Comparative Study of CDST & Multiplex PCR to Detect MBL Producing Gram-Negative Bacilli among VAP Patients Admitted in a Public Medical College Hospital of Bangladesh
Abstract
1. Introduction
2. Objectives of the Study
3. Materials and Methods
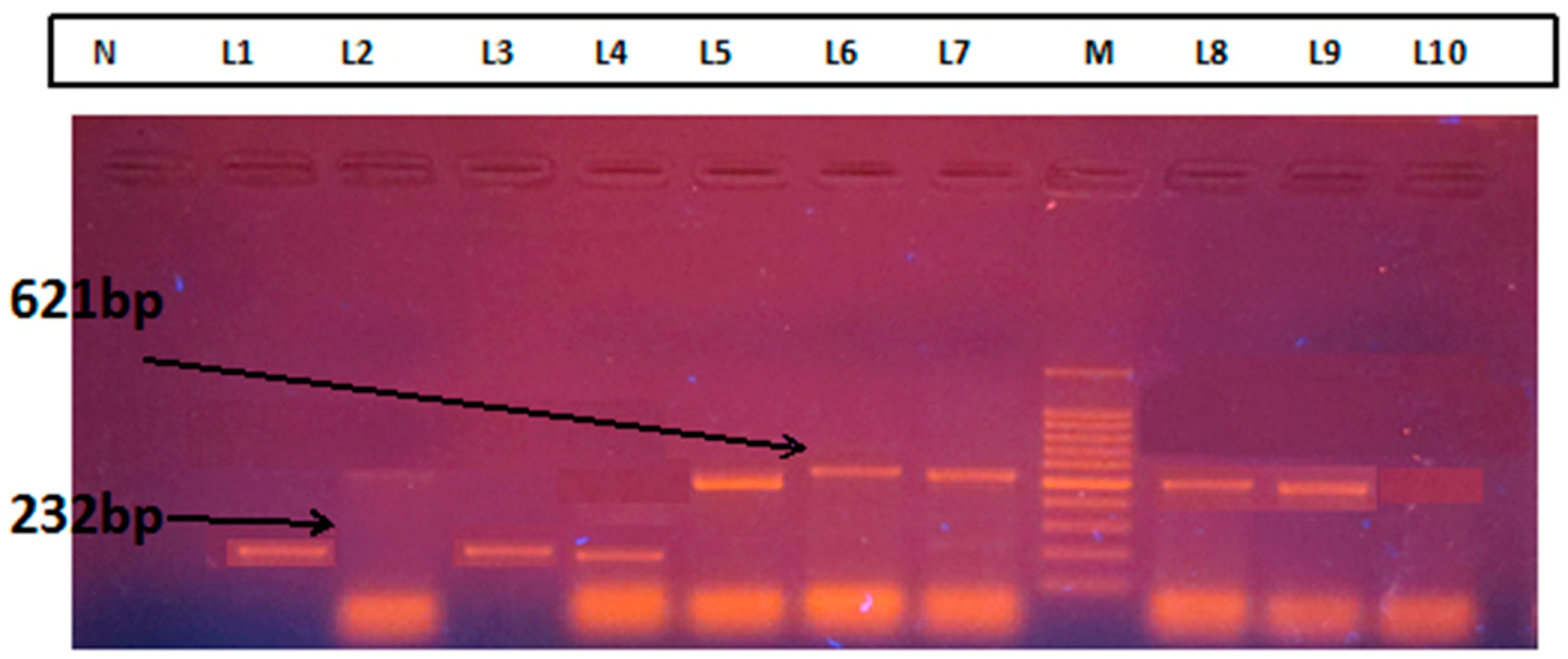
Laboratory Method
4. Results
5. Discussion
6. Conclusions
7. Recommendations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kalanuria, A.A.; Ziai, W.; Mirski, M. Ventilator-associated pneumonia in the ICU. Crit. Care. 2014, 18. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M. Healthcare-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Bhadade, R.; Harde, M.; De Souza, R.; More, A.; Bharmal, R. Emerging trends of nosocomial pneumonia in intensive care unit of a tertiary care public teaching hospital in Western India. Ann. Afr. Med. 2017, 16, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.M.; Wahsheh, M.A. Knowledge level of nurses in Jordan on ventilator-associated pneumonia and preventive measures. Nurs. Crit. Care. 2017, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Bairy, I. Incidence of multidrug-resistant organisms causing ventilator-associated pneumonia in a tertiary care hospital: A nine months’ prospective study. Ann. Thorac. Med. 2007, 2, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.M.; Aruj, P.; Niederman, M.S.; Garzón, J.; Violi, D.; Prignoni, A.; Ríos, F.; Baquero, S.; Gando, S.; Grupo Argentino de Estudio de la Neumonía Asociada al Respirador Group. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur. Respir. 2006, 27, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Scholte, J.B.; van Dessel, H.A.; Linssen, C.F.; Bergmans, D.C.; Savelkoul, P.H.; Roekaerts, P.M.; van Mook, W.N. Endotracheal aspirate and bronchoalveolar lavage fluid analysis: Interchangeable diagnostic modalities in suspected ventilator-associated pneumonia? J. Clin. Microbiol. 2014, 52, 3597–3604. [Google Scholar] [CrossRef] [PubMed]
- Khilnani, G.C.; Arafath, T.K.; Hadda, V.; Kapil, A.; Sood, S.; Sharma, S.K. Comparison of bronchoscopic and non-bronchoscopic techniques for diagnosis of ventilator-associated pneumonia. Indian J. Crit. Care Med. 2011, 15. [Google Scholar] [CrossRef]
- Koenig, S.M.; Truwit, J.D. Ventilator-associated pneumonia: Diagnosis, treatment, and prevention. Clin. Microbiol. Rev. 2006, 19, 637–657. [Google Scholar] [CrossRef]
- Shin, Y.M.; Oh, Y.M.; Kim, M.N.; Shim, T.S.; Lim, C.M.; Lee, S.D.; Koh, Y.; Kim, W.S.; Kim, D.S.; Hong, S.B. Usefulness of quantitative endotracheal aspirate cultures in intensive care unit patients with suspected pneumonia. J. Korean Med. Sci. 2011, 26, 865–869. [Google Scholar] [CrossRef]
- Zaccard, C.R.; Schell, R.F.; Spiegel, C.A. Efficacy of bilateral bronchoalveolar lavage for diagnosis of ventilator-associated pneumonia. J. Clin. Microbiol. 2009, 47, 2918–2924. [Google Scholar] [CrossRef]
- Prats, E.; Dorca, J.; Pujol, M.; Garcia, L.; Barreiro, B.; Verdaguer, R.; Gudiol, F.; Manresa, F. Endotracheal aspirate, and bronchoalveolar lavage fluid analysis: Interchangeable diagnostic modalities in suspected ventilator-associated pneumonia? Eur. Respir. J. 2002, 19, 944–951. [Google Scholar] [CrossRef]
- Li, Y.; Gong, Z.; Lu, Y.; Hu, G.; Cai, R.; Chen, Z. Impact of nosocomial infections surveillance on nosocomial infection rates: A systematic review. Int. J. Surg. 2017, 42, 164–169. [Google Scholar] [CrossRef] [PubMed]
- McCann, E.; Srinivasan, A.; De Ryke, C.A.; Gang, Y.; DePestel, D.D.; Murray, J.; Gupta, V. Carbapenem-nonsusceptible gram-negative pathogens in ICU and non-ICU settings in US hospitals in 2017: A Multicenter Study. Open Forum Infect. Dis. 2018, 5, ofy241. [Google Scholar] [CrossRef] [PubMed]
- Porwal, R.; Gopalakrishnan, R.; Rajesh, N.J.; Ramasubramanian, V. Carbapenem-resistant gram-negative bacteremia in an Indian intensive care unit: A review of the clinical profile and treatment outcome of 50 patients. Indian J. Crit. Care Med. 2014, 18, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Poirel, L.; Nordmann, P. Worldwide dissemination of the NDM-type carbapenemases in gram-negative bacteria. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: A review. Med. Sci. 2017, 6. [Google Scholar] [CrossRef]
- Petersen-Morfin, S.; Bocanegra-Ibarias, P.; Morfin-Otero, R.; Garza-Gonzales, E.; Perez-Gomez, H.R.; Gonzales-Diaz, E.; Esparza-Ahumada, S.; León-Garnica, G.; Amezcua-Salazar, G.; Rodriguez-Noriega, E. New Delhi metallo-beta-lactamase (NDM-1)-producing klebsiella pneumoniae isolated from a burned patient. Am. J. Case Rep. 2017, 18, 805–809. [Google Scholar] [CrossRef]
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A.; et al. Better tests, better care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013, 57, S139–S170. [Google Scholar] [CrossRef]
- Aydemir, O.; Aydemir, Y.; Ozdemir, M. The role of multiplex PCR test in identification of bacterial pathogens in lower respiratory tract infections. Pak. J. Med. Sci. 2014, 30, 1011–1016. [Google Scholar] [CrossRef]
- Dogonchi, A.A.; Ghaemi, E.A.; Ardebili, A.; Yazdansetad, S.; Pournajaf, A. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi 2018, 30, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.; Petersen, M.B.; Jørgensen, A.L.; Paulmann, D.; Wang, M. Rapid real-time PCR for the detection of IMP, NDM, VIM, KPC and OXA-48 carbapenemase genes in isolates and spiked stool samples. Diagn. Microbiol. Infect. Dis. 2018, 92, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Fallah, F.; Borhan, R.S.; Hashemi, A. Detection of bla (IMP) and bla (VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int. J. Burns Trauma 2013, 3, 122–124. [Google Scholar] [PubMed]
- Hong, D.J.; Bae, I.K.; Jang, I.H.; Jeong, S.H.; Kang, H.K.; Lee, K. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect. Chemother. 2015, 47, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Craven, D.E.; Chroneou, A.; Zias, N.; Hjalmarson, K.I. Ventilator-associated tracheobronchitis: The impact of targeted antibiotic therapy on patient outcomes. Chest 2009, 135, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Medcalculator, Free Statistical Calculator: Diagnostic Test Evaluation Calculator. Available online: https://www.medcalc.org/calc/diagnostic_test.php (accessed on 30 August 2019).
- Ali, S.Q.; Zehra, A.; Naqvi, B.S.; Shah, S.; Bushra, R. Resistance pattern of ciprofloxacin against different pathogens. Oman. Med. J. 2010, 25, 294–298. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 29th ed.; CLSI Supplement M100: Wayne, PA, USA, 2019. [Google Scholar]
- Jing, X.; Zhou, H.; Min, X.; Zhang, X.; Yang, Q.; Du, S.; Li, Y.; Yu, F.; Jia, M.; Zhan, Y. The simplified carbapenem inactivation method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative bacilli. Front. Microbiol. 2018, 9, 2391. [Google Scholar] [CrossRef] [PubMed]
- Hodiwala, A.; Dhoke, R.; Urhekar, A.D. Incidence of mettalo-beta-lactamase producing pseudomonas, Acinetobacter & enterobacterial isolates in hospitalized patients. IJPBS. 2013, 3, 79–83. [Google Scholar]
- Khosravi, Y.; Loke, M.; Chua, E.G.; Tay, S.T.; Vadivelu, J. Phenotypic detection of metallo-β-lactamase in imipenem-resistant Pseudomonas aeruginosa. Sci. World J. 2012. [Google Scholar] [CrossRef]
- Yong, D.; Lee, K.; Yum, J.H.; Shin, H.B.; Rossolini, G.M.; Chong, Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2002, 40, 3798–3801. [Google Scholar] [CrossRef]
- Sachdeva, R.; Sharma, B.; Sharma, R. Evaluation of different phenotypic tests for detection of metallo-β-lactamases in imipenem-resistant Pseudomonas aeruginosa. J. Lab. Physicians. 2017, 9, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Prabhu, T. Detection and characterization of metallo-β-lactamases producing Pseudomonas aeruginosa clinical isolates at a tertiary care. Int. J. Res. Med. Sci. 2016, 4, 4084–4088. [Google Scholar] [CrossRef][Green Version]
- Mohanam, L.; Menon, T. Coexistence of metallo-beta-lactamase-encoding genes in Pseudomonas aeruginosa. Indian J. Med. Res. 2017, 146, S46–S52. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Upadhyay, S.; Sen, M.R.; Maurya, A.P.; Choudhury, D.; Bhattacharjee, A. Genetic acquisition of NDM gene offers sustainability among clinical isolates of Pseudomonas aeruginosa in clinical settings. PLoS ONE 2015, 10, e0116611. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.R.; Achariya, M.; Lalshaptai, T.; Leungtong, U.; Thummeepak, R.; Sitthisak, S. Co-existence of blaOXA -23 and blaNDM -1 gene of Acinetobacter baumannii isolated from Nepal: Antimicrobial resistance and clinical significance. Antimicrob. Resist. Infect. Control 2017, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Feldman, C.; Richards, G. Appropriate antibiotic management of bacterial lower respiratory tract infections. F1000 Res. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Baselski, V.; Klutts, J.S.; Baselski, V.; Klutts, J.S. Quantitative cultures of bronchoscopically obtained specimens should be performed for optimal management of ventilator-associated pneumonia. J. Clin. Microbiol. 2013, 51, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Jourdain, B.; Novara, A.; Joly-Guillou, M.L.; Dombret, M.C.; Calvat, S.; Trouillet, J.L.; Gibert, C.; Chastre, J. Role of quantitative cultures of endotracheal aspirates in the diagnosis of nosocomial pneumonia. Am. J. Respir. Crit. Care Med. 1995, 152, 241–246. [Google Scholar] [CrossRef]
- Pugain, J.; Auckenthaler, R.; Mili, N.; Janssens, J.P.; Lew, P.D.; Suter, P.M. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic blind bronchoalveolar lavage fluid. Am. Rev. Respir. Dis. 1991, 143, 1121–1129. [Google Scholar] [CrossRef]
- Rajasekhar, T.; Anuradha, K.; Suhasini, T.; Lakshmi, V. The role of quantitative cultures of non-bronchoscopic samples in ventilator-associated pneumonia. Indian J. Med. Microbiol. 2006, 24, 107–113. [Google Scholar]
- Azarudeen, M.; Sharma, B.S.; Jain, P.K.; Goyal, A.K.; Malhotra, B. Study of quantitative bacterial cultures of non-bronchoscopic samples in ventilator-associated pneumonia. Int. J. Contemp. Pediatr. 2018, 5, 1837–1843. [Google Scholar] [CrossRef]
- Lipovy, B.; Rihová, H.; Gregorova, N.; Hanslianova, M.; Zaloudikova, Z.; Kaloudova, Y.; Brychta, P. Epidemiology of ventilator-associated Tracheobronchitis and ventilator pneumonia in patients with inhalation injury at a burn center in Brno (Czech Republic). Ann. Burns Fire Disasters 2011, 24, 120–125. [Google Scholar] [PubMed]
- Goel, V.; Hogade, S.A.; Karadesai, S. Ventilator-associated pneumonia in a medical intensive care unit: Microbial etiology, susceptibility patterns of isolated microorganisms and outcome. Indian J. Anaesth. 2012, 56, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, N.; Chaudhary, U.; Chaudhry, D.; Ranjan, K.P. Ventilator-associated pneumonia in a tertiary care intensive care unit: Analysis of incidence, risk factors, and mortality. Indian J. Crit. Care Med. 2014, 18, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Bonten, M.J.; Kollef, M.H.; Hall, J.B. Risk factors for ventilator-associated pneumonia: From epidemiology to patient management. Clin. Infect. Dis. 2004, 38, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, S.K.; Mudali, I.; Strandvik, G.; El-Menyar, A.; Al-Hassani, A.; Al-Thani, H. Risk factors for ventilator-associated pneumonia in trauma patients: A descriptive analysis. World J. Emerg. Med. 2018, 9, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.S.; Khan, F.Y.; George, S.; Shaikh, N.; Al-Ajmi, J. Epidemiology and outcome of ventilator-associated Pneumonia in a heterogeneous ICU population in Qatar. Biomed Res. Int. 2016, 8231787. [Google Scholar] [CrossRef] [PubMed]
- Panchal, C.A.; Oza, S.S.; Mehta, S.J. Comparison of four phenotypic methods for detection of metallo-β-lactamase-producing Gram-negative bacteria in rural teaching hospital. J. Lab. Physicians 2017, 9, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Solanki, R.; Vanjari, L.; Subramanian, S.; Aparna, B.; Nagapriyanka, E.; Vemu, L. Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of Carbapenemases among gram-negative bacilli. J. Clin. Diagn. Resp. 2014, 8, 23–26. [Google Scholar]
- Picão, R.C.; Andrade, S.S.; Nicoletti, A.G. Metallo-beta-lactamase detection: Comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J. Clin. Microbiol. 2008, 46, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, S.; Banashankari, G.S.; Babu, P.R. Evaluation of phenotypic tests and screening markers for detection of metallo-β-lactamases in clinical isolates of Pseudomonas aeruginosa: A prospective study. Med. J. DY Patil Univ. 2015, 8, 599–605. [Google Scholar] [CrossRef]
- Meletis, G.; Exindari, M.; Vavatsi, N.; Sofianou, D.; Diza, E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia 2012, 16, 303–307. [Google Scholar] [PubMed]
- Martins, A.F.; Borges, A.; Pagano, M.; Dalla-Costa, L.M.; Barth, A.L. False-positive results in screening for metallo-β-lactamase are observed in isolates of Acinetobacter baumannii due to production of Oxacillinases. Braz. J. Infect. Dis. 2013, 17, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Shamsuzzaman, S.M. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Ci Ji Yi Xue Za Zhi 2016, 28, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Lagrutta, E.; Taylor, P.; Pham, J.; Nordmann, P. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 2010, 54, 4914–4916. [Google Scholar] [CrossRef] [PubMed]
- Nasrin, T.; Jilani, M.S.A.; Barai, L.; Haq, J.A. Metallo-ß-lactamase producing Pseudomonas species in a tertiary care hospital of Dhaka City. Bangladesh J. Med. Microbiol. 2010, 4, 43–45. [Google Scholar] [CrossRef]
- Islam, M.A.; Talukdar, P.K.; Hoque, A.; Huq, M.; Nabi, A.; Ahmed, D.; Talukder, K.A.; Pietroni, M.A.; Hays, J.P.; Cravioto, A.; et al. Emergence of multidrug-resistant NDM-1-producing gram-negative bacteria in Bangladesh. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Akeda, Y.; Hagiya, H.; Sakamoto, N.; Takeuchi, D.; Shanmugakani, R.K.; Motooka, D.; Nishi, I.; Zin, K.N.; Aye, M.M.; et al. Spreading patterns of NDM-producing enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Ahmad, N.; Khalid, S.; Ali, S.M.; Khan, A.U. Occurrence of blaNDM variants among enterobacteriaceae from a neonatal intensive care unit in a Northern India hospital. Front Microbiol. 2018, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Khatun, M.N.; Farzana, R.; Lopes, B.S.; Shamsuzzaman, S.M. Molecular characterization and resistance profile of nosocomial Acinetobacter baumannii in intensive care unit of tertiary care hospital in Bangladesh. Bangladesh Med. Res. Counc. Bull. 2015, 41, 101–107. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Age Group (Years) | Sex (%) | Suspected VAP Cases | |
|---|---|---|---|
| Male | Female | ||
| 5–15 | 3 | 1 | 4 |
| 16–25 | 5 | 4 | 9 |
| 26–35 | 6 | 4 | 10 |
| 36–45 | 6 | 1 | 7 |
| 46–55 | 11 | 4 | 15 |
| 56–65 | 18 | 2 | 20 |
| 66–75 | 20 | 10 | 30 |
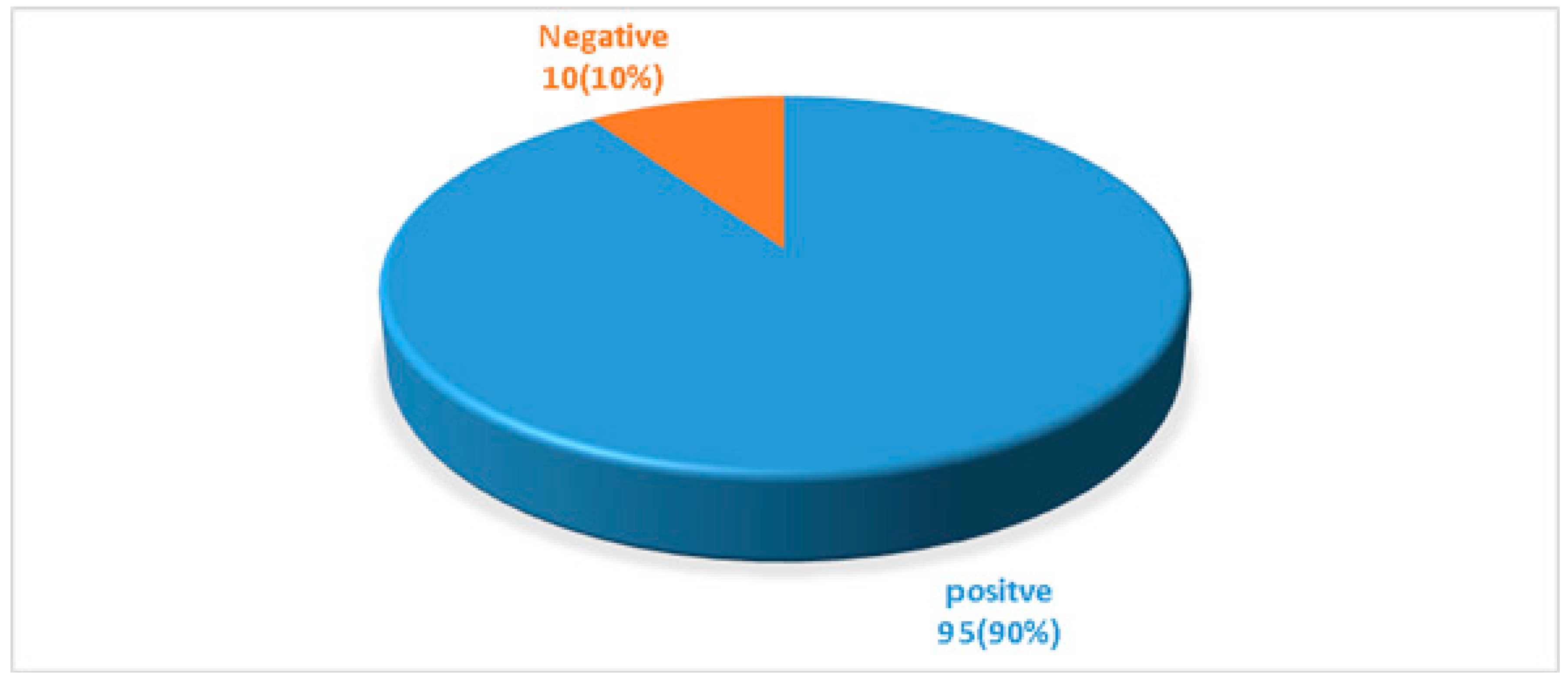
| Total | 69 (72.63%) | 26 (27.37%) | 95 |
| Name of the Organism | Imipenem-Resistant Screening Test (%) | |
|---|---|---|
| Positive | Negative | |
| Acinetobacter spp. (41) | 23 (56.1) | 18 |
| Klebsiella spp. (24) | 13 (54.17) | 11 |
| Pseudomonas spp. (18) | 6 (33.33) | 12 |
| E. coli (9) | 3 (33.33) | 6 |
| Proteus spp. (4) | 1 (25) | 3 |
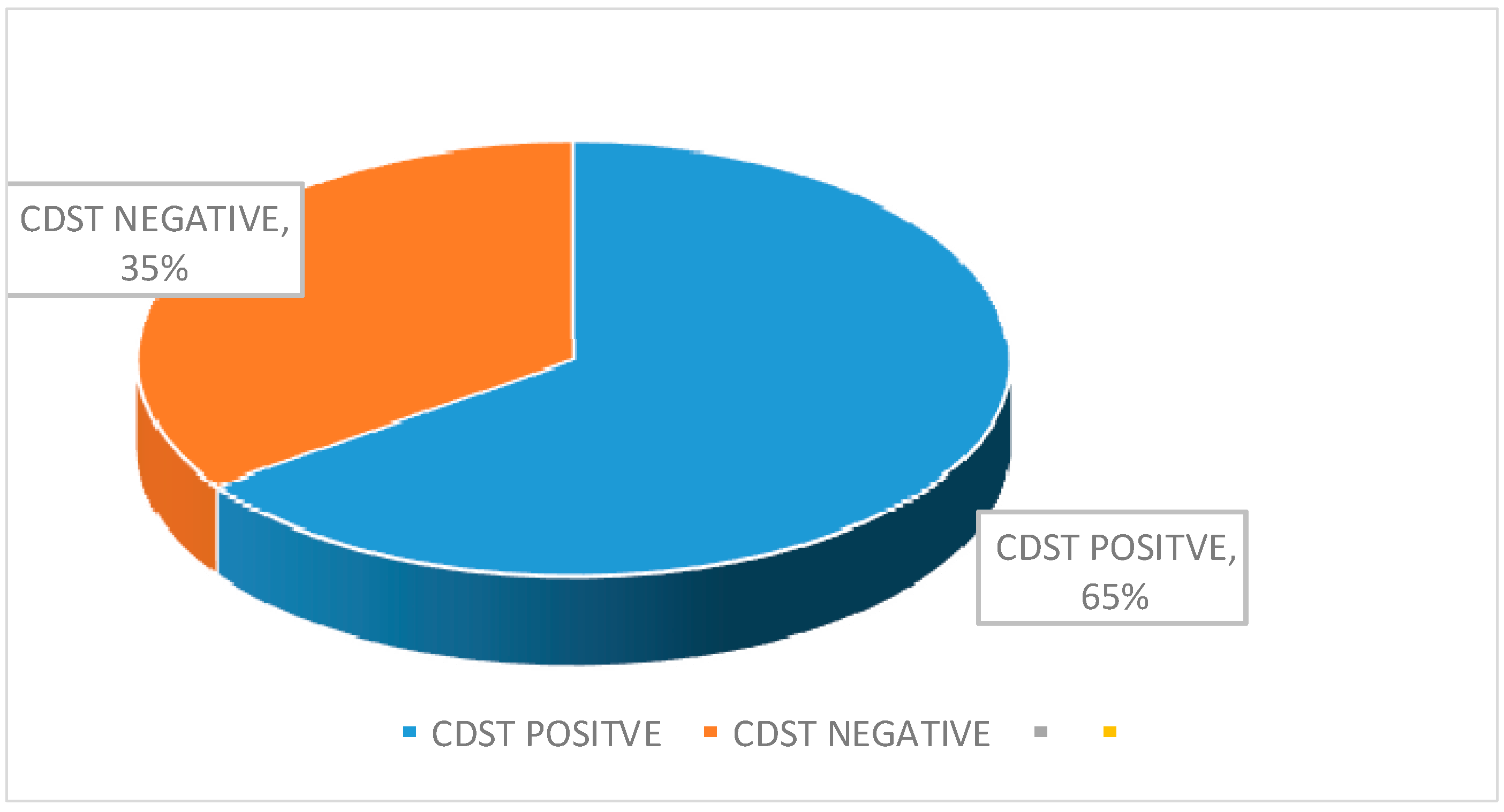
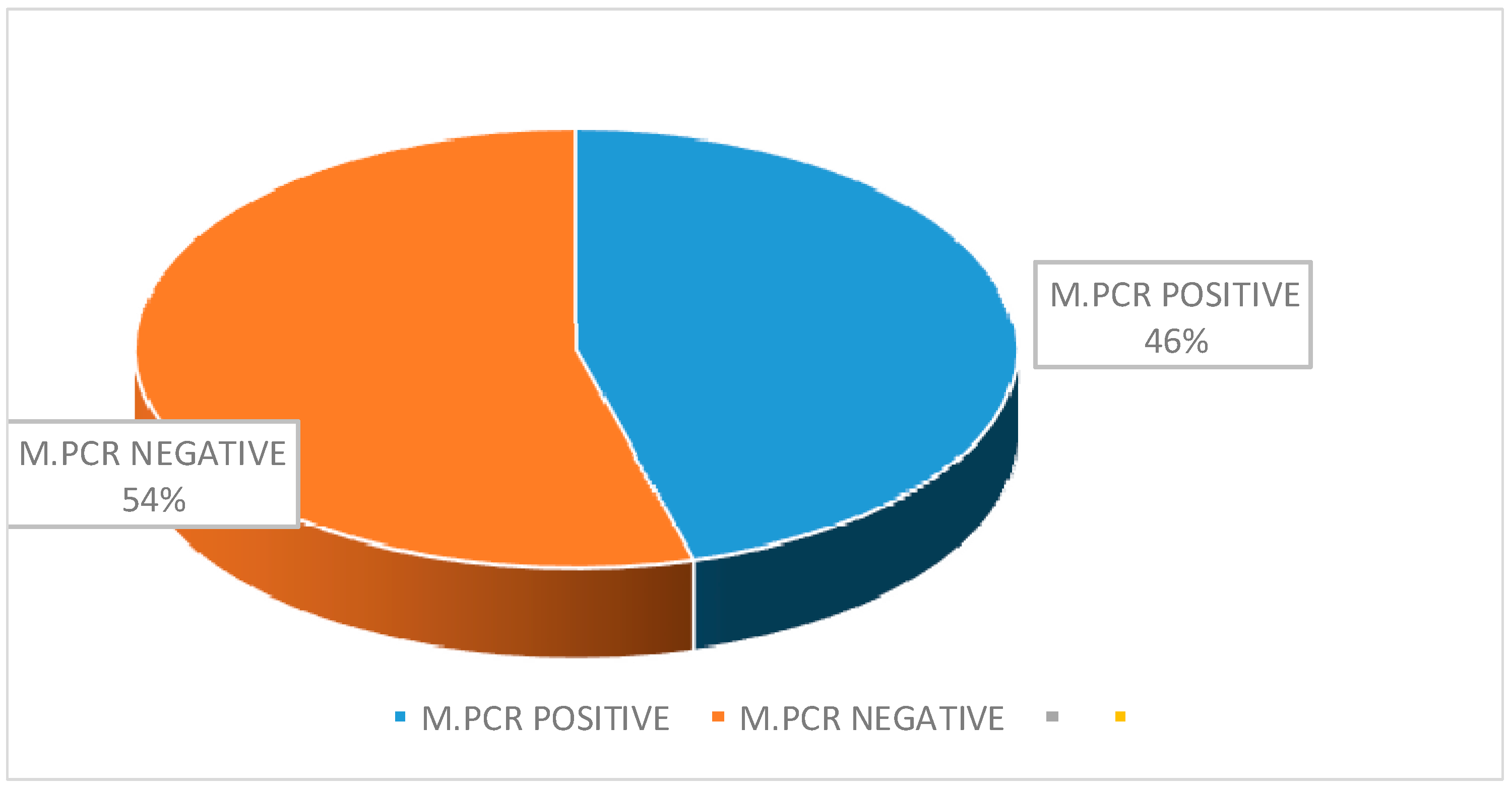
| Total | 46 (48.42) | 49 (51.60) |
| Test | PCR | Total | ||
|---|---|---|---|---|
| Positive | Negative | |||
| CDST | Positive | 18 | 12 | 30 |
| Negative | 3 | 13 | 16 | |
| Total | 21 | 25 | 46 | |
| Statistic | Value | 95% Confidence Interval |
|---|---|---|
| Sensitivity | 85.7% | 63.7–97.0% |
| Specificity | 52.0% | 31.3–72.2% |
| Positive predictive value | 60.0% | 49.0–70.0% |
| Negative predictive value | 81.3% | 58.7–93.0% |
| Accuracy | 67.4% | 52.0–80.5% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nusrat, T.; Akter, N.; Haque, M.; Rahman, N.A.A.; Dewanjee, A.K.; Ahmed, S.; Rozario, D.T.D. Comparative Study of CDST & Multiplex PCR to Detect MBL Producing Gram-Negative Bacilli among VAP Patients Admitted in a Public Medical College Hospital of Bangladesh. Pathogens 2019, 8, 151. https://doi.org/10.3390/pathogens8030151
Nusrat T, Akter N, Haque M, Rahman NAA, Dewanjee AK, Ahmed S, Rozario DTD. Comparative Study of CDST & Multiplex PCR to Detect MBL Producing Gram-Negative Bacilli among VAP Patients Admitted in a Public Medical College Hospital of Bangladesh. Pathogens. 2019; 8(3):151. https://doi.org/10.3390/pathogens8030151
Chicago/Turabian StyleNusrat, Tanzina, Nasima Akter, Mainul Haque, Nor Azlina A. Rahman, Arup Kanti Dewanjee, Shakeel Ahmed, and Diana Thecla D. Rozario. 2019. "Comparative Study of CDST & Multiplex PCR to Detect MBL Producing Gram-Negative Bacilli among VAP Patients Admitted in a Public Medical College Hospital of Bangladesh" Pathogens 8, no. 3: 151. https://doi.org/10.3390/pathogens8030151
APA StyleNusrat, T., Akter, N., Haque, M., Rahman, N. A. A., Dewanjee, A. K., Ahmed, S., & Rozario, D. T. D. (2019). Comparative Study of CDST & Multiplex PCR to Detect MBL Producing Gram-Negative Bacilli among VAP Patients Admitted in a Public Medical College Hospital of Bangladesh. Pathogens, 8(3), 151. https://doi.org/10.3390/pathogens8030151





