sp99_4038 (serotype 3, GenBank Accession No. FQ312041) (98.4% identity) [43], respectively. These findings suggest that the fusion types of Pht may be commonly distributed to clinical isolates of pneumococci with various serotypes.
Adamou et al. first reported pneumococcal pht genes [44] and described PCR primers to detect phtA, phtB, phtD, and phtE, which were used for their detection in a previous study [41]. However, hybrid types are not detected by uniplex PCR with these primers, and it is also possible that the hybrid types may be misclassified as phtA, phtB, or phtD by nonspecific PCR amplification due to sequence diversity in the pht genes. In contrast, there have been few studies that have determined full-length pht genes that determine Pht types. These may be possible reasons that fusion-type Pht has been rarely reported.
The most significant finding in the present study is that the fusion types PhtA/B and PhtA/D had almost identical sequences to B cell epitopes that have been reported for PhtD previously [27], despite overall sequence diversity in Pht. This finding suggests that S. pneumoniae with the fusion type Pht may also be recognized by antibodies to PhtD. These B cell epitopes of PhtD are also conserved in PhtA and PhtB [27]. In the present study, 26.3% of isolates (n = 15) had only fusion-type pht genes (phtA/B or phtA/D), except for phtE. These isolates could be judged as negative for phtA, phtB, and phtD by using the previously reported PCR scheme [41] as described above, and therefore these isolates are not regarded as being protected by an immune response to PhtD. However, our present study revealed that all of the S. pneumoniae isolates examined (belonging to various serotypes) possessed one or more of phtA, phtB, phtD, phtA/B, or phtA/D. This finding may suggest the possibility that PhtD is useful as a broadly protective pneumococcal vaccine.
In the present study, the most common serotypes were 15A, 3, 6C, and 35B. The prevalence of MDR was 70.2%, and high rates of nonsusceptibility to penicillin were notable for non-PCV13 serotypes 6C, 15A, and 35B. In studies in the United Kingdom [15] and Germany [14], an increase of MDR serotype 15A was observed in the PCV vaccination era, and the major MDR serotypes 15A, 6C, and 35B were noted in the USA [45]. Further, in Canada, the high prevalence has been reported for MDR serotypes 15A and 35B, which are related to the Sweden15A-25 PMEN clone and the Utah35B-24 PMEN clone, respectively [15]. The recent trends from various countries were also observed in our present study in Japan, suggesting concerns about the dissemination of MDR clones with non-PCV13 serotypes. Taken together, the worldwide spread of non-PCV13 serotypes with multidrug resistance may lead to a limit in the effectiveness of antimicrobial therapy and current vaccination; thus, the development of novel effective vaccines that are irrespective of prevailing serotypes is anticipated.
The limitations of this study were its small sample size and the fact that most isolates were collected from noninvasive infections or colonization in a single hospital. PhtD is one of the promising vaccine candidate proteins and has been one of the most well studied [25,34,36,37]. In this regard, for basic information, further epidemiological studies are necessary on the prevalence of pneumococcal proteins in clinical isolates, especially Pht proteins, in various regions and countries.
4. Materials and Methods
4.1. Pneumococcal Isolates
From March 2016 to February 2018, 57 nonduplicate S. pneumoniae clinical isolates from consecutive patients with pneumococcal diseases (either invasive (four isolates from blood) or noninvasive (53 isolates from sputum, nasal discharges, or other nonsterile sites) infections) were collected at the Sapporo Medical University Hospital, Hokkaido, on the northern main island of Japan. Among the isolates studied, 10 and 47 isolates were obtained from children (age < 16 years) and adults (age ≥ 16 years), respectively, and the male/female ratio was 1.1 (27/25). S. pneumoniae characteristics were identified by an automated bacterial identification and susceptibility testing system (MicroScan® WalkAway 96 plus system MicroScan; SIEMENS Healthcare Diagnostics) and were confirmed by the detection of the lytA gene using PCR, as described previously [46]. Isolates were stored in a Microbank (Pro-lab Diagnostics, Richmond Hill, Canada) at −80 °C. Frozen isolates were inoculated onto a blood agar base supplemented with 5% sheep blood (Nippon Becton Dickinson) and were incubated at 37 °C with 5% CO2 for 24 h before further analysis.
In the present study, no human participants were involved directly. Hence, human ethics clearance was not required. We analyzed bacterial isolates as study subjects, which had already been isolated from clinical samples through routine bacteriological examination in our university hospital.
4.2. Total DNA Extraction and Sequencing
Genomic DNA was extracted from each isolate as described preciously [47] and was used as a template in all PCR reactions. For determination of the nucleotide sequence, the purified PCR products were sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an automated DNA sequencer (ABI PRISM 3130).
4.3. Serotyping, Virulence Gene Detection, and Multilocus Sequence Typing (MLST)
All pneumococcal isolates were subjected to serotyping, virulence gene identification, and genotyping through an MLST scheme. The serotyping of pneumococcal isolates was performed by PCR-based deduction protocols [48] with serogroup/serotype-specific primers (described on the CDC website (http://www.cdc.gov/streplab/pcr.html)). After the PCRs, additional subtyping was performed by PCR-based sequencing methods, as described in our previous studies [11,49,50,51,52]. Fourteen virulence-associated genes, pht (A, B, D, and E), pspA (Family 1, 2, or 3), pspC, pspC.4, nan (A, B, and C), pcpA, psrp, ply, and pavA, were examined by uniplex PCR with the specific primers reported previously [34,41]. MLST was performed as described on the PubMLST website (http://pubmlst.org/spneumoniae) with modified primers (http://www.cdc.gov/streplab/alt-mlst-primers.html). Subsequently, the obtained STs were compared to Pneumococcal Molecular Epidemiology Network (PMEN) international clones (http://www.sph.emory.edu/PMEN). Allelic numbers/locus sequences of untypable STs were submitted to the PubMLST database curator for assignment of new STs.
4.4. Antimicrobial Resistance Determinants
The minimum inhibitory concentrations (MICs) of all isolates against 10 antimicrobial agents (penicillin (PEN), erythromycin (ERY), tetracycline (TET), clindamycin (CLI), trimethoprim-sulfamethoxazole (SXT), ceftriaxone (CRO), cefaclor (CEC), imipenem (IPM), levofloxacin (LVX), and vancomycin (VAN)) were measured through the broth microdilution method using a Dry Plate (Eiken, Tokyo, Japan) as described previously [49] and were interpreted as susceptible (S), intermediate (I), or resistant (R) according to Clinical and Laboratory Standards Institute guidelines (CLSI 2015). The CLSI provides breakpoints as follows: PEN (I = 0.12–1 μg/mL, R ≥ 2 μg/mL), ERY (I = 0.5 μg/mL, R ≥ 1 μg/mL), TET (I = 2 μg/mL, R ≥ 4 μg/mL), CLI (I = 0.5 μg/mL, R ≥ 1 μg/mL), SXT (I = 1/19–2/38 μg/mL, R ≥ 4/76 μg/mL), CRO (I = 2 μg/mL, R ≥ 4 μg/mL), CEC (I = 2 μg/mL, R ≥ 4 μg/mL), IPM (I = 0.25–0.5 μg/mL, R ≥ 1 μg/mL), LVX (I = 4 μg/mL, R ≥ 8 μg/mL), and VAN (S ≤ 1 μg/mL). Multidrug resistance (MDR) was defined as resistance to three or more different antimicrobial agent classes (penicillin resistance was defined using the CLSI breakpoint for oral penicillin V, MIC ≥ 2 μg /mL) [53,54].
For all isolates, alterations of the PBP genes (pbp1a, pbp2x, and pbp2b) and the presence of macrolide (erm(B), mef(A/E))- and tetracycline (tetM)- resistant genes were confirmed by a PCR or multiplex PCR assay [46,55,56]. For three isolates that showed resistance to LVX (MIC of ≥ 8 μg/mL), quinolone resistance-determining region (QRDR) mutations of the DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) genes were investigated by direct sequencing with PCR products, as described previously [57].
4.5. Detection and Sequence Analysis of PhtA/B and PhtA/D Fusion Types
Fusion types of PhtA/B and PhtA/D genes were detected, and their sequences were analyzed as follows. First, to identify the fusion type-associated phtA gene, all of the isolates were subjected to two PCRs to detect 5′- and 3′-half regions of the phtA gene. For these PCRs, two primer pairs were designed on the basis of the published phtA sequence (AF291695): PhtA-5’F (5′-ACATCGTGAAGGTGGAACTCC-3′) and PhtA-5′R (5′-GTGTTATCGCTATTTTGTCG-3′) for the 5′-end region (product size 273 bp) and PhtA-3′F (5′-GCCAGTAGAGGAAACACCTGC-3′) and PhtA-3′R (5′-TATCCATAATTTGAAGAGTC-3′) for the 3′-end region (product size 186 bp). The PCR program consisted of the following steps: initial denaturation at 94 °C for 2 min, followed by 35 cycles at 94 °C for 15 s, 55 °C for 150 s, and 72 °C for 15 s, followed by a final extension step at 72 °C for 3 min. Among all isolates, 28 isolates (49.1%) were positive for phtA in the two PCRs as well as in the initial PCR for phtA gene detection.
Second, for all of the remaining 29 isolates (50.9%) that had only a 5’-end region of phtA (negative for 3’-end region of phtA), PCR was attempted with a forward primer of phtA (phtA primer-F) and reverse primers specific to phtB (phtB primer-R) or phtD (phtD primer-R) with the PCR program and conditions described previously [34]. Using the obtained RCR products, pht gene sequences were determined through the Sanger method. Finally, 5’-end and 3’-end portions of pht gene sequences that were not covered by the above PCR were determined by PCR and direct sequencing using the primers PhtA.F-5’outer (5’-AAGTCCAACCTTGAAAAAGTAGTGG-3’, for phtA), PhtB.R-3’outer (5’-GAACTAGAACTCACATTCTGC-3’, for phtB), and phtD.R-3’outer (5’-TAACAGCTGATCCAGCTGC-3’, for phtD). Sequence data (full-length, 5’-end, and 3’-end half regions) of the presumptive fusion type genes were further analyzed for their highly similar sequences in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) by using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple alignments of nucleotide and amino acid sequences for the fusion type pht and authentic phtA, phtB, phtD, and phtE were performed by the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/), which was used for the calculation of sequence identities between them. In addition, we determined sequences of phtB for a representative five isolates because only a few phtB genes have been deposited into the GenBank database.
4.6. GenBank Accession Numbers
The nucleotide sequences of Pht fusion type phtA/B and phtA/D genes and the phtB gene were deposited into the GenBank database under accession numbers MN206792 to MN206804, and they are listed in the Supplementary Materials, Table S3.
5. Conclusions
In conclusion, our study revealed the prevalence of 14 vaccine candidate protein genes in clinical isolates of S. pneumoniae and demonstrated the existence of PhtA/B and PhtA/D fusion types in various pneumococcal serotypes. These fusion types, as well as PhtA and PhtB, contained sequences of B cell epitopes similar to those previously reported for PhtD, which is included in the investigational protein-based pneumococcal vaccine presently. Despite the small number and limited source of isolates, PhtD and PhtA/D were detected in 61.4% and 14.0% of all isolates. However, all of the isolates with various serotypes had one or more of PhtA, PhtB, PhtD, and fusion types PhtA/B and PhtA/D, suggesting that an immune response to PhtD may confer protective immunity to S. pneumoniae irrespective of serotype. Further epidemiological studies on higher numbers of isolates from various sources in various regions are required to determine the prevalence and profiles of pht genes among pneumococci as basic information for the development of Pht-based vaccines.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/162/s1, Figure S1: Alignment of PhtA/B (fusion type) and PhtA (a)/PhtB (b) amino acid sequences; Figure S2: Alignment of PhtA/D (fusion type) and PhtA (a)/PhtD (b) amino acid sequences; Figure S3: Amino acid sequence alignment of three B cell epitope regions (I, II, III) of PhtA, PhtB, PhtD, and Pht fusion types (PhtA/B, PhtA/D); Table S1: Resistance gene profiles and antimicrobial susceptibility of the 57 isolates among individual serotypes; Table S2: Percent identity matrix based on nucleotide (upper right) and amino acid (lower left) sequences of phtA, phtB, phtD, phtE, and pht fusion types; Table S3: GenBank accession numbers assigned for phtA/B, phtA/D, and phtB genes of representative pneumococcal isolates analyzed in the present study.
Author Contributions
Conceptualization, M.K. and N.K.; methodology, M.K and N.K.; investigation, M.K., N.U., and M.S.A.; resources, M.S. and S.T.; data curation, M.K.; writing—original draft preparation, N.K. and M.K.; writing—review and editing, M.K. and N.K.; supervision, N.K.; funding acquisition, M.K.
Funding
This research was partially supported by JSPS (Japan Society for the Promotion of Science) KAKENHI, Grant Nos. 16K09101 and 19K10603.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Weiser, J.N.; Ferreira, D.M.; Paton, J.C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 2018, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Kavalari, I.D.; Fuursted, K.; Krogfelt, K.A.; Slotved, H.C. Molecular characterization and epidemiology of Streptococcus pneumoniae serotype 24F in Denmark. Sci. Rep. 2019, 9, 5481. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, A.L.; Rumke, L.W.; Arp, K.; Bosch, A.; Bruin, J.P.; Rots, N.Y.; Wijmenga-Monsuur, A.J.; Sanders, E.A.M.; Trzcinski, K. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci. Rep. 2016, 6, 34888. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.C.; Rogers, P.D.; Shelton, C.M. A Review of Pneumococcal Vaccines: Current Polysaccharide Vaccine Recommendations and Future Protein Antigens. J. Pediatric Pharmacol. Ther. 2016, 21, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Moseley, M.A.; Burton, R.L.; Nahm, M.H. Pneumococcal vaccine and opsonic pneumococcal antibody. J. Infect. Chemother. 2013, 19, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Feikin, D.R.; Kagucia, E.W.; Loo, J.D.; Link-Gelles, R.; Puhan, M.A.; Cherian, T.; Levine, O.S.; Whitney, C.G.; O’Brien, K.L.; Moore, M.R. Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: A pooled analysis of multiple surveillance sites. PLoS Med. 2013, 10, e1001517. [Google Scholar] [CrossRef] [PubMed]
- Gladstone, R.A.; Devine, V.; Jones, J.; Cleary, D.; Jefferies, J.M.; Bentley, S.D.; Faust, S.N.; Clarke, S.C. Pre-vaccine serotype composition within a lineage signposts its serotype replacement—A carriage study over 7 years following pneumococcal conjugate vaccine use in the UK. Microb. Genom. 2017, 3, e000119. [Google Scholar] [CrossRef] [PubMed]
- Quirk, S.J.; Haraldsson, G.; Erlendsdottir, H.; Hjalmarsdottir, M.A.; van Tonder, A.J.; Hrafnkelsson, B.; Sigurdsson, S.; Bentley, S.D.; Haraldsson, A.; Brueggemann, A.B.; et al. Effect of Vaccination on Pneumococci Isolated from the Nasopharynx of Healthy Children and the Middle Ear of Children with Otitis Media in Iceland. J. Clin. Microbiol. 2018, 56, e01046-18. [Google Scholar] [CrossRef]
- Ladhani, S.N.; Collins, S.; Djennad, A.; Sheppard, C.L.; Borrow, R.; Fry, N.K.; Andrews, N.J.; Miller, E.; Ramsay, M.E. Rapid increase in non-vaccine serotypes causing invasive pneumococcal disease in England and Wales, 2000–2017: A prospective national observational cohort study. Lancet Infect. Dis. 2018, 18, 441–451. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Aung, M.S.; Morimoto, S.; Ito, M.; Kudo, K.; Sumi, A.; Kobayashi, N. Emerging non-PCV13 serotypes of noninvasive Streptococcus pneumoniae with macrolide resistance genes in northern Japan. New Microbes New Infect. 2016, 9, 66–72. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Ghosh, S.; Kuwahara, O.; Morimoto, S.; Ito, M.; Kudo, K.; Kobayashi, N. Serotype distribution and susceptibility to penicillin and erythromycin among noninvasive or colonization isolates of Streptococcus pneumoniae in northern Japan: A cross-sectional study in the pre-PCV7 routine immunization period. Microb. Drug Resist. 2014, 20, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Southern, J.; Andrews, N.; Sandu, P.; Sheppard, C.L.; Waight, P.A.; Fry, N.K.; Van Hoek, A.J.; Miller, E. Pneumococcal carriage in children and their household contacts six years after introduction of the 13-valent pneumococcal conjugate vaccine in England. PLoS ONE 2018, 13, e0195799. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, E.; Sahin-Toth, J.; Tothpal, A.; Kristof, K.; van der Linden, M.; Tirczka, T.; Dobay, O. Vaccine-driven serotype-rearrangement is seen with latency in clinical isolates: Comparison of carried and clinical pneumococcal isolates from the same time period in Hungary. Vaccine 2019, 37, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Van der Linden, M.; Perniciaro, S.; Imohl, M. Increase of serotypes 15A and 23B in IPD in Germany in the PCV13 vaccination era. BMC Infect. Dis. 2015, 15, 207. [Google Scholar] [CrossRef] [PubMed]
- Golden, A.R.; Adam, H.J.; Gilmour, M.W.; Baxter, M.R.; Martin, I.; Nichol, K.A.; Demczuk, W.H.; Hoban, D.J.; Zhanel, G.G. Assessment of multidrug resistance, clonality and virulence in non-PCV-13 Streptococcus pneumoniae serotypes in Canada, 2011–2013. J. Antimicrob. Chemother. 2015, 70, 1960–1964. [Google Scholar] [PubMed]
- Sheppard, C.; Fry, N.K.; Mushtaq, S.; Woodford, N.; Reynolds, R.; Janes, R.; Pike, R.; Hill, R.; Kimuli, M.; Staves, P.; et al. Rise of multidrug-resistant non-vaccine serotype 15A Streptococcus pneumoniae in the United Kingdom, 2001 to 2014. Eurosurveillance 2016, 21, 30423. [Google Scholar] [CrossRef] [PubMed]
- Briles, D.E.; Tart, R.C.; Swiatlo, E.; Dillard, J.P.; Smith, P.; Benton, K.A.; Ralph, B.A.; Brooks-Walter, A.; Crain, M.J.; Hollingshead, S.K.; et al. Pneumococcal diversity: Considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 1998, 11, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Miyaji, E.N.; Ferreira, D.M.; Lopes, A.P.; Brandileone, M.C.; Dias, W.O.; Leite, L.C. Analysis of serum cross-reactivity and cross-protection elicited by immunization with DNA vaccines against Streptococcus pneumoniae expressing PspA fragments from different clades. Infect. Immun. 2002, 70, 5086–5090. [Google Scholar] [CrossRef]
- Moreno, A.T.; Oliveira, M.L.; Ho, P.L.; Vadesilho, C.F.; Palma, G.M.; Ferreira, J.M., Jr.; Ferreira, D.M.; Santos, S.R.; Martinez, M.B.; Miyaji, E.N. Cross-reactivity of antipneumococcal surface protein C (PspC) antibodies with different strains and evaluation of inhibition of human complement factor H and secretory IgA binding via PspC. Clin. Vaccine Immunol. 2012, 19, 499–507. [Google Scholar] [CrossRef]
- Vadesilho, C.F.; Ferreira, D.M.; Gordon, S.B.; Briles, D.E.; Moreno, A.T.; Oliveira, M.L.; Ho, P.L.; Miyaji, E.N. Mapping of epitopes recognized by antibodies induced by immunization of mice with PspA and PspC. Clin. Vaccine Immunol. 2014, 21, 940–948. [Google Scholar] [CrossRef]
- Visan, L.; Rouleau, N.; Proust, E.; Peyrot, L.; Donadieu, A.; Ochs, M. Antibodies to PcpA and PhtD protect mice against Streptococcus pneumoniae by a macrophage- and complement-dependent mechanism. Hum. Vaccines Immunother. 2018, 14, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Simell, B.; Jaakkola, T.; Lahdenkari, M.; Briles, D.; Hollingshead, S.; Kilpi, T.M.; Kayhty, H. Serum antibodies to pneumococcal neuraminidase NanA in relation to pneumococcal carriage and acute otitis media. Clin. Vaccine Immunol. 2006, 13, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Janapatla, R.P.; Chen, C.L.; Hsu, M.H.; Liao, W.T.; Chiu, C.H. Immunization with pneumococcal neuraminidases NanA, NanB and NanC to generate neutralizing antibodies and to increase survival in mice. J. Med. Microbiol. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Surendran, N.; Ochs, M.; Pichichero, M.E. Human antibodies to PhtD, PcpA, and Ply reduce adherence to human lung epithelial cells and murine nasopharyngeal colonization by Streptococcus pneumoniae. Infect. Immun. 2014, 82, 5069–5075. [Google Scholar] [CrossRef] [PubMed]
- Godfroid, F.; Hermand, P.; Verlant, V.; Denoel, P.; Poolman, J.T. Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect. Immun. 2011, 79, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Melin, M.; Di Paolo, E.; Tikkanen, L.; Jarva, H.; Neyt, C.; Kayhty, H.; Meri, S.; Poolman, J.; Vakevainen, M. Interaction of pneumococcal histidine triad proteins with human complement. Infect. Immun. 2010, 78, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Lagousi, T.; Routsias, J.; Piperi, C.; Tsakris, A.; Chrousos, G.; Theodoridou, M.; Spoulou, V. Discovery of Immunodominant B Cell Epitopes within Surface Pneumococcal Virulence Proteins in Pediatric Patients with Invasive Pneumococcal Disease. J. Biol. Chem. 2015, 290, 27500–27510. [Google Scholar] [CrossRef] [PubMed]
- Kallio, A.; Sepponen, K.; Hermand, P.; Denoel, P.; Godfroid, F.; Melin, M. Role of Pht proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect. Immun. 2014, 82, 1683–1691. [Google Scholar] [CrossRef]
- Odutola, A.; Ota, M.O.C.; Antonio, M.; Ogundare, E.O.; Saidu, Y.; Foster-Nyarko, E.; Owiafe, P.K.; Ceesay, F.; Worwui, A.; Idoko, O.T.; et al. Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: A phase 2, randomized, controlled, observer-blind study. Vaccine 2017, 35, 2531–2542. [Google Scholar] [CrossRef]
- Odutola, A.; Ota, M.O.C.; Antonio, M.; Ogundare, E.O.; Saidu, Y.; Owiafe, P.K.; Worwui, A.; Idoko, O.T.; Owolabi, O.; Kampmann, B.; et al. Immunogenicity of pneumococcal conjugate vaccine formulations containing pneumococcal proteins, and immunogenicity and reactogenicity of co-administered routine vaccines—A phase II, randomised, observer-blind study in Gambian infants. Vaccine 2019, 37, 2586–2599. [Google Scholar] [CrossRef]
- Odutola, A.; Ota, M.O.; Ogundare, E.O.; Antonio, M.; Owiafe, P.; Worwui, A.; Greenwood, B.; Alderson, M.; Traskine, M.; Verlant, V.; et al. Reactogenicity, safety and immunogenicity of a protein-based pneumococcal vaccine in Gambian children aged 2–4 years: A phase II randomized study. Hum. Vaccines Immunother. 2016, 12, 393–402. [Google Scholar] [CrossRef]
- Leroux-Roels, G.; Maes, C.; De Boever, F.; Traskine, M.; Ruggeberg, J.U.; Borys, D. Safety, reactogenicity and immunogenicity of a novel pneumococcal protein-based vaccine in adults: A phase I/II randomized clinical study. Vaccine 2014, 32, 6838–6846. [Google Scholar] [CrossRef] [PubMed]
- Denoel, P.; Philipp, M.T.; Doyle, L.; Martin, D.; Carletti, G.; Poolman, J.T. A protein-based pneumococcal vaccine protects rhesus macaques from pneumonia after experimental infection with Streptococcus pneumoniae. Vaccine 2011, 29, 5495–5501. [Google Scholar] [CrossRef] [PubMed]
- Adamou, J.E.; Heinrichs, J.H.; Erwin, A.L.; Walsh, W.; Gayle, T.; Dormitzer, M.; Dagan, R.; Brewah, Y.A.; Barren, P.; Lathigra, R.; et al. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 2001, 69, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Rioux, S.; Neyt, C.; Di Paolo, E.; Turpin, L.; Charland, N.; Labbe, S.; Mortier, M.C.; Mitchell, T.J.; Feron, C.; Martin, D.; et al. Transcriptional regulation, occurrence and putative role of the Pht family of Streptococcus pneumoniae. Microbiology 2011, 157, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Holmlund, E.; Quiambao, B.; Ollgren, J.; Jaakkola, T.; Neyt, C.; Poolman, J.; Nohynek, H.; Kayhty, H. Antibodies to pneumococcal proteins PhtD, CbpA, and LytC in Filipino pregnant women and their infants in relation to pneumococcal carriage. Clin. Vaccine Immunol. 2009, 16, 916–923. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seiberling, M.; Bologa, M.; Brookes, R.; Ochs, M.; Go, K.; Neveu, D.; Kamtchoua, T.; Lashley, P.; Yuan, T.; Gurunathan, S. Safety and immunogenicity of a pneumococcal histidine triad protein D vaccine candidate in adults. Vaccine 2012, 30, 7455–7460. [Google Scholar] [CrossRef]
- Berglund, J.; Vink, P.; Tavares Da Silva, F.; Lestrate, P.; Boutriau, D. Safety, immunogenicity, and antibody persistence following an investigational Streptococcus pneumoniae and Haemophilus influenzae triple-protein vaccine in a phase 1 randomized controlled study in healthy adults. Clin. Vaccine Immunol. 2014, 21, 56–65. [Google Scholar] [CrossRef]
- Nabors, G.S.; Braun, P.A.; Herrmann, D.J.; Heise, M.L.; Pyle, D.J.; Gravenstein, S.; Schilling, M.; Ferguson, L.M.; Hollingshead, S.K.; Briles, D.E.; et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 2000, 18, 1743–1754. [Google Scholar] [CrossRef]
- Bologa, M.; Kamtchoua, T.; Hopfer, R.; Sheng, X.; Hicks, B.; Bixler, G.; Hou, V.; Pehlic, V.; Yuan, T.; Gurunathan, S. Safety and immunogenicity of pneumococcal protein vaccine candidates: Monovalent choline-binding protein A (PcpA) vaccine and bivalent PcpA-pneumococcal histidine triad protein D vaccine. Vaccine 2012, 30, 7461–7468. [Google Scholar] [CrossRef]
- Blumental, S.; Granger-Farbos, A.; Moisi, J.C.; Soullie, B.; Leroy, P.; Njanpop-Lafourcade, B.M.; Yaro, S.; Nacro, B.; Hallin, M.; Koeck, J.L. Virulence Factors of Streptococcus pneumoniae. Comparison between African and French Invasive Isolates and Implication for Future Vaccines. PLoS ONE 2015, 10, e0133885. [Google Scholar] [CrossRef] [PubMed]
- Dopazo, J.; Mendoza, A.; Herrero, J.; Caldara, F.; Humbert, Y.; Friedli, L.; Guerrier, M.; Grand-Schenk, E.; Gandin, C.; de Francesco, M.; et al. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 2001, 7, 99–125. [Google Scholar] [CrossRef] [PubMed]
- Donati, C.; Hiller, N.L.; Tettelin, H.; Muzzi, A.; Croucher, N.J.; Angiuoli, S.V.; Oggioni, M.; Dunning Hotopp, J.C.; Hu, F.Z.; Riley, D.R.; et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010, 11, R107. [Google Scholar] [CrossRef] [PubMed]
- Adam, H.J.; Golden, A.R.; Karlowsky, J.A.; Baxter, M.R.; Nichol, K.A.; Martin, I.; Demczuk, W.; Mulvey, M.R.; Gilmour, M.W.; Hoban, D.J.; et al. Analysis of multidrug resistance in the predominant Streptococcus pneumoniae serotypes in Canada: The SAVE study, 2011–2015. J. Antimicrob. Chemother. 2018, 73, vii12–vii19. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.S.; Diekema, D.J.; Heilmann, K.P.; Dohrn, C.L.; Riahi, F.; Doern, G.V. Changes in pneumococcal serotypes and antimicrobial resistance after introduction of the 13-valent conjugate vaccine in the United States. Antimicrob. Agents Chemother. 2014, 58, 6484–6489. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Shibasaki, Y.; Hasegawa, K.; Davies, T.A.; Jacobs, M.R.; Ubukata, K.; Appelbaum, P.C. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and beta-lactam resistance, and to detect common macrolide resistance determinants. J. Antimicrob. Chemother. 2001, 48, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Wu, H.; Kojima, K.; Taniguchi, K.; Urasawa, S.; Uehara, N.; Omizu, Y.; Kishi, Y.; Yagihashi, A.; Kurokawa, I. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol. Infect. 1994, 113, 259–266. [Google Scholar] [CrossRef]
- Pai, R.; Gertz, R.E.; Beall, B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 2006, 44, 124–131. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Aung, M.S.; Habadera, S.; Ito, M.; Kudo, K.; Kobayashi, N. Association Between Pneumococcal Surface Protein A Family and Genetic/Antimicrobial Resistance Traits of Non-Invasive Pneumococcal Isolates from Adults in Northern Japan. Microb. Drug Resist. 2019, 25, 744–751. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Aung, M.S.; Morimoto, S.; Ito, M.; Kudo, K.; Kobayashi, N. Genetic diversity of pneumococcal surface protein A (PspA) in paediatric isolates of non-conjugate vaccine serotypes in Japan. J. Med. Microbiol. 2018, 67, 1130–1138. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Aung, M.S.; Shinagawa, M.; Takahashi, S.; Kobayashi, N. Serotype distribution, antimicrobial resistance and prevalence of pilus islets in pneumococci following the use of conjugate vaccines. J. Med. Microbiol. 2017, 66, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchiya, M.; Urushibara, N.; Kobayashi, N. High prevalence of genotype 6E (putative serotype 6E) among noninvasive/colonization isolates of Streptococcus pneumoniae in northern Japan. Microb. Drug Resist. 2015, 21, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Golden, A.R.; Rosenthal, M.; Fultz, B.; Nichol, K.A.; Adam, H.J.; Gilmour, M.W.; Baxter, M.R.; Hoban, D.J.; Karlowsky, J.A.; Zhanel, G.G. Characterization of MDR and XDR Streptococcus pneumoniae in Canada, 2007–2013. J. Antimicrob. Chemother. 2015, 70, 2199–2202. [Google Scholar] [CrossRef] [PubMed]
- Choe, Y.J.; Lee, H.J.; Lee, H.; Oh, C.E.; Cho, E.Y.; Choi, J.H.; Kang, H.M.; Yoon, I.A.; Jung, H.J.; Choi, E.H. Emergence of antibiotic-resistant non-vaccine serotype pneumococci in nasopharyngeal carriage in children after the use of extended-valency pneumococcal conjugate vaccines in Korea. Vaccine 2016, 34, 4771–4776. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, C.H.; Mouton, J.W. Molecular detection of the macrolide efflux gene: To discriminate or not to discriminate between mef(A) and mef(E). Antimicrob. Agents Chemother. 2005, 49, 1271–1278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doherty, N.; Trzcinski, K.; Pickerill, P.; Zawadzki, P.; Dowson, C.G. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 2979–2984. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.S.; Ambler, J.; Mehtar, S.; Fisher, L.M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1996, 40, 2321–2326. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).