New Taxon-Specific Heterobasidion PCR Primers Detect and Differentiate North American Heterobasidion spp. in Various Substrates and Led to the Discovery of Heterobasidion irregulare in British Columbia, Canada
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Sites and Isolates
4.2. DNA Extraction
4.3. PCR, Sequencing and Primer Design
4.4. Primer Testing and Validation of the Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Woodward, S.; Stenlid, J.; Karjalainen, R.; Hüttermann, A. Heterobasidion Annosum: Biology, Ecology, Impact and Control; CABI (Cab International): Wallingford, UK, 1998. [Google Scholar]
- Gonthier, P.; Anselmi, N.; Capretti, P.; Bussotti, F.; Feducci, M.; Giordano, L.; Honorati, T.; Lione, G.; Luchi, N.; Michelozzi, M.; et al. An integrated approach to control the introduced forest pathogen Heterobasidion irregulare in Europe. Forestry 2014, 87, 471–481. [Google Scholar] [CrossRef]
- La Porta, N.; Capretti, P.; Thomsen, I.M.; Kasanen, R.; Hietala, A.M.; Von Weissenberg, K. Forest pathogens with higher damage potential due to climate change in Europe. Can. J. Plant Pathol. 2008, 30, 177–195. [Google Scholar] [CrossRef]
- Chen, J.J.; Cui, B.K.; Zhou, L.W.; Korhonen, K.; Dai, Y.C. Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 2015, 71, 185–200. [Google Scholar] [CrossRef]
- Sillo, F.; Gonthier, P.; Lockman, B.; Kasuga, T.; Garbelotto, M. Molecular analyses identify hybridization-mediated nuclear evolution in newly discovered fungal hybrids. Ecol. Evol. 2019. [Google Scholar] [CrossRef]
- Otrosina, W.J.; Garbelotto, M. Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov.: A disposition of North American Heterobasidion biological species. Fungal Biol. 2010, 114, 16–25. [Google Scholar] [CrossRef]
- Niemelä, T.; Korhonen, K. Taxonomy of the genus Heterobasidion. In Heterobasidion annosum Biology, Ecology, Impact and Control; Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A., Eds.; CAB International, University Press: Cambridge, UK, 1998; pp. 27–33. [Google Scholar]
- Buchanan, P. A new species of Heterobasidion (Polyporaceae) from Australia. Mycotaxon 1988, 32, 325–337. [Google Scholar]
- Linzer, R.E.; Otrosina, W.J.; Gonthier, P.; Bruhn, J.; Laflamme, G.; Bussières, G.; Garbelotto, M. Inferences on the phylogeography of the fungal pathogen Heterobasidion annosum, including evidence of interspecific horizontal genetic transfer and of human-mediated, long-range dispersal. Mol. Phylogenet. Evol. 2008, 46, 844–862. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Biology, epidemiology, and control of Heterobasidion species worldwide. Annu. Rev. Phytopathol. 2013, 51, 39–59. [Google Scholar] [CrossRef]
- Gonthier, P.; Warner, R.; Nicolotti, G.; Mazzaglia, A.; Garbelotto, M.M. Pathogen introduction as a collateral effect of military activity. Mycol. Res. 2004, 108, 468–470. [Google Scholar] [CrossRef]
- Giordano, L.; Gonthier, P.; Lione, G.; Capretti, P.; Garbelotto, M. The saprobic and fruiting abilities of the exotic pathogen Heterobasidion irregulare may explain its invasiveness. Biol. Invasions 2013. [Google Scholar] [CrossRef]
- Sillo, F.; Garbelotto, M.; Friedman, M.; Gonthier, P. Comparative genomics of sibling fungal pathogenic taxa identifies adaptive evolution without divergence in pathogenicity genes or genomic structure. Genome Biol. Evol. 2015, 7, 3190–3206. [Google Scholar] [CrossRef]
- Garbelotto, M.; Friedman, M.; Bedell, W.; Henkel, T. First report of Heterobasidion occidentale on Sequoia sempervirens in Northern California. Plant Dis. 2017, 101, 2152. [Google Scholar] [CrossRef]
- Harrington, T.C.; Worrall, J.J.; Rizzo, D.M. Compatibility among host-specialized isolates of Heterobasidion annosum from western North America. Phytopathology 1989, 79, 290–296. [Google Scholar] [CrossRef]
- Garbelotto, M.; Ratcliff, A.; Bruns, T.D.; Cobb, F.W.; Otrosina, W.J. Use of taxon-specific competitive-priming PCR to study host specificity, hybridization, and intergroup gene flow in intersterility groups of Heterobasidion annosum. Phytopathology 1996, 66, 543–551. [Google Scholar] [CrossRef]
- Lockman, B.; Mascheretti, S.; Schechter, S.; Garbelotto, M. A first generation Heterobasidion hybrid discovered in Larix lyalli in Montana. Plant Dis. 2014, 98, 1003. [Google Scholar] [CrossRef]
- Gonthier, P.; Garbelotto, M.; Varese, G.C.; Nicolotti, G. Relative abundance and potential dispersal range of intersterility groups of Heterobasidion annosum in pure and mixed forests. Can. J. Bot. 2001, 79, 1057–1065. [Google Scholar]
- Gonthier, P.; Guglielmo, F.; Sillo, F.; Giordano, L.; Garbelotto, M. A molecular diagnostic assay for the detection and identification of wood decay fungi of conifers. For. Pathol. 2015, 45, 89–101. [Google Scholar] [CrossRef]
- Lamarche, J.; Potvin, A.; Stewart, D.; Blais, M.; Pelletier, G.; Shamoun, S.F.; Hamelin, R.C.; Tanguay, P. Real-time PCR assays for the detection of Heterobasidion irregulare, H. occidentale, H. annosum sensu stricto and the Heterobasidion annosum complex. For. Pathol. 2016. [Google Scholar] [CrossRef]
- Hantula, J.; Vainio, E. Specific primers for the differentiation of Heterobasidion annosum (s. str.) and H. parviporum infected stumps in northern Europe. Silva Fenn. 2003, 37, 181–187. [Google Scholar] [CrossRef]
- Ðíipars, V.; Ruòìis, D. Detection of Heterobasidion annosum in Scots pine trees using a polymerase chain reaction based method. Balt. For. 2011, 17, 2–7. [Google Scholar]
- Nowakowska, J.A.; Borys, M.; Oszako, T. DNA markers used to identify pathogens causing root rot disease in spruce (Picea abies L. Krast) for forensic purposes. J. Forensic Sci. 2013, 93, 465–474. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Johannesson, H.; Stenlid, J. Molecular markers reveal genetic isolation and phylogeography of the S and F intersterility groups of the wood-decay fungus Heterobasidion annosum. Mol. Phylogenet. Evol. 2003, 29, 94–101. [Google Scholar] [CrossRef]
- Heinze, B. A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods 2007, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, F.; Gonthier, P.; Garbelotto, M.; Nicolotti, G. Optimization of sampling procedures for DNA-based diagnosis of wood decay fungi in standing trees. Lett. Appl. Microbiol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Banik, M.T.; Lindner, D.L.; Juzwik, J.; Glaeser, J.A. Development of a DNA sampling kit to detect pathogenic, saprotrophic, and stain fungi in sapwood of declining red pine (Pinus resinosa) in the upper midwest. In Browning, John; Palacios, Patsy, comps. Proceedings of the 60th Annual Western International Forest Disease Work Conference, Lake Tahoe, CA, USA, 8–12 October 2012; pp. 101–110. [Google Scholar]
- Särkinen, T.; Staats, M.; Richardson, J.E.; Cowan, R.S.; Bakker, F.T. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS ONE 2012, 7, e43808. [Google Scholar] [CrossRef]
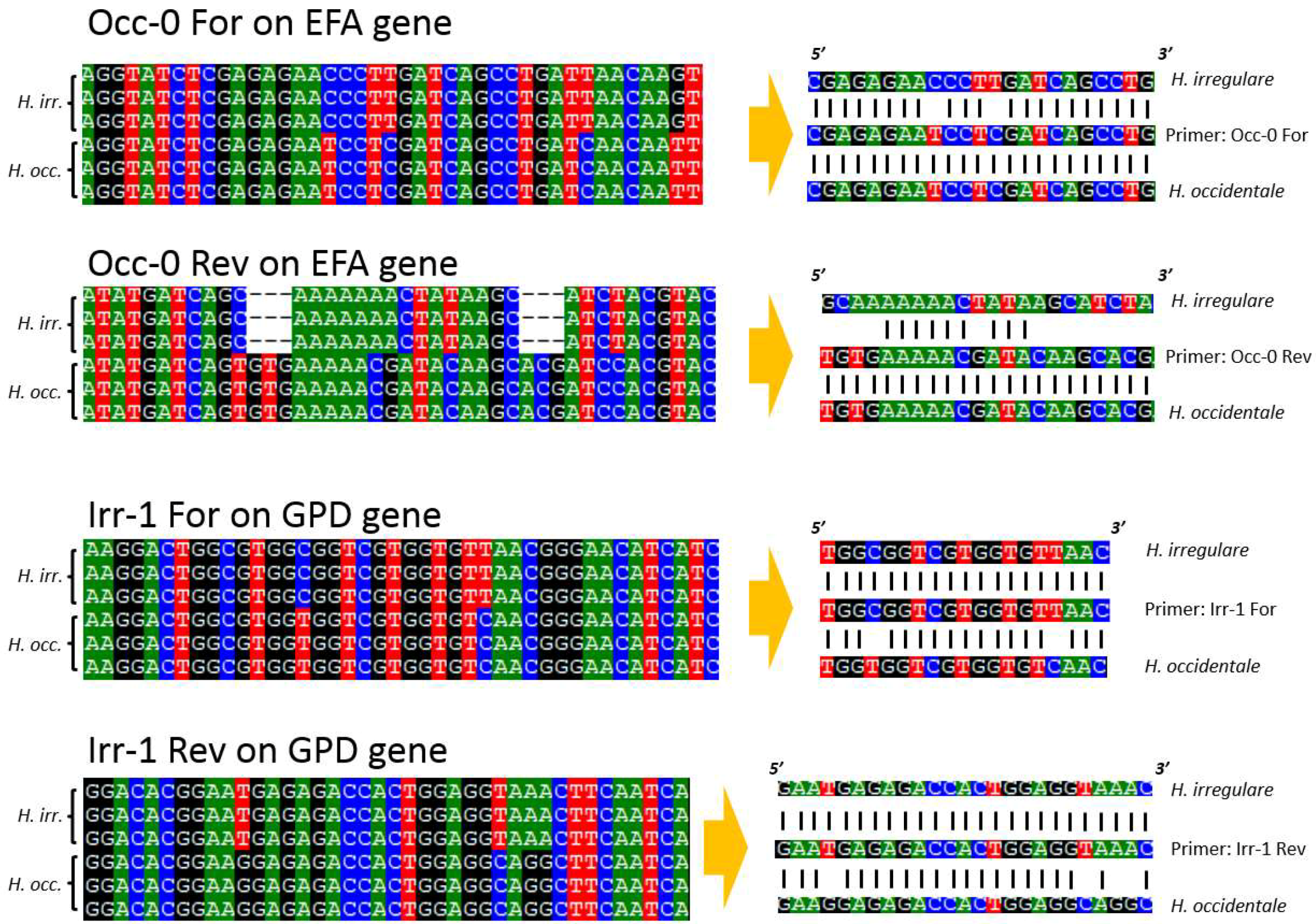
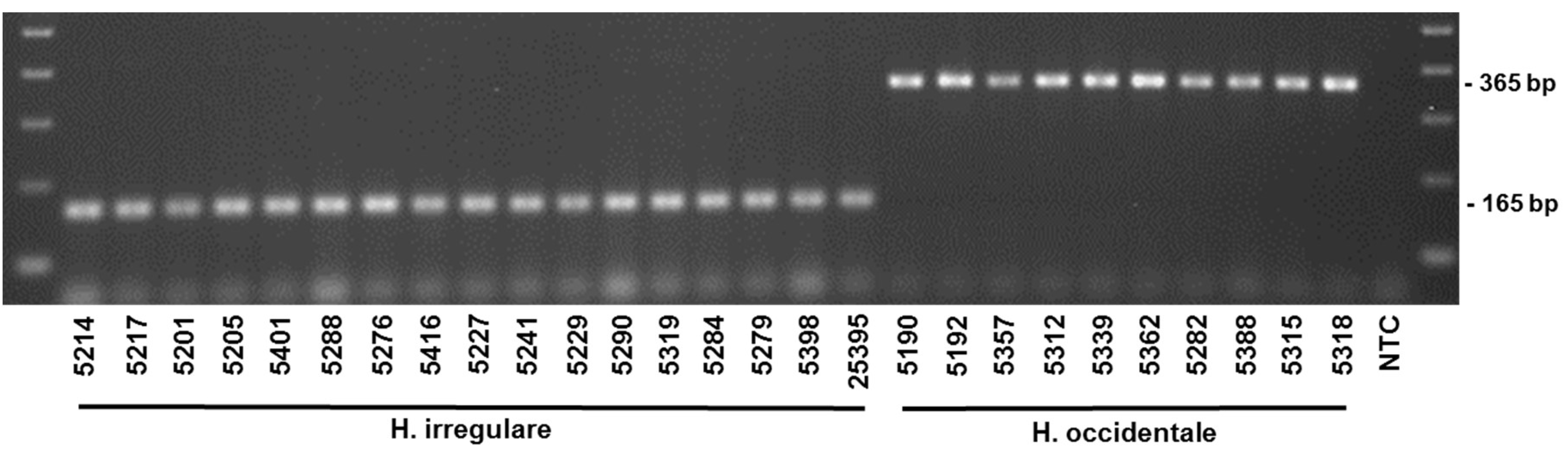
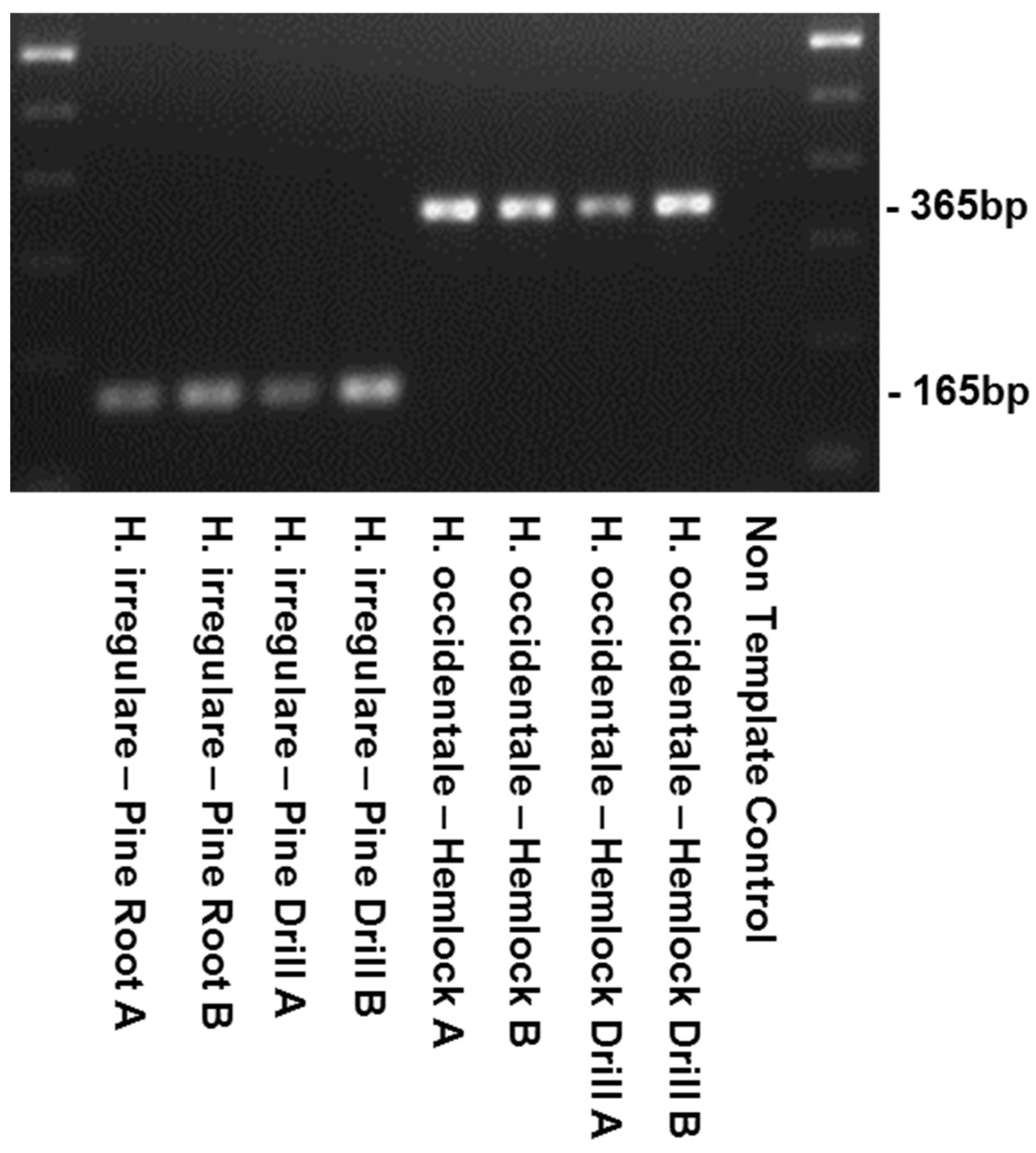
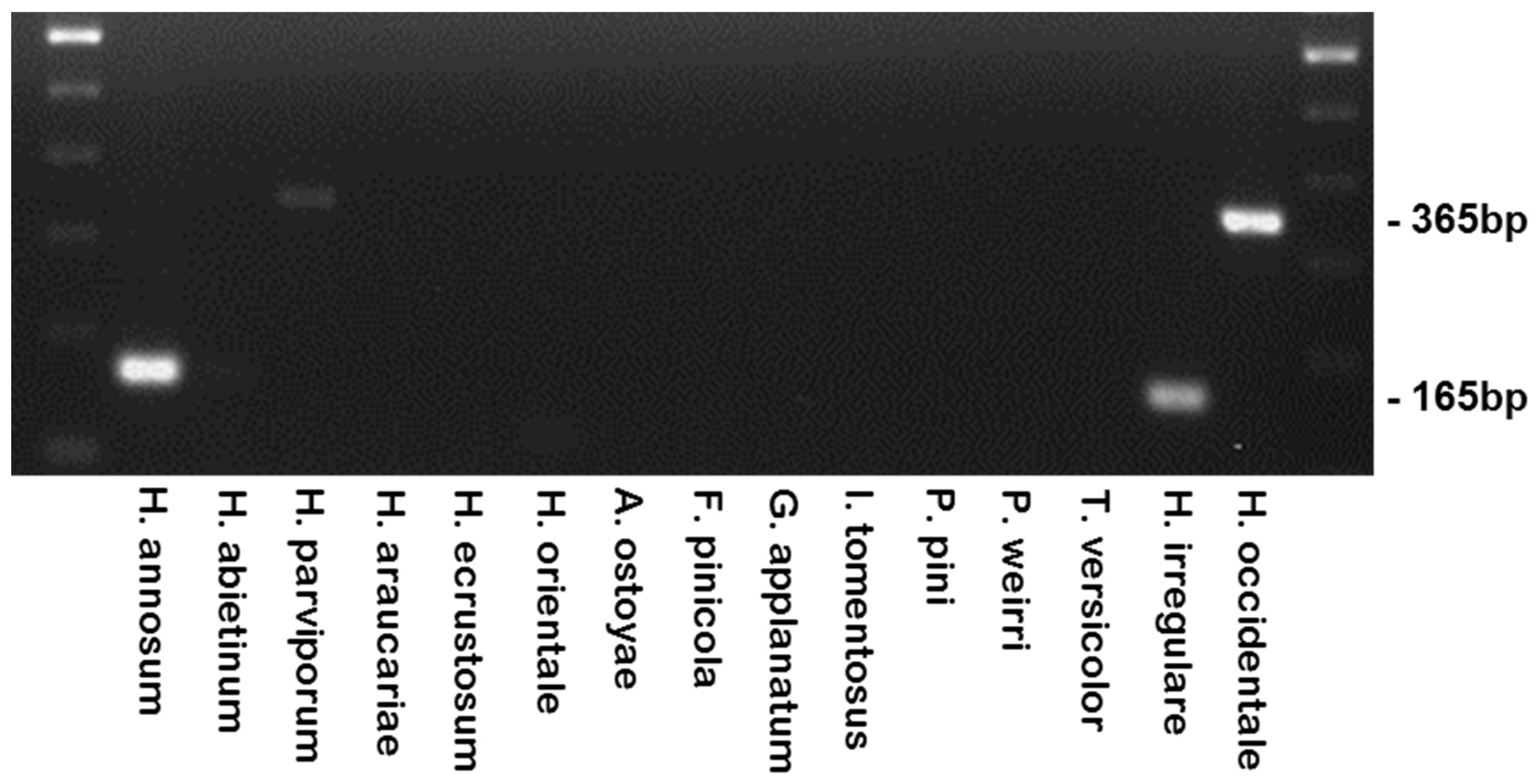
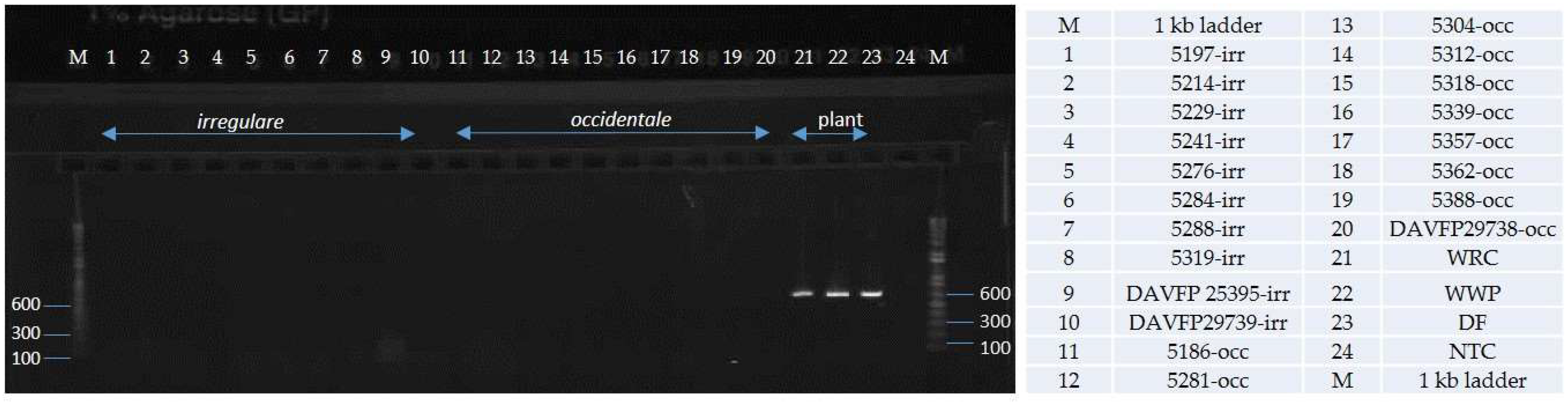

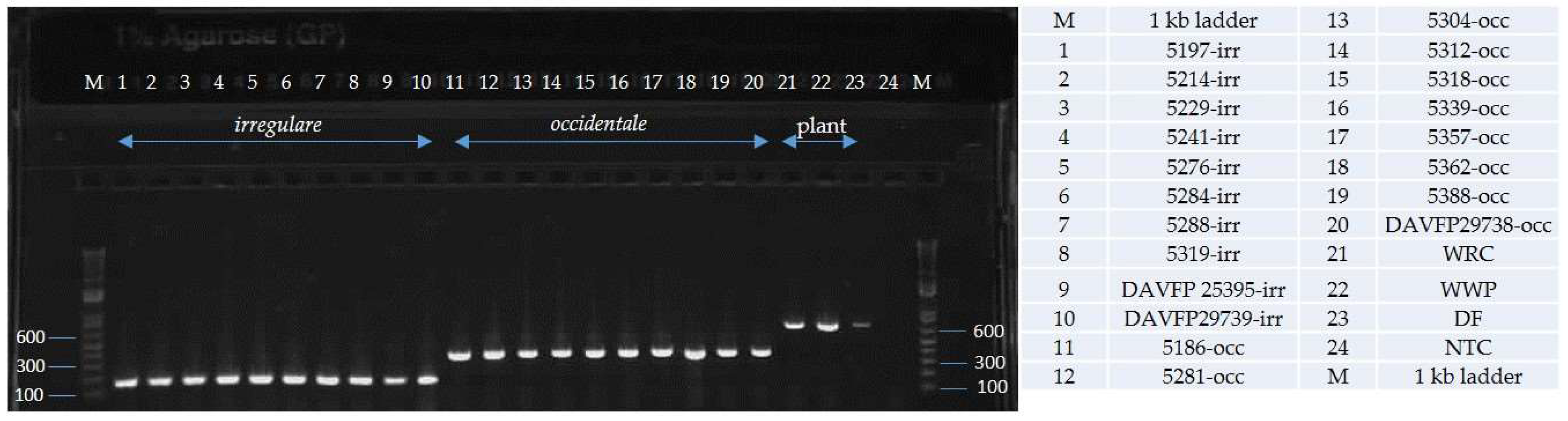
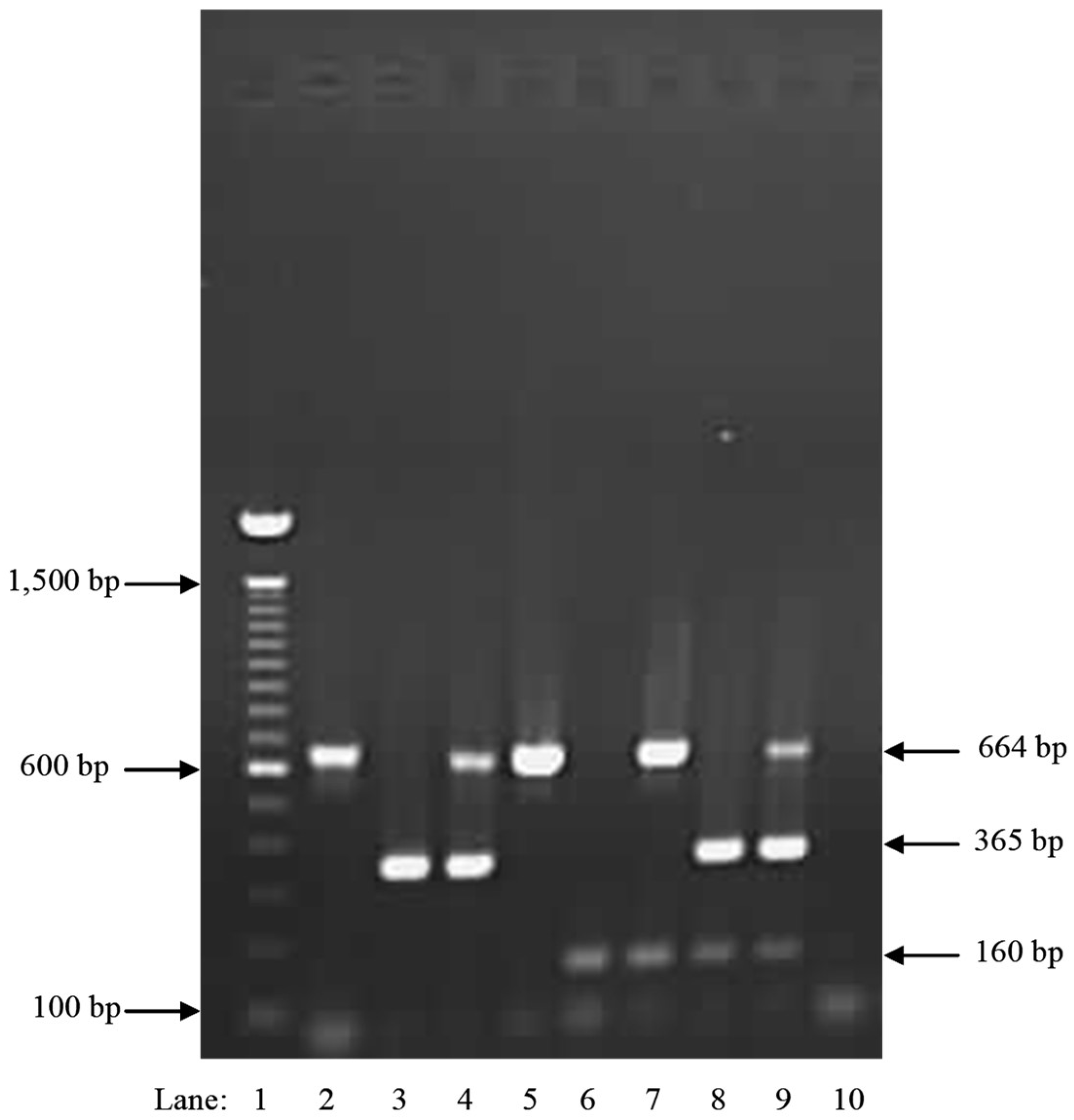
| Species | Isolate or Herbarium Collection Number | Geographic Origin | Host | Year Collected | Source/Collector | ITS | EFA | GPD |
|---|---|---|---|---|---|---|---|---|
| irregulare | PFC5201 | Limerick, ON, Canada | Pinus resinosa | 2003 | M. Dumas | KP863563 | KP863594 | KP863623 |
| irregulare | PFC5205 | York, ON, Canada | P. resinosa | 2005 | M. Dumas | KP863564 | KP863596 | KP863625 |
| irregulare | PFC5214 | St. Philippe, QC, Canada | P. resinosa | 2002 | G. Laflamme | KP863565 | KP863597 | KP863626 |
| irregulare | PFC5217 | Lac La Blanche, QC, Canada | P. resinosa | 2002 | G. Laflamme | KP863566 | KP863598 | KP863627 |
| irregulare | PFC5227 | Iowa Co, WI, USA | P. resinosa or P. strobus | 1994 | G. Stanosz | KP863567 | KP863599 | KP863629 |
| irregulare | PFC5229 | Union Co, IL, USA | P. echinata | 1994 | G. Stanosz | KP863568 | KP863600 | KP863630 |
| irregulare | PFC5241 | Portage Co, WI, USA | Abies balsamea | 2010 | G. Stanosz | KP863571 | KP863602 | KP863634 |
| irregulare | PFC5276 | S. Pines, NC, USA | P. taeda | 1967 | J.S. Boyle | KP863572 | KP863603 | KP863635 |
| irregulare | PFC5279 | Lassen Natl. Forest, CA, USA | P. ponderosa | 1981 | J. Worrall | KP863573 | KP863604 | KP863636 |
| irregulare | PFC5284 | San Bernardino Mtns., CA, USA | P. jeffreyii | 1975 | J. Worrall | KP863574 | KP863605 | KP863639 |
| irregulare | PFC5288 | Durham College Woods, NH, USA | Juniperus virginiana | 1987 | T. Harrington | KP863575 | KP863606 | KP863641 |
| irregulare | PFC5290 | Nebraska, USA | P. ponderosa | J. Blodgett | KP863576 | KP863607 | KP863642 | |
| irregulare | PFC5319 | Montrose Co, CO, USA | P. ponderosa | 2006 | J. Worrall | KP863581 | KP863611 | KP863647 |
| irregulare | PFC5401 | St. Williams, ON, Canada | Thuja plicata | D.C. Constable | KP863587 | KP863617 | KP863653 | |
| irregulare | PFC5416 | Warren Co, GA, USA | Spore trap | 2007 | M. Cram | KP863589 | KP863619 | KP863655 |
| irregulare | DAVFP25395 | Okanagan Falls, BC, Canada | P. ponderosa | 1997 | J. Hodges | KP863588 | KP863618 | KP863654 |
| irregulare | DAVFP29739 | Summarland, BC, Canada | P. strobus | 2013 | J.H. Ginns | |||
| irregulare | DAVFP29740 | Summerland, BC, Canada | P. strobus | 2013 | J.H. Ginns | |||
| occidentale | PFC5190 | Ladysmith, BC, Canada | Tsuga heterophylla | B. Callan | KP863561 | KP863592 | KP863621 | |
| occidentale | PFC5192 | Jordan River, BC, Canada | T. heterophylla | B. Callan | KP863562 | KP863593 | KP863622 | |
| occidentale | PFC5282 | Modoc Natl Forest, CA, USA | A. concolor | 1981 | J. Worrall | KP492941.1 | KP571672.1 | KP863638 |
| occidentale | PFC5312 | King Co, WA, USA | T. heterophylla | 2011 | R. Edmonds | KP863578 | KP863608 | KP863644 |
| occidentale | PFC5315 | Ouray Co, CO, USA | A. concolor | 2005 | J. Worrall | KP863579 | KP863609 | KP863645 |
| occidentale | PFC5318 | Mineral CO, CO, USA | A. concolor | 2005 | J. Worrall | KP863580 | KP863610 | KP863646 |
| occidentale | PFC5339 | OR, USA | A. concolor | 2009 | E. Goheen | KP863582 | KP863612 | KP863648 |
| occidentale | PFC5357 | Clallam Co, WA, USA | T. heterophylla | 2011 | R. Edmonds | KP863583 | KP863613 | KP863649 |
| occidentale | PFC5362 | Lincoln Co, OR, USA | T. heterophylla | 2011 | M. Elliott | KP863584 | KP863614 | KP863650 |
| occidentale | PFC5388 | San Bernardino, CA, USA | A. concolor | 2011 | P. Zambino | KP863585 | KP863615 | KP863651 |
| occidentale | DAVFP29738 | Summerland, BC, Canada | Abies stump | 2013 | J.H. Ginns |
| Species | Isolate or Herbarium Collection Number | Geographic Origin | Host | Year Collected | Source/Collector | ITS | EFA | GPD |
|---|---|---|---|---|---|---|---|---|
| abietinum | PFC5247 | Poland | Abies alba | H. Solheim | KC492895.1 | KC571636.1 | KP863657 | |
| abietinum | PFC5249 | Austria | Picea abies | H. Solheim | KC492896.1 | KC571637.1 | KP863658 | |
| abietinum | PFC5373 | Greece | Abies cephallonica | 1993 | P. Tsopelas | KC492956.1 | KC571687.1 | KP863664 |
| annosum | PFC5252 | Norway | Pinus sylvestris | 1937 | R.H. Roll-Hansen | KC492906.1 | KC571646.1 | KP863659 |
| annosum | PFC5257 | Italy | Pinus pinaster | 2008 | A. Biraghi | KC492909.1 | KC571649.1 | KP863660 |
| annosum | PFC5260 | Serbia | Pinus nigra | 2003 | D. Dubak | KC492911.1 | KC571651.1 | KP863661 |
| araucariae | PFC5434 | New Zealand | Agathis australis | 1958 | J.W. Gilmour | KX130098 | KX130101 | KX130104 |
| ecrustosum | PFC5438 | Japan | Pinus thunbergii | P.K. Buchanan | KX130099 | KX130102 | KX130105 | |
| orientale | PFC5439 | Japan | Tsuga sp. | P.K. Buchanan | KX130100 | KX130103 | KX130106 | |
| parviporum | PFC5262 | Norway | Picea abies | 2004 | R. Saursaunet | KC492957.1 | KC571688.1 | KP863662 |
| parviporum | PFC5269 | Japan | Abies mayriana | 1942 | S. Kamei | KC492951.1 | KC571682.1 | KP863663 |
| parviporum | PFC5293 | Russia | Abies sibirica | K. Korhonen | KC492913.1 | KC571653.1 | KP863665 |
| Primer | Sequence | Gene | Product (bp) | Tm (°C) | Reference |
|---|---|---|---|---|---|
| Irr-1 For | TGGCGGTCGTGGTGTTAAC | GPD | 165 | 64 | This study |
| Irr-1 Rev | GAATGAGAGACCACTGGAGGTAAAC | GPD | 165 | 64 | This study |
| Occ-0 For | CGAGAGAATCCTCGATCAGCCTG | EFA | 365 | 64 | This study |
| Occ-0 Rev | TGTGAAAAACGATACAAGCACG | EFA | 365 | 64 | This study |
| ITS1-F | CTTGGTCATTTAGAGGAAGTAA | ITS | 486/515 | 55 | [24] |
| ITS4 | TCCTCCGCTTATTGATATGC | ITS | 486/515 | 55 | [25] |
| EFA For | TCAACGTGGTCGGTGAGCAGGTA | EFA | 447-54 | 66 | [26] |
| EFA Rev | AAGTCACGATGTCCAGGAGCATC | EFA | 447-54 | 66 | [26] |
| GPD-Seq For | CAGAGCCTCTGCCCACTTGAAGG | GPD | 666/754 | 59 | This study |
| GPD-Seq Rev | GCCGGGTGGCCGACAAAGTC | GPD | 666 | 59 | This study |
| PC-UPC7 For | GGATTRCGTATGGGMAATATTGAAAC | CHLOROPLAST | 664 | 64 | [27] |
| PC-UPC7 Rev | CCCCTTGGACTRCTACGAAAAACACC | CHLOROPLAST | 664 | 64 | [27] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamoun, S.F.; Hammett, C.; Sumampong, G.; Li, X.; Garbelotto, M. New Taxon-Specific Heterobasidion PCR Primers Detect and Differentiate North American Heterobasidion spp. in Various Substrates and Led to the Discovery of Heterobasidion irregulare in British Columbia, Canada. Pathogens 2019, 8, 156. https://doi.org/10.3390/pathogens8030156
Shamoun SF, Hammett C, Sumampong G, Li X, Garbelotto M. New Taxon-Specific Heterobasidion PCR Primers Detect and Differentiate North American Heterobasidion spp. in Various Substrates and Led to the Discovery of Heterobasidion irregulare in British Columbia, Canada. Pathogens. 2019; 8(3):156. https://doi.org/10.3390/pathogens8030156
Chicago/Turabian StyleShamoun, Simon Francis, Craig Hammett, Grace Sumampong, Xiang Li, and Matteo Garbelotto. 2019. "New Taxon-Specific Heterobasidion PCR Primers Detect and Differentiate North American Heterobasidion spp. in Various Substrates and Led to the Discovery of Heterobasidion irregulare in British Columbia, Canada" Pathogens 8, no. 3: 156. https://doi.org/10.3390/pathogens8030156
APA StyleShamoun, S. F., Hammett, C., Sumampong, G., Li, X., & Garbelotto, M. (2019). New Taxon-Specific Heterobasidion PCR Primers Detect and Differentiate North American Heterobasidion spp. in Various Substrates and Led to the Discovery of Heterobasidion irregulare in British Columbia, Canada. Pathogens, 8(3), 156. https://doi.org/10.3390/pathogens8030156






