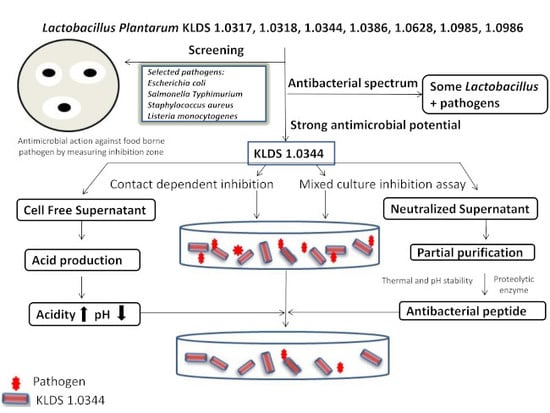
Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Potentiality Screening by Oxford Cup Technique
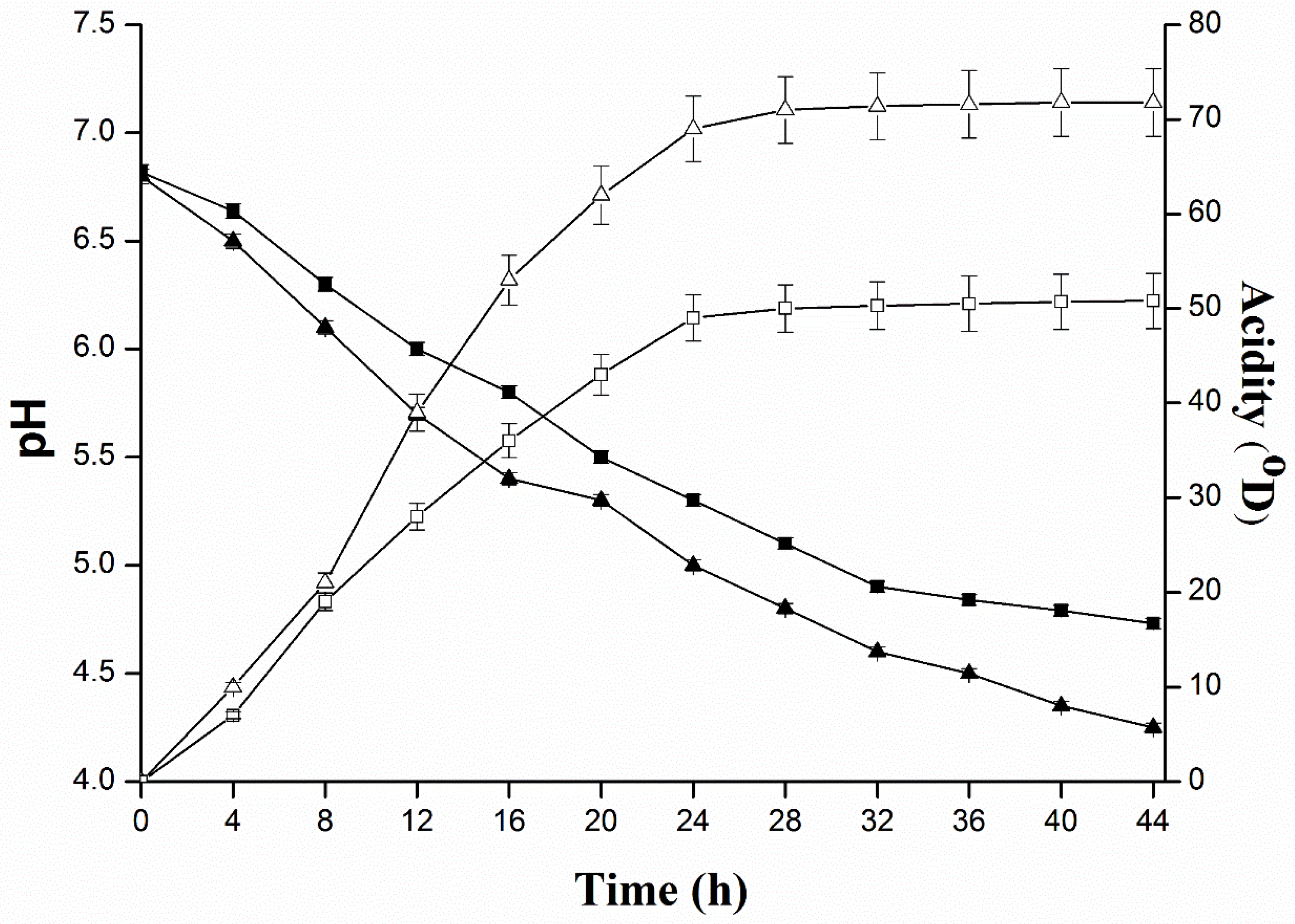
2.2. Determination of Inhibitory Substances of L. plantarum KLDS 1.0344 and Evaluate the Effect of Protease Enzymes on Their Activity
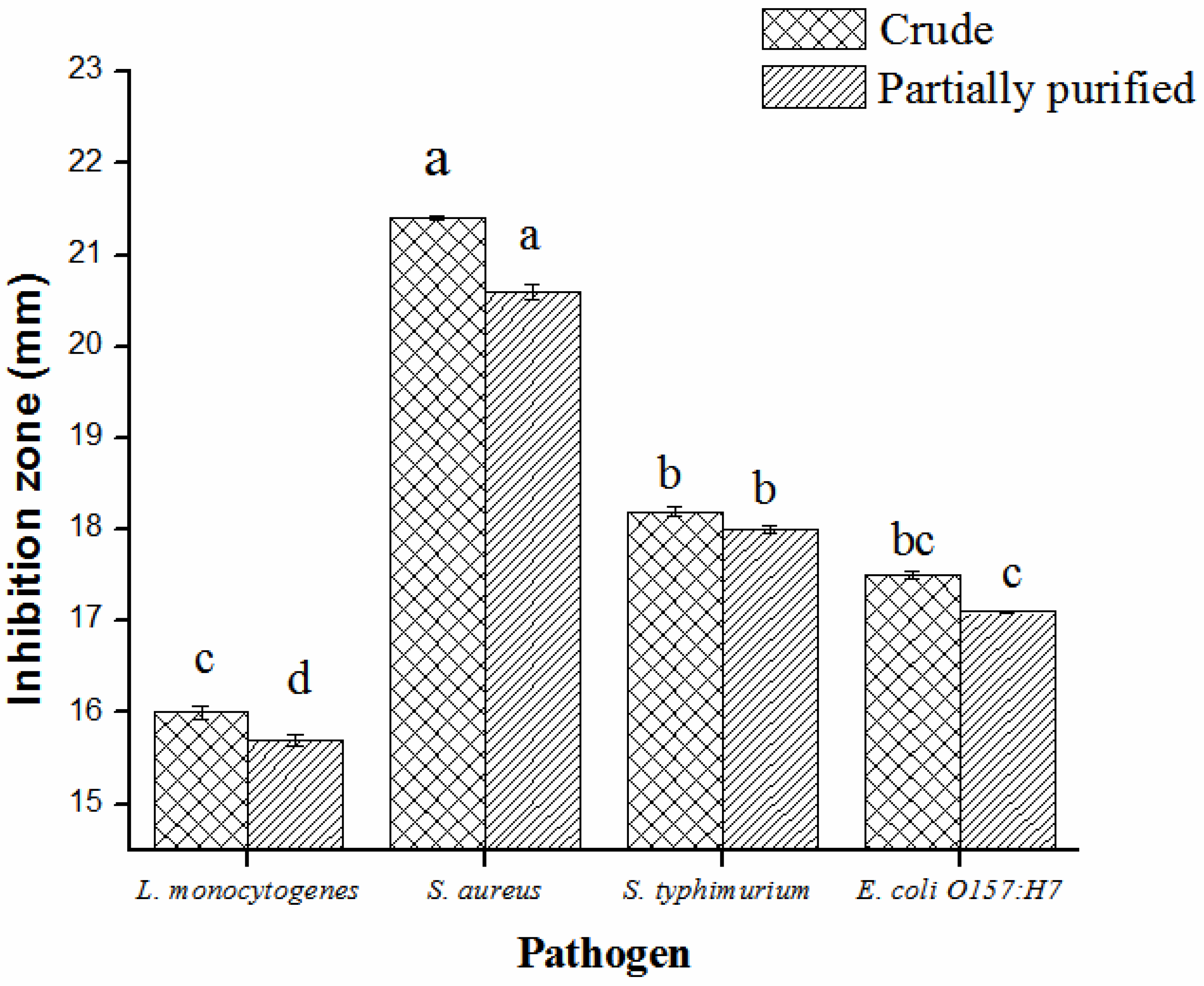
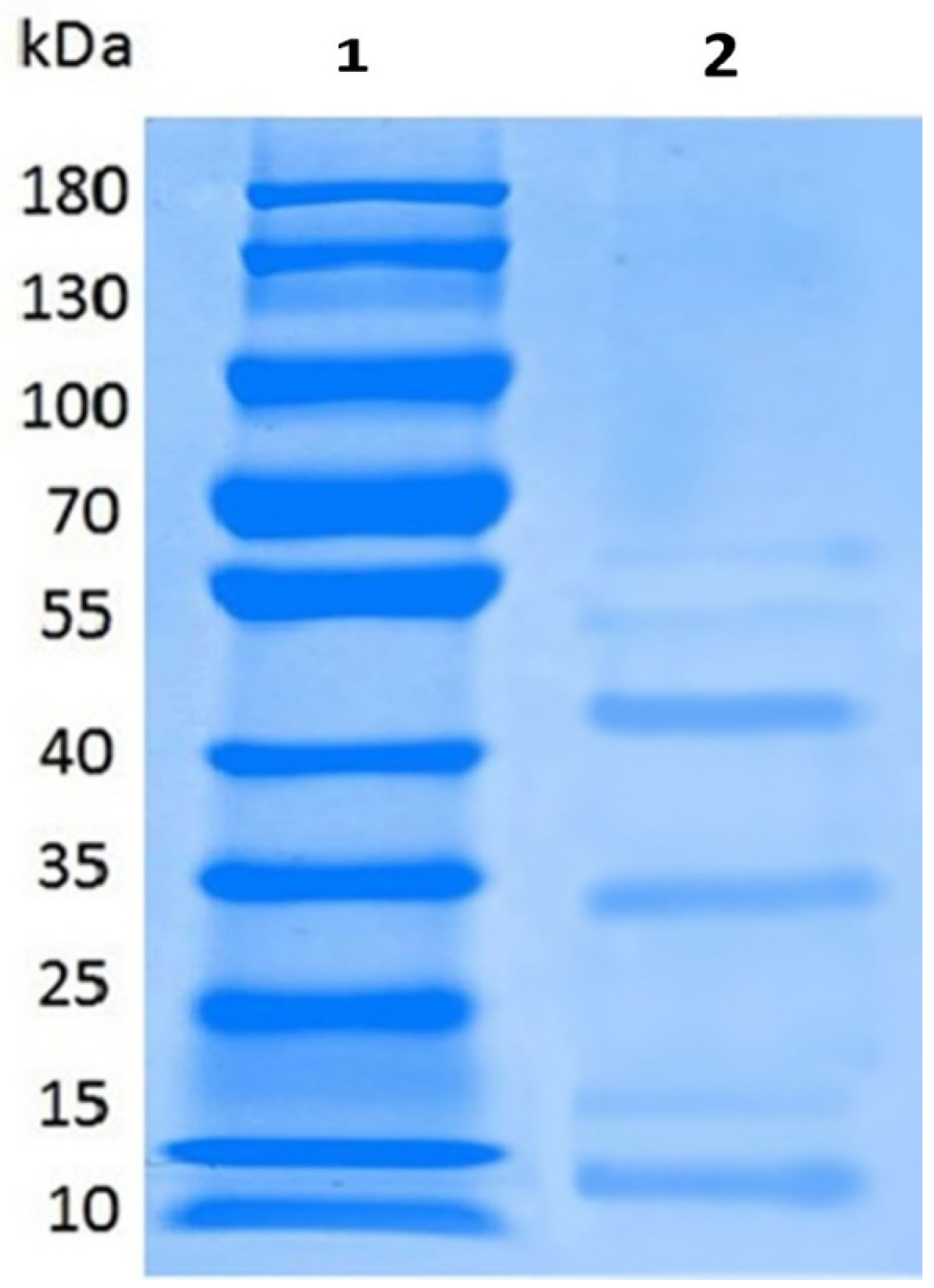
2.3. Partial Purification, Molecular Weight, Thermal and pH Stability of Bacteriocin
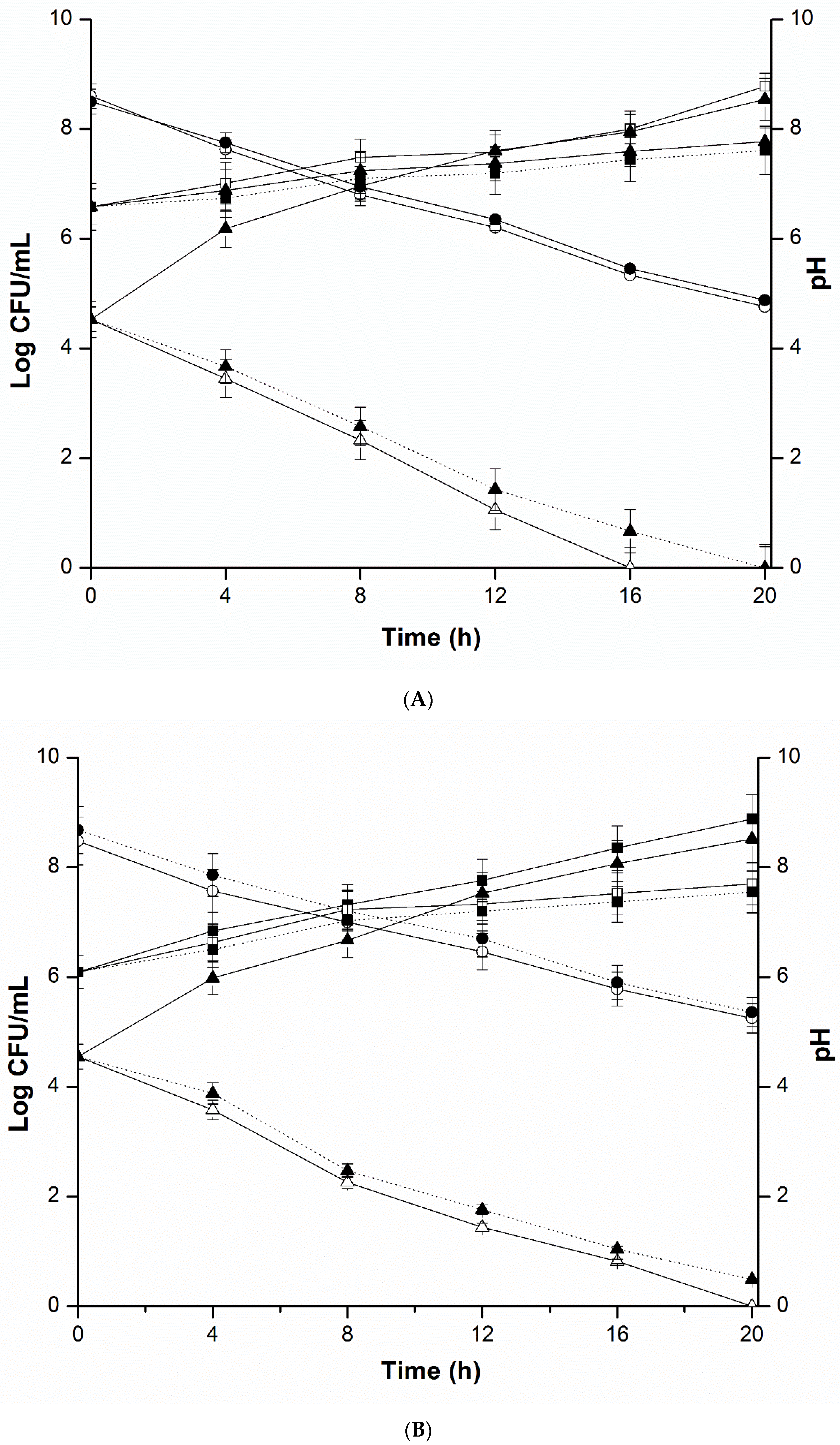
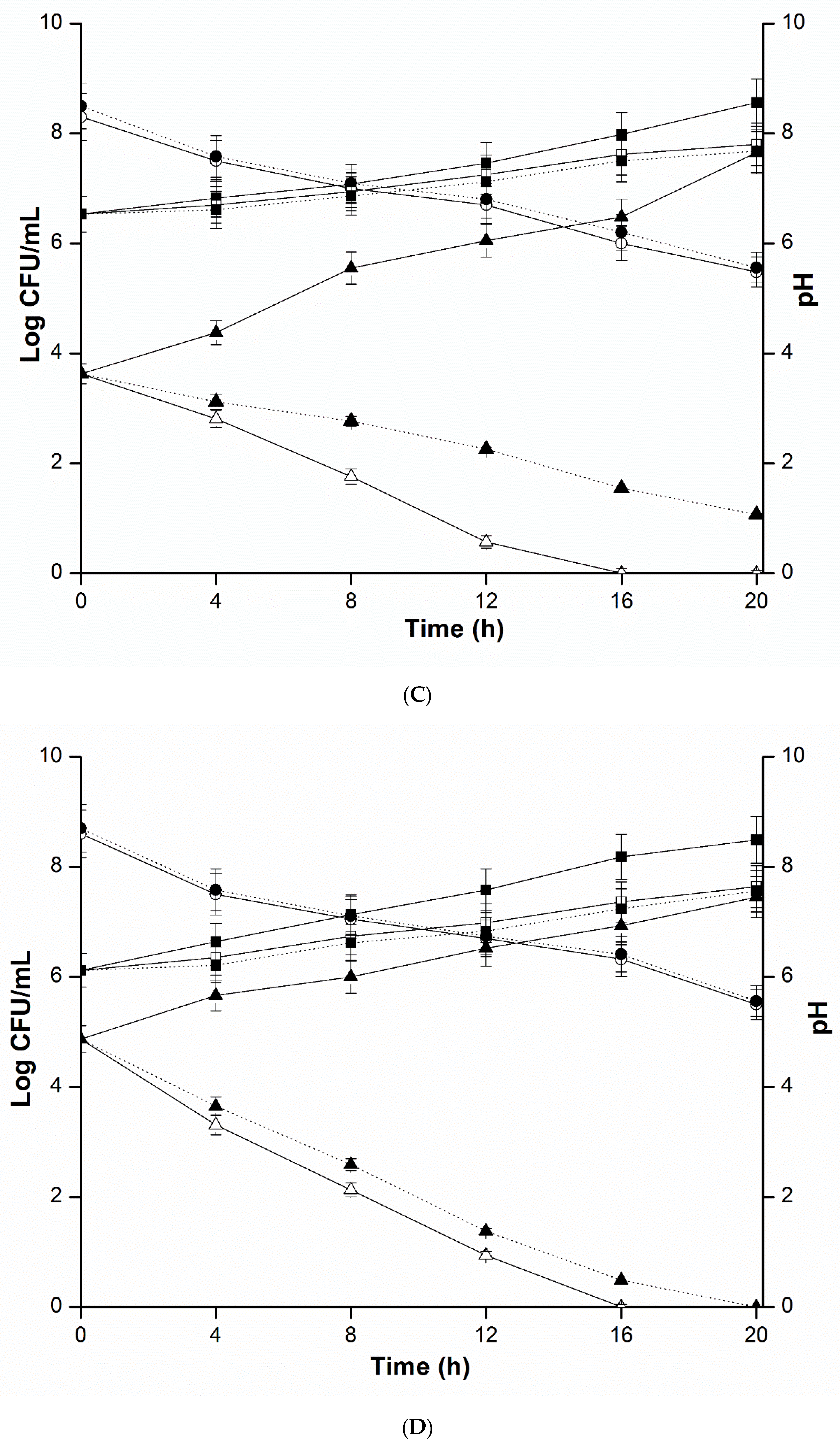
2.4. Mixed Culture Inhibition Assay
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Preparation Cell Free Supernatants of L. plantarum and Screening for Antimicrobial Potentiality
4.3. Antibacterial Ambit of L. plantarum KLDS 1.0344
4.4. Quantification of Acid Production of L. plantarum KLDS 1.0344
4.5. Mixed Culture Inhibition Assay
4.6. Antimicrobial Peptide Production
4.7. Antimicrobial Fractional Refinement by Dialysis and Ammonium Sulfate Precipitation
4.7.1. The Sensitivity of Antimicrobial Substances from L. plantarum LKDS 1.0344 to Proteolytic Enzymes
4.7.2. Thermal Stability of Partially Purified Antimicrobial Compounds
4.7.3. pH Stability of Partially Purified Antimicrobial Compounds
4.8. Electrophoresis Analysis
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- WHO. WHO Methods and Data Sources for Country-Level Causes of Death 2000-2015; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- The World Health Organization. Foodborne Disease Outbreaks: Guidelines for Investigation and Control WHO Library Cataloguing-in-Publication Data; WHO: Geneva, Switzerland, 2008. [Google Scholar]
- Flint, J.A.; Van Duynhoven, Y.T.; Angulo, F.J.; DeLong, S.M.; Braun, P.; Kirk, M.; Scallan, E.; Fitzgerald, M.; Adak, G.K.; Sockett, P.; et al. Estimating the burden of acute gastroenteritis, foodborne disease, and pathogens commonly transmitted by food: An International Review. Clin. Infect. Dis. 2005, 41, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Anyogu, A.; Awamaria, B.; Sutherland, J.P.; Ouoba, L.I.I. Molecular characterisation and antimicrobial activity of bacteria associated with submerged lactic acid cassava fermentation. Food Control 2014, 39, 119–127. [Google Scholar] [CrossRef]
- da Silva Sabo, S.; Vitolo, M.; González, J.M.D.; de Souza Oliveira, R.P. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014, 64, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Šušković, J.; Kos, B.; Beganović, J.; Pavunc, A.L.; Habjanič, K.; Matoć, S. Antimicrobial activity - The most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 2010, 48, 296–307. [Google Scholar]
- Guidone, A.; Zotta, T.; Ross, R.P.; Stanton, C.; Rea, M.C.; Parente, E.; Ricciardi, A. Functional properties of Lactobacillus plantarum strains: A multivariate screening study. LWT Food Sci. Technol. 2014, 56, 69–76. [Google Scholar] [CrossRef]
- Gaur, Y.; Narayan, K.; Chauhan, S.; Ali, A. Bacteriocinogeny: Concept, nomenclature, prevalence and application. Indian J. Microbiol. 2004, 44, 1–30. [Google Scholar]
- Jia, F.; Zhang, L.; Pang, X.; Gu, X.; Abdelazez, A.; Liang, Y. Genomics Complete genome sequence of bacteriocin-producing Lactobacillus plantarum KLDS1. 0391, a probiotic strain with gastrointestinal tract resistance and adhesion to the intestinal epithelial cells. Genomics 2017, 109, 6–11. [Google Scholar] [CrossRef]
- Kalita, D.; Saikia, S.; Gautam, G.; Mukhopadhyay, R. LWT - Food Science and Technology Characteristics of synbiotic spray dried powder of litchi juice with Lactobacillus plantarum and different carrier materials. LWT Food Sci. Technol. 2018, 87, 351–360. [Google Scholar] [CrossRef]
- Oguntoyinbo, F.A.; Narbad, A. Multifunctional properties of Lactobacillus plantarum strains isolated from fermented cereal foods. J. Funct. Foods 2015, 17, 621–631. [Google Scholar] [CrossRef]
- Rodríguez-Pazo, N.; Vázquez-Araújo, L.; Pérez-Rodríguez, N.; Cortés-Diéguez, S.; Domínguez, J.M. Cell-free supernatants obtained from fermentation of cheese whey hydrolyzates and phenylpyruvic acid by Lactobacillus plantarum as a source of antimicrobial compounds, bacteriocins, and natural aromas. Appl. Biochem. Biotechnol. 2013, 171, 1042–1060. [Google Scholar] [CrossRef]
- Tejero-Sariñena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Neal-McKinney, J.M.; Lu, X.; Duong, T.; Larson, C.L.; Call, D.R.; Shah, D.H.; Konkel, M.E. Production of organic acids by probiotic Lactobacilli can be used to reduce pathogen load in poultry. PLoS ONE 2012, 7, e43928. [Google Scholar] [CrossRef] [PubMed]
- Al Kassaa, I.; Hamze, M.; Hober, D.; Chihib, N.E.; Drider, D. Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microb. Ecol. 2014, 67, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Kos, B.; Beganović, J.; Jurašić, L.; Švađumović, M. Coculture-inducible bacteriocin biosynthesis of different probiotic strains by dairy starter culture Lactococcus lactis. Mljekarstvo 2011, 61, 273–282. [Google Scholar]
- Cortés-Zavaleta, O.; López-Malo, A.; Hernández-Mendoza, A.; García, H.S. Antifungal activity of lactobacilli and its relationship with 3-phenyllactic acid production. Int. J. Food Microbiol. 2014, 173, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic acid bacteria antimicrobial compounds: Characteristics and applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Varsha, K.K.; Nampoothiri, K.M. Appraisal of lactic acid bacteria as protective cultures. Food Control 2016, 69, 61–64. [Google Scholar] [CrossRef]
- Ma, Q.; Fu, Y.; Sun, H.; Huang, Y.; Li, L.; Yu, Q.; Dinnyes, A.; Sun, Q. Antimicrobial resistance of Lactobacillus spp. from fermented foods and human gut. LWT Food Sci. Technol. 2017, 86, 201–208. [Google Scholar] [CrossRef]
- Demirbaş, F.; İspirli, H.; Kurnaz, A.A.; Yilmaz, M.T.; Dertli, E. Antimicrobial and functional properties of lactic acid bacteria isolated from sourdoughs. LWT Food Sci. Technol. 2017, 79, 361–366. [Google Scholar] [CrossRef]
- Elviso, S.B.; Iordano, M.G.; Olci, P.D.; Eppa, G.Z. Original article in vitro cholesterol-lowering activity of Lactobacillus plantarum and Lactobacillus paracasei strains isolated from the Italian Castelmagno PDO cheese. Dairy Sci. Technol. 2009, 89, 169–176. [Google Scholar] [CrossRef]
- Sip, A.; Wieckowicz, M.; Olejnik-Schmidt, A.; Grajek, W. Anti-Listeria activity of lactic acid bacteria isolated from golka, a regional cheese produced in Poland. Food Control 2012, 26, 117–124. [Google Scholar] [CrossRef]
- Papanikolaou, Z.; Hatzikamari, M.; Georgakopoulos, P.; Yiangou, M.; Litopoulou-tzanetaki, E.; Tzanetakis, N. Selection of dominant NSLAB from a mature traditional cheese according to their technological properties and in vitro intestinal challenges. J. Food Sci. 2012, 77, M298–M306. [Google Scholar] [CrossRef] [PubMed]
- Turgut, T.; Erdoğan, A. Karın Kaymağı Peynirinden İzole Edilen Laktobasillerin Tanımlanması Makale Kodu (Article Code): KVFD-2011-5272 Identification of Lactobacilli Isolated from Cheese Karın Kaymak. Kafkas Üniversitesi Veteriner Fakültesi Dergisi 2012, 18, 209–213. [Google Scholar]
- Veljovic, K.; Vukasinovic, M.; Strahinic, I.; Begovic, J.; Lozo, J.; Ostojic, M. Preliminary characterization of lactic acid bacteria isolated from Zlatar cheese. J. Appl. Microbiol. 2007, 103, 2142–2152. [Google Scholar] [CrossRef] [PubMed]
- Nanda, D.K.; Tomar, S.K.; Singh, R.; Mal, G.; Singh, P.; Arora, D.K.; Joshi, B.K.; Chaudhary, R.; Kumar, D. Phenotypic and genotypic characterisation of Lactobacilli isolated from camel cheese produced in India. Int. J. Dairy Technol. 2011, 64, 437–443. [Google Scholar] [CrossRef]
- Maris, S.; Meira, M.; Helfer, V.E.; Velho, R.V.; Lopes, F.C.; Brandelli, A. Probiotic potential of Lactobacillus spp. isolated from Brazilian regional ovine cheese. J. Dairy Res. 2012, 79, 119–127. [Google Scholar]
- Duan, Y.; Tan, Z.; Wang, Y.; Li, Z.; Li, Z.; Qin, G.; Huo, Y.; Cai, Y. Identification and characterization of lactic acid bacteria isolated from Tibetan Qula cheese. J. Gen. Appl. Microbiol. 2008, 60, 51–60. [Google Scholar] [CrossRef]
- Adegoke, G.O. Bacteriocin and cellulose production by lactic acid bacteria isolated from West African soft cheese. Afr. J. Biotechnol. 2007, 6, 2616–2619. [Google Scholar]
- Zhang, J.; Zhang, X.; Zhang, L.; Zhao, Y.; Niu, C.; Yang, Z.; Li, S. Potential probiotic characterization of Lactobacillus plantarum strains isolated from Inner Mongolia “Hurood” Cheese. J. Microbiol. Biotechnol. 2014, 24, 225–235. [Google Scholar] [CrossRef]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with functional properties: An approach to increase safety and shelf-life of fermented foods. BioMed Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. Correction: World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, Z.; Sun, X.; Pan, Y.; Zhao, Y. Development of a quantitative real-time PCR assay for viable Salmonella spp. without enrichment. Food Control 2015, 57, 185–189. [Google Scholar] [CrossRef]
- Omiccioli, E.; Amagliani, G.; Brandi, G.; Magnani, M. A new platform for Real-Time PCR detection of Salmonella spp., Listeria monocytogenes and Escherichia coli O157 in milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Y.; Yu, L.R.; Jones, R.C.; Elkins, C.A.; Hart, M.E. Inhibition of Staphylococcus aureus by lysostaphin-expressing Lactobacillus plantarum WCFS1 in a modified genital tract secretion medium. Appl. Environ. Microbiol. 2011, 77, 8500–8508. [Google Scholar] [CrossRef] [PubMed]
- Kase, J.A.; Zhang, G.; Chen, Y. Recent foodborne outbreaks in the United States linked to atypical vehicles—Lessons learned. Curr. Opin. Food Sci. 2017, 18, 56–63. [Google Scholar] [CrossRef]
- Ly, V.; Parreira, V.R.; Farber, J.M. Current understanding and perspectives on Listeria monocytogenesin low-moisture foods. Curr. Opin. Food Sci. 2019, 26, 18–24. [Google Scholar] [CrossRef]
- Thakur, M.; Asrani, R.K.; Patial, V. Listeria monocytogenes: A food-borne pathogen. In Foodborne Diseases; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780128114964. [Google Scholar]
- Kim, Y.C.; Won, H.K.; Lee, J.W.; Sohn, K.H.; Kim, M.H.; Kim, T.B.; Chang, Y.S.; Lee, B.J.; Cho, S.H.; Bachert, C.; et al. Staphylococcus aureus nasal colonization and asthma in adults: Systematic review and meta-analysis. J. Allergy Clin. Immunol. Pract. 2019, 7, 606–615. [Google Scholar] [CrossRef]
- Oestergaard, L.B.; Schmiegelow, M.D.S.; Bruun, N.E.; Skov, R.; Andersen, P.S.; Larsen, A.R.; Gerds, T.A.; Dahl, A.; Petersen, A.; Lauridsen, T.K.; et al. Staphylococcus aureus bacteremia in children aged 5-18 years—Risk factors in the new Millennium. J. Pediatr. 2018, 203, 108–115. [Google Scholar] [CrossRef]
- Tanveer, F.; Bhargava, A.; Riederer, K.; Johnson, L.B.; Khatib, R. Low frequency of Staphylococcus aureus in lower extremity skin and soft tissue infections. Am. J. Med. Sci. 2018, 356, 528–530. [Google Scholar] [CrossRef]
- Wakabayashi, Y.; Umeda, K.; Yonogi, S.; Nakamura, H.; Yamamoto, K.; Kumeda, Y.; Kawatsu, K. Staphylococcal food poisoning caused by Staphylococcus argenteus harboring staphylococcal enterotoxin genes. Int. J. Food Microbiol. 2018, 265, 23–29. [Google Scholar] [CrossRef]
- Beutin, L.; Martin, A. Outbreak of Shiga Toxin–Producing Escherichia coli (STEC) O104:H4 Infection in germany causes a paradigm shift with regard to human pathogenicity of STEC Sstrains. J. Food Prot. 2012, 75, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Larraínzar, M.; Rúa, J.; Caro, I.; de Castro, C.; de Arriaga, D.; García-Armesto, M.R.; del Valle, P. Evaluation of antimicrobial and antioxidant activities of natural phenolic compounds against foodborne pathogens and spoilage bacteria. Food Control 2012, 26, 555–563. [Google Scholar] [CrossRef]
- Cálix-Lara, T.F.; Rajendran, M.; Talcott, S.T.; Smith, S.B.; Miller, R.K.; Castillo, A.; Sturino, J.M.; Taylor, T.M. Inhibition of Escherichia coli O157: H7 and Salmonella enterica on spinach and identification of antimicrobial substances produced by a commercial lactic acid bacteria food safety intervention. Food Microbiol. 2014, 38, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Son, S.H.; Jeon, H.L.; Yang, S.J.; Lee, N.K.; Paik, H.D. In vitro characterization of Lactobacillus brevis KU15006, an isolate from kimchi, reveals anti-adhesion activity against foodborne pathogens and antidiabetic properties. Microb. Pathog. 2017, 112, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Sato, Y.; Kudo, S.; Sato, S.; Nakajima, H.; Toba, T. The screening of hydrogen peroxide-producing lactic acid bacteria and their application to inactivating psychrotrophic food-borne pathogens. Curr. Microbiol. 2003, 47, 231–236. [Google Scholar]
- Chiu, H.H.; Tsai, C.C.; Hsih, H.Y.; Tsen, H.Y. Screening from pickled vegetables the potential probiotic strains of lactic acid bacteria able to inhibit the Salmonella invasion in mice. J. Appl. Microbiol. 2008, 104, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Tharmaraj, N.; Shah, N.P. Antimicrobial effects of probiotics against selected pathogenic and spoilage bacteria in cheese-based dips. Int. Food Res. J. 2009, 16, 261–276. [Google Scholar]
- Herreros, M.A.; Sandoval, H.; González, L.; Castro, J.M.; Fresno, J.M.; Tornadijo, M.E. Antimicrobial activity and antibiotic resistance of lactic acid bacteria isolated from Armada cheese (a Spanish goats’ milk cheese). Food Microbiol. 2005, 22, 455–459. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Abushelaibi, A.; Al-Mahadin, S.; El-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT Food Sci. Technol. 2017, 79, 316–325. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Requena, T.; Peláez, C.; Bartolomé, B.; Moreno-Arribas, M.V.; Martínez-Cuesta, M.C. Antimicrobial activity of lacticin 3147 against oenological lactic acid bacteria. Combined effect with other antimicrobial agents. Food Control 2013, 32, 477–483. [Google Scholar] [CrossRef]
- Hernández, D.; Cardell, E.; Zárate, V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: Initial characterization of plantaricin TF711, a bacteriocin-like substance produced by Lactobacillus plantarum TF711. J. Appl. Microbiol. 2005, 99, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Bezares, B.; Sáenz, Y.; Navarro, L.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Coculture-inducible bacteriocin activity of Lactobacillus plantarum strain J23 isolated from grape must. Food Microbiol. 2007, 24, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.K.; Tsai, H.C.; Lin, P.P.; Tsen, H.Y.; Tsai, C.C. Lactobacillus acidophilus LAP5 able to inhibit the Salmonella choleraesuis invasion to the human Caco-2 epithelial cell. Anaerobe 2008, 14, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Kong, B.; Chen, Q.; Sun, F.; Zhang, H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J. Funct. Foods 2017, 32, 391–400. [Google Scholar] [CrossRef]
- Katina, K.; Sauri, M.; Alakomi, H.-L.; Mattila-Sandholm, T. Potential of lactic acid bacteria to inhibit rope spoilage in wheat sourdough bread. LWT Food Sci. Technol. 2002, 35, 38–45. [Google Scholar] [CrossRef]
- Anas, M.; Jamal Eddine, H.; Mebrouk, K. Antimicrobial activity of Lactobacillus species isolated from Algerian raw goat’s milk against Staphylococcus aureus. World J. Dairy Food Sci. 2008, 3, 39–49. [Google Scholar]
- Pei, J.; Li, X.; Han, H.; Tao, Y. Purification and characterization of plantaricin SLG1, a novel bacteriocin produced by Lb. plantarum isolated from yak cheese. Food Control 2018, 84, 111–117. [Google Scholar] [CrossRef]
- Yi, L.; Dang, J.; Zhang, L.; Wu, Y.; Liu, B.; Lü, X. Purification, characterization and bactericidal mechanism of a broad spectrum bacteriocin with antimicrobial activity against multidrug-resistant strains produced by Lactobacillus coryniformis XN8. Food Control 2016, 67, 53–62. [Google Scholar] [CrossRef]
- Ben Omar, N.; Abriouel, H.; Keleke, S.; Sánchez Valenzuela, A.; Martínez-Cañamero, M.; Lucas López, R.; Ortega, E.; Gálvez, A. Bacteriocin-producing Lactobacillus strains isolated from poto poto, a Congolese fermented maize product, and genetic fingerprinting of their plantaricin operons. Int. J. Food Microbiol. 2008, 127, 18–25. [Google Scholar] [CrossRef]
- Abo-Amer, A.E. Characterization of a bacteriocin-like inhibitory substance produced by Lactobacillus plantarum isolated from Egyptian home-made yogurt. ScienceAsia 2007, 33, 313–319. [Google Scholar] [CrossRef]
- Martinez, R.C.R.; Wachsman, M.; Torres, N.I.; LeBlanc, J.G.; Todorov, S.D.; de Melo Franco, B.D.G. Biochemical, antimicrobial and molecular characterization of a noncytotoxic bacteriocin produced by Lactobacillus plantarum ST71KS. Food Microbiol. 2013, 34, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Ho, P.; Vaz-Velho, M.; Dicks, L.M.T. Characterization of bacteriocins produced by two strains of Lactobacillus plantarum isolated from Beloura and Chouriço, traditional pork products from Portugal. Meat Sci. 2010, 84, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Chun, C.C.; Chen, K.C.; Pin Der, D.; Peng, S.W.; Shu, C.W. Antibacterial properties of Lactobacillus plantarum isolated from fermented mustards against Streptococcus mutans. Afr. J. Microbiol. Res. 2013, 7, 4787–4793. [Google Scholar] [CrossRef]
- Huang, R.; Tao, X.; Wan, C.; Li, S.; Xu, H.; Xu, F.; Shah, N.P.; Wei, H. In vitro probiotic characteristics of Lactobacillus plantarum ZDY 2013 and its modulatory effect on gut microbiota of mice. J. Dairy Sci. 2015, 98, 5850–5861. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, D. Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobe 2015, 33, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Cordisco, L.; Tarasco, V.; Locatelli, E.; Di Gioia, D.; Oggero, R.; Matteuzzi, D. Antagonistic effect of Lactobacillus strains against gas-producing coliforms isolated from colicky infants. BMC Microbiol. 2011, 11, 157. [Google Scholar] [CrossRef] [PubMed]
- Varma, P.; Dinesh, K.R.; Menon, K.K.; Biswas, R. Lactobacillus fermentum Iisolated from human colonic mucosal biopsyinhibits the growth and adhesion of enteric and foodborne pathogens. J. Food Sci. 2010, 75, M546–M551. [Google Scholar] [CrossRef]
- Delbes-Paus, C.; Dorchies, G.; Chaabna, Z.; Callon, C.; Montel, M.C. Contribution of hydrogen peroxide to the inhibition of Staphylococcus aureus by Lactococcus garvieae in interaction with raw milk microbial community. Food Microbiol. 2010, 27, 924–932. [Google Scholar] [CrossRef]
- Turner, M.S.; Lo, R.; Giffard, P.M. Inhibition of Staphylococcus aureus growth on tellurite-containing media by Lactobacillus reuteri is dependent on CyuC and thiol production. Appl. Environ. Microbiol. 2007, 73, 1005–1009. [Google Scholar] [CrossRef]
- Laughton, J.M.; Devillard, E.; Heinrichs, D.E.; Reid, G.; McCormick, J.K. Inhibition of expression of a Staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology 2006, 152, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Davoodabadi, A.; Soltan Dallal, M.M.; Rahimi Foroushani, A.; Douraghi, M.; Sharifi Yazdi, M.K.; Amin Harati, F. Antibacterial activity of Lactobacillus spp. isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe 2015, 34, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Davoodabadi, A.; Dallal, M.M.S.; Lashani, E.; Ebrahimi, M.T. Antimicrobial activity of Lactobacillus spp. isolated from fecal flora of healthy breast-fed infants against diarrheagenic Escherichia coli. Jundishapur J. Microbiol. 2015, 8, e27852. [Google Scholar] [CrossRef] [PubMed]
- Han, K.S.; Kim, Y.; Kim, S.H.; Oh, S. Characterization and purification of acidocin 1B, a bacteriocin produced by Lactobacillus acidophilus GP1B. J. Microbiol. Biotechnol. 2007, 17, 774–783. [Google Scholar] [PubMed]
- Atassi, F.; Brassart, D.; Grob, P.; Graf, F.; Servin, A.L. In vitro antibacterial activity of Lactobacillus helveticus strain KS300 against diarrhoeagenic, uropathogenic and vaginosis-associated bacteria. J. Appl. Microbiol. 2006, 101, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Saraoui, T.; Fall, P.A.; Leroi, F.; Antignac, J.P.; Chéreau, S.; Pilet, M.F. Inhibition mechanism of Listeria monocytogenes by a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 2016, 53, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Bavananthasivam, J.; Dassanayake, R.P.; Kugadas, A.; Shanthalingam, S.; Call, D.R.; Knowles, D.P.; Srikumaran, S. Proximity-dependent inhibition of growth of Mannheimia haemolytica by Pasteurella multocida. Appl. Environ. Microbiol. 2012, 78, 6683–6688. [Google Scholar] [CrossRef]
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D. A contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309, 1245–1248. [Google Scholar] [CrossRef]
- Zechner, E.L.; Lang, S.; Schildbach, J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2012, 367, 1073–1087. [Google Scholar] [CrossRef]
- Schmidt, A.; Kochanowski, K.; Vedelaar, S.; Ahrné, E.; Volkmer, B.; Callipo, L.; Knoops, K.; Bauer, M.; Aebersold, R.; Heinemann, M. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016, 34, 104–110. [Google Scholar] [CrossRef]
- Sutrina, S.L.; Daniel, K.; Lewis, M.; Charles, N.T.; Anselm, C.K.E.; Thomas, N.; Holder, N. Biofilm growth of Escherichia coli is subject to cAMP-dependent and cAMPindependent inhibition. J. Mol. Microbiol. Biotechnol. 2015, 25, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Diner, E.J.; Beck, C.M.; Webb, J.S.; Low, D.A.; Hayes, C.S. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev. 2012, 26, 515–525. [Google Scholar] [CrossRef]
- Jabbari, V.; Khiabani, M.S.; Mokarram, R.R.; Hassanzadeh, A.M.; Ahmadi, E.; Gharenaghadeh, S.; Karimi, N.; Kafil, H.S. Lactobacillus plantarum as a probiotic potential from Kouzeh cheese (Traditional Iranian Cheese) and its antimicrobial activity. Probiotics Antimicrob. Proteins 2017, 9, 189–193. [Google Scholar] [CrossRef]
- Bian, X.; Evivie, S.E.; Muhammad, Z.; Luo, G.-W.; Liang, H.-Z.; Wang, N.-N.; Huo, G.-C. In vitro assessment of the antimicrobial potentials of Lactobacillus helveticus strains isolated from traditional cheese in Sinkiang China against food-borne pathogens. Food Funct. 2016, 7, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chi, Z.; Yue, L.; Li, J. Purification and characterization of killer toxin from a marine yeast Pichia anomala YF07b against the pathogenic yeast in crab. Curr. Microbiol. 2007, 55, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Z.; Wu, H.; Lv, F. Study on the antibiotic activity of microcapsule curcumin against foodborne pathogens. Int. J. Food Microbiol. 2009, 136, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Yin, R.; Yu, L.; Wang, G.; Tian, F.; Yu, R.; Zhao, J.; Liu, X.; Chen, Y.Q.; Zhang, H.; et al. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control 2015, 54, 23–30. [Google Scholar] [CrossRef]
- Srinivasan, P.; Khan, K.A.; Perumal, U.R.; Kumar, R.V.; Suganya, K.; Rajalakshmi, M. In vitro antibacterial activity of Lactobacillus plantarum isolated from soy milk. Int. J. Pharma Bio Sci. 2012, 3, 209–219. [Google Scholar]
- Gao, Y.; Jia, S.; Gao, Q.; Tan, Z. A novel bacteriocin with a broad inhibitory spectrum produced by Lactobacillus sake C2, isolated from traditional Chinese fermented cabbage. Food Control 2010, 21, 76–81. [Google Scholar] [CrossRef]
| KLDS 1.0317 | KLDS 1.0318 | KLDS 1.0344 | KLDS 1.0386 | KLDS 1.0628 | KLDS 1.0985 | KLDS 1.0986 | |
| pH | 4.6 ± 0.0 E | 4.9 ± 0.0 C | 3.4 ± 0.1 G | 5.5 ± 0.1 A | 5.3 ± 0.0 B | 4.7 ± 0.0 D | 4.4 ± 0.0 F |
| Pathogens | Culture | Culture | Culture | Culture | Culture | Culture | Culture |
| L. monocytogenes | 11.2 ± 0.1 aA | 5.6 ± 0.1 abE | 9.2 ± 0.1 bB | 7.9 ± 0.0 aD | - | 8.8 ± 0.1 aC | 9.3 ± 0.1 aB |
| S. aureus | - | 6.9 ± 0.2 aC | 15.8 ± 0.1 aA | - | 5.7 ± 0.0 aB | - | - |
| S. typhimurium | 7.8 ± 0.0 bB | - | 14.2 ± 0.0 aA | - | - | 4.9 ± 0.2 bC | - |
| E. coli O157:H7 | - | - | 12.7 ± 0.0 abA | - | - | - | - |
| KLDS 1.0317 | KLDS 1.0318 | KLDS 1.0344 | KLDS 1.0386 | KLDS 1.0628 | KLDS 1.0985 | KLDS 1.0986 | |
| pH | 5.6 ± 0.1 A | 4.8 ± 0.0 D | 3.3 ± 0.1 G | 5.4 ± 0.0 B | 5.3 ± 0.0 C | 4.7 ± 0.0 E | 4.3 ± 0.1 F |
| Pathogens | CFS | CFS | CFS | CFS | CFS | CFS | CFS |
| L. monocytogenes | 8.4 ± 0.2 aA | 2.4 ± 0.0 cE | 8.5 ± 0.0 bA | 4.6 ± 0.1 bD | - | 5.3 ± 0.1 aC | 6.3 ± 0.0 aB |
| S. aureus | - | 3.7 ± 0.0 aC | 13.8 ± 0.0 aA | - | 3.0 ± 0.0 bB | - | - |
| S. typhimurium | 4.9 ± 0.0 bB | - | 12.4 ± 0.1 aA | - | - | 3.0 ± 0.9 bC | - |
| E. coli O157:H7 | - | - | 10.4 ± 0.1 abA | - | - | - | - |
| Indicator Bacterium | Medium | Temperature | Sensitivity by Culture | Sensitivity by CFS |
|---|---|---|---|---|
| pH | 3.23 ± 0.06 | 3.19 ± 0.08 | ||
| Lactobacillus paracasei KLDS1.0201 | mMRS | 37 °C | + | + |
| Lactobacillus plantarum KLDS 1.0628 | mMRS | 37 °C | + | + |
| Lactobacillus helveticus KLDS 1.9202 | mMRS | 37 °C | + | + |
| Lactobacillus helveticus KLDS 1.9204 | mMRS | 37 °C | + | + |
| Lactococcus lactis KLDS 4.0325 | M17 | 37 °C | + | − |
| Sterptococcus thermophilus KLDS 3.0207 | M17 | 37 °C | − | + |
| Escherichia coli ATCC 43889 | BHI | 37 °C | +++ | +++ |
| Salmonella typhimurium ATCC 14028 | BHI | 37 °C | +++ | +++ |
| Staphylococcus aureus ATCC 25923 | BHI | 37 °C | +++ | +++ |
| Listeria monocytogenes ATCC 19115 | BHI | 37 °C | ++ | ++ |
| Lactobacillus plantarum KLDS 1.0986 | mMRS | 37 °C | − | − |
| Enzyme | L. monocytogenes | S. aureus | S. typhimurium | E. coli O157:H7 |
|---|---|---|---|---|
| Control | 15.2 ± 0.6 a | 21.7 ± 0.9 a | 18.3 ± 0.7 a | 17.8 ± 0.7 a |
| Pepsin | 12.7 ± 0.5 e | 18.5 ± 0.8 d | 16.3 ± 0.7 d | 15.6 ± 0.6 d |
| Protease | 14.5 ± 0.5 b | 20.4 ± 0.8 b | 17.9 ± 0.7 ab | 17.2 ± 0.7 b |
| Pronase | 13.3 ± 0.5 de | 18.7 ± 0.8 cd | 16.8 ± 0.6 cd | 16.2 ± 0.6 cd |
| Bromelain | 12.5 ± 0.5 e | 18.2 ± 0.7 e | 16.0 ± 0.6 e | 15.3 ± 0.6 e |
| Ficin | 14.0 ± 0.6 c | 19.8 ± 0.8 bc | 17.5 ± 0.7 b | 16.7 ± 0.7 bc |
| α-Chymotrypsin | 13.5 ± 0.6 d | 19.3 ± 0.8 c | 17.0 ± 0.6 c | 16.4 ± 0.7 c |
| pH | L. monocytogenes | S. aureus | S. typhimurium | E. coli O157:H7 |
|---|---|---|---|---|
| 2 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 3 | 82.3 ± 0.3 b | 78.1 ± 0.2 b | 73.2 ± 0.3 b | 71.4 ± 0.2 b |
| 4 | 65.4 ± 0.2 d | 60.0 ± 0.2 c | 56.4 ± 0.2 c | 54.0 ± 0.2 c |
| 5 | 50.0 ± 0.2 e | 42.4 ± 0.1 d | 39.1 ± 0.1 d | 33.0 ± 0.1 d |
| 6 | 29.2 ± 0.1 f | 20.2 ± 0.1 f | 16.0 ± 0.0 f | 15.2 ± 0.0 f |
| 7 | 0.0 ± 0.0 g | 0.0 ± 0.0 g | 0.0 ± 0.0 g | 0.0 ± 0.0 g |
| 8 | 0.0 ± 0.0 g | 0.0 ± 0.0 g | 0.0 ± 0.0 g | 0.0 ± 0.0 g |
| Temperature/Time | L. monocytogenes | S. aureus | S. typhimurium | E. coli O157:H7 |
|---|---|---|---|---|
| 80/20 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 80/30 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 80/40 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 100/20 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 100/30 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 100/40 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 a |
| 120/20 | 85.0 ± 0.7 b | 86.5 ± 0.3 b | 83.0 ± 0.1 b | 81.0 ± 0.2 b |
| 120/30 | 74.3 ± 0.4 c | 78.0 ± 0.2 c | 71.5 ± 0.1 c | 69.0 ± 0.0 c |
| 120/40 | 61.2 ± 0.1 d | 54.5 ± 0.1 d | 47.0 ± 0.3 d | 40.5 ± 0.0 d |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, Z.; Ramzan, R.; Abdelazez, A.; Amjad, A.; Afzaal, M.; Zhang, S.; Pan, S. Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens. Pathogens 2019, 8, 71. https://doi.org/10.3390/pathogens8020071
Muhammad Z, Ramzan R, Abdelazez A, Amjad A, Afzaal M, Zhang S, Pan S. Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens. Pathogens. 2019; 8(2):71. https://doi.org/10.3390/pathogens8020071
Chicago/Turabian StyleMuhammad, Zafarullah, Rabia Ramzan, Amro Abdelazez, Adnan Amjad, Muhammad Afzaal, Shanshan Zhang, and Siyi Pan. 2019. "Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens" Pathogens 8, no. 2: 71. https://doi.org/10.3390/pathogens8020071
APA StyleMuhammad, Z., Ramzan, R., Abdelazez, A., Amjad, A., Afzaal, M., Zhang, S., & Pan, S. (2019). Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens. Pathogens, 8(2), 71. https://doi.org/10.3390/pathogens8020071