Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe
Abstract
:1. Introduction
2. Results
3. Discussion
4. Material and Methods
4.1. S. Derby Genomes Selection
4.2. Whole Genome Sequencing
4.3. Phylogenetic Analysis
4.4. Statistical Analyses
4.5. Identification of Acquired Resistance Genes
4.6. Detection and Characterization of Phages and Prophages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, e04634. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, e05077. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, 5182. [Google Scholar] [CrossRef]
- Hayward, M.R.; AbuOun, M.; La Ragione, R.M.; Tchorzewska, M.A.; Cooley, W.A.; Everest, D.J.; Petrovska, L.; Jansen, V.A.; Woodward, M.J. SPI-23 of S. Derby: Role in adherence and invasion of porcine tissues. PLoS ONE 2014, 9, e107857. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.R.; Petrovska, L.; Jansen, V.A.; Woodward, M.J. Population structure and associated phenotypes of Salmonella enterica serovars Derby and Mbandaka overlap with host range. BMC Microbiol. 2016, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Zhou, Z.; Sergeant, M.J.; Achtman, M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018, 14, e1007261. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Analysis of the baseline survey on the prevalence of Salmonella in holdings with breeding pigs in the EU, 2008 - Part A: Salmonella prevalence estimates. EFSA J. 2009, 7, 1377. [Google Scholar] [CrossRef]
- Valdezate, S.; Vidal, A.; Herrera-Leon, S.; Pozo, J.; Rubio, P.; Usera, M.A.; Carvajal, A.; Echeita, M.A. Salmonella Derby clonal spread from pork. Emerg. Infect. Dis. 2005, 11, 694–698. [Google Scholar] [CrossRef]
- Simon, S.; Trost, E.; Bender, J.; Fuchs, S.; Malorny, B.; Rabsch, W.; Prager, R.; Tietze, E.; Flieger, A. Evaluation of WGS based approaches for investigating a food-borne outbreak caused by Salmonella enterica serovar Derby in Germany. Food Microbiol. 2017. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- WHO. World Health Day 2015: Food safety. Available online: http://www.who.int/campaigns/world-health-day/2015/event/en/ (accessed on 4 February 2018).
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013; p. 1535. [Google Scholar] [CrossRef]
- Weigel, L.M.; Steward, C.D.; Tenover, F.C. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 1998, 42, 2661–2667. [Google Scholar] [CrossRef]
- Barçante, L.; Vale, M.M.; Alves, M.A.S. Altitudinal migration by birds: A review of the literature and a comprehensive list of species. J. Field Ornithol. 2017, 88, 321–335. [Google Scholar] [CrossRef]
- Tizard, I. Salmonellosis in wild birds. Semin. Avian Exotic. Pet. Med. 2004, 13, 50–66. [Google Scholar] [CrossRef]
- Jourdain, E.; Gauthier-Clerc, M.; Bicout, D.J.; Sabatier, P. Bird migration routes and risk for pathogen dispersion into western Mediterranean wetlands. Emerg. Infect. Dis. 2007, 13, 365–372. [Google Scholar] [CrossRef]
- ITAVI. Structures et Organisation des Filières Volailles de Chair en Europe. 2013. Available online: https://www.viandesetproduitscarnes.fr/index.php/fr/economie2/615-structures-et-organisation-des-filieres-volailles-de-chair-en-europe (accessed on 5 February 2019).
- Malpel, G.P.; Marigeaud, M.; Marty, S. La Filière Volaille de Chair. 2014. Available online: http://www.igf.finances.gouv.fr/files/live/sites/igf/files/contributed/IGF%20internet/2.RapportsPublics/2014/2013-M-099.pdf (accessed on 1 February 2019).
- Beutlich, J.; Jahn, S.; Malorny, B.; Hauser, E.; Huhn, S.; Schroeter, A.; Rodicio, M.R.; Appel, B.; Threlfall, J.; Mevius, D.; et al. Antimicrobial resistance and virulence determinants in European Salmonella genomic island 1-positive Salmonella enterica isolates from different origins. Appl. Environ. Microbiol. 2011, 77, 5655–5664. [Google Scholar] [CrossRef]
- Sévellec, Y.; Vignaud, M.L.; Granier, S.A.; Lailler, R.; Feurer, C.; Le Hello, S.; Mistou, M.Y.; Cadel-Six, S. Polyphyletic Nature of Salmonella enterica Serotype Derby and Lineage-Specific Host-Association Revealed by Genome-Wide Analysis. Front. Microbiol. 2018, 9, 891. [Google Scholar] [CrossRef]
- Yasuike, M.; Nishiki, I.; Iwasaki, Y.; Nakamura, Y.; Fujiwara, A.; Sugaya, E.; Kawato, Y.; Nagai, S.; Kobayashi, T.; Ototake, M.; et al. Full-genome sequence of a novel myovirus, GF-2, infecting Edwardsiella tarda: Comparison with other Edwardsiella myoviral genomes. Arch. Virol. 2015, 160, 2129–2133. [Google Scholar] [CrossRef]
- Suttle, C. The Significance of Viruses to Mortality in Aquatic Microbial Communities. Microb. Ecol. 1994, 28, 237–243. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. MMBR 2000, 64, 69–114. [Google Scholar] [CrossRef]
- Bettarel, Y.; Amblard, C.; Sime-Ngando, T.; Carrias, J.F.; Sargos, D.; Garabetian, F.; Lavandier, P. Viral lysis, flagellate grazing potential, and bacterial production in Lake Pavin. Microb. Ecol. 2003, 45, 119–127. [Google Scholar] [CrossRef]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. 2014. Available online: https://www.researchgate.net/publication/281238574_FastQC_A_Quality_Control_tool_for_High_Throughput_Sequence_Data (accessed on 1 February 2019).
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Petrovska, L.; Mather, A.E.; AbuOun, M.; Branchu, P.; Harris, S.R.; Connor, T.; Hopkins, K.L.; Underwood, A.; Lettini, A.A.; Page, A.; et al. Microevolution of Monophasic Salmonella Typhimurium during Epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 2016, 22, 617–624. [Google Scholar] [CrossRef]
- Ung, A.; Baidjoe, A.Y.; Van Cauteren, D.; Fawal, N.; Fabre, L.; Guerrisi, C.; Danis, K.; Morand, A.; Donguy, M.P.; Lucas, E.; et al. Disentangling a complex nationwide Salmonella Dublin outbreak associated with raw-milk cheese consumption, France, 2015 to 2016. Eurosurveillance 2019, 24. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Royston, P. Remark AS R94: A Remark on Algorithm AS 181: The W-test for Normality. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1995, 44, 547–551. [Google Scholar] [CrossRef]
- Markowski, C.A.; Markowski, E.P. Conditions for the Effectiveness of a Preliminary Test of Variance. Am. Stat. 1990, 44, 322–326. [Google Scholar] [CrossRef]
- Huang, X.; Huang, Q.; Dun, Z.; Huang, W.; Wu, S.; Liang, J.; Deng, X.; Zhang, Y. Nontyphoidal Salmonella Infection, Guangdong Province, China, 2012. Emerg. Infect. Dis. 2016, 22, 726–729. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
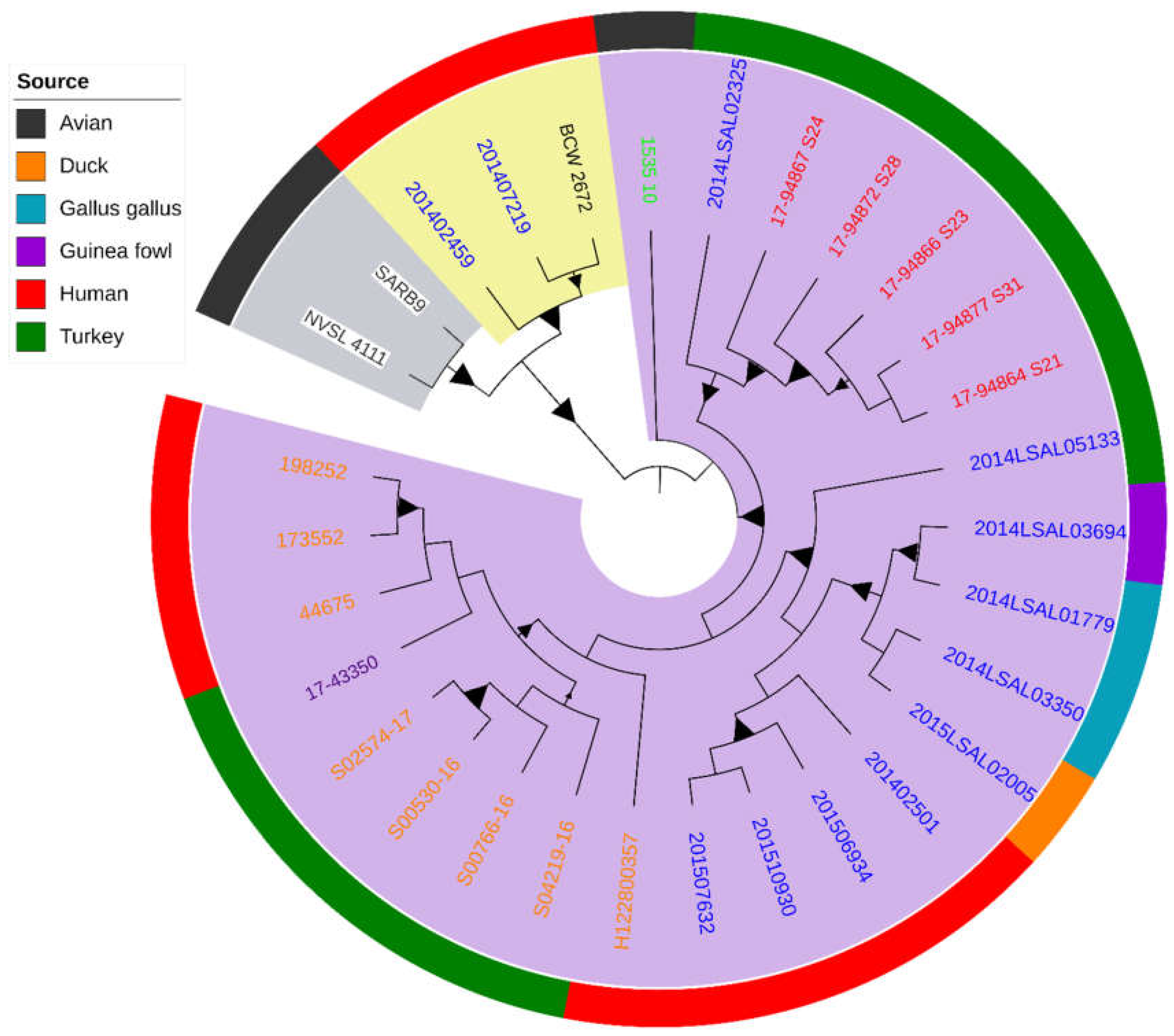
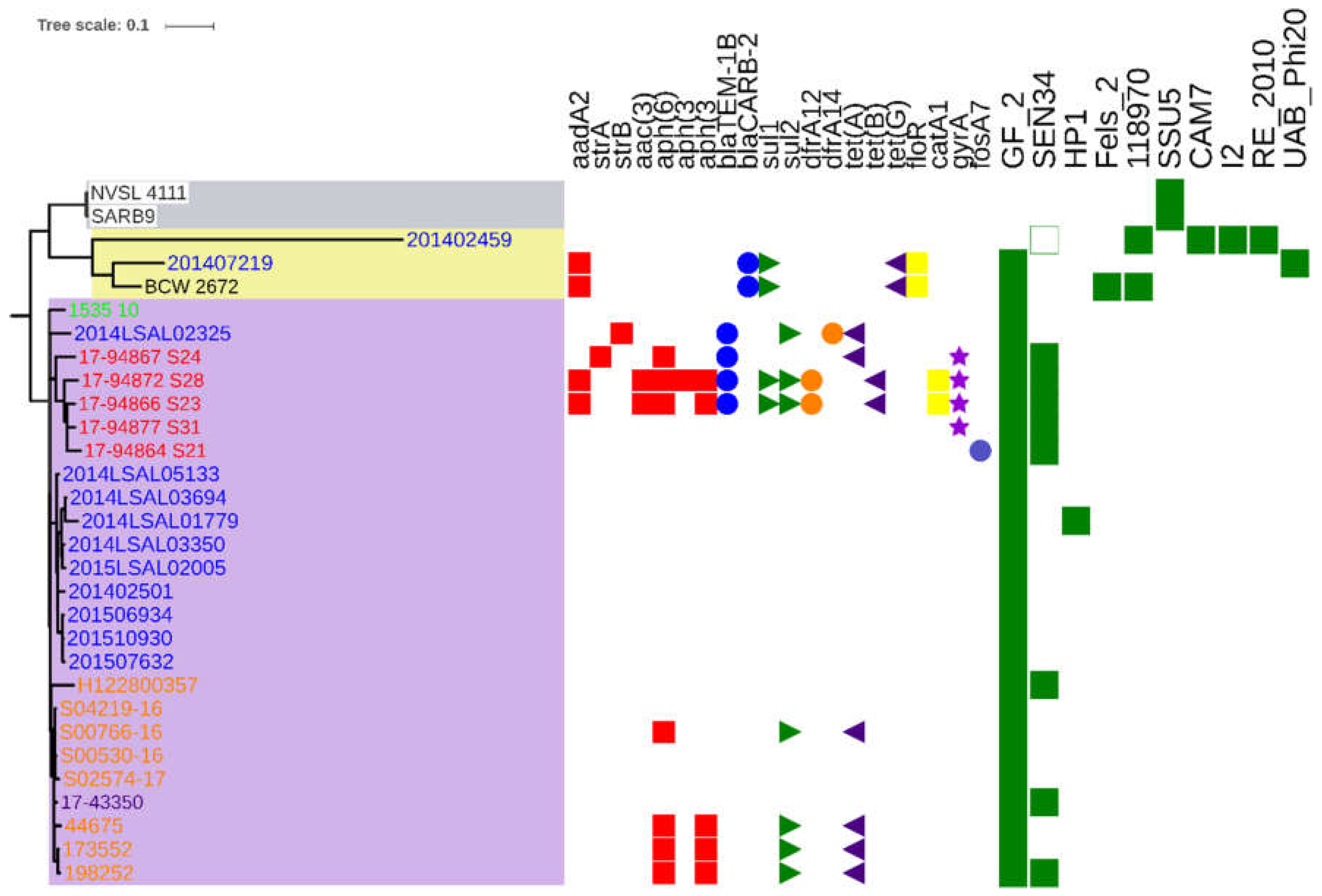
| Name | Database/Collection | Source Niche | Source Type | Source Details | Collection Year | Continent | Country | Sample ID | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 17-43350_S13 | Enterobase | Food | Avian | Meleagris; Food, turkey meat | 2015 | Europe | Germany | SAMEA104379658 | NS |
| 17-94867_S24 | Enterobase | Poultry | Avian | Meleagris; Animal, animal | 2011 | Europe | Italy | SAMEA104379733 | NS |
| 17-94864_S21 | Enterobase | Poultry | Avian | Meleagris; Animal, swab | 2011 | Europe | Italy | SAMEA104379730 | NS |
| 17-94877_S31 | Enterobase | Wild animal | Avian | Meleagris; Animal, feces | 2013 | Europe | Italy | SAMEA104379740 | NS |
| 17-94872_S28 | Enterobase | Wild animal | Avian | Meleagris; Animal, feces | 2012 | Europe | Italy | SAMEA104379737 | NS |
| 17-94866_S23 | Enterobase | Wild animal | Avian | Meleagris; Animal, feces | 2011 | Europe | Italy | SAMEA104379732 | NS |
| 1535_10 | Enterobase | Poultry | Avian | NS | 2010 | Europe | Poland | SAMEA104437530 | NS |
| BCW_2672 | Enterobase | Human | Human | Human; Homo sapiens | 2000 | Asia | Taiwan | SAMN02368552 | NS |
| 44675 | Enterobase | Human | Human | Human; Homo sapiens | 2014 | Europe | UK | SAMN03465613 | NS |
| H122800357 | Enterobase | Human | Human | Human; Homo sapiens | 2012 | Europe | UK | SAMN03168996 | NS |
| 173552 | Enterobase | Human | Human | human; Homo sapiens | 2015 | Europe | UK | SAMN06680388 | NS |
| 198252 | Enterobase | Human | Human | human; Homo sapiens | 2015 | Europe | UK | SAMN06680393 | NS |
| SARB9 (Fidelma Boyd) | Enterobase | NS | Avian | Bird | 1986 | North America | United States | NA | NS |
| NVSL 4111 | Enterobase | Wild animal | Avian | NS | 1986 | North America | United States | SAMN02367710 | NS |
| 2014LSAL02325 | ANSES | Poultry | Avian | Meat from turkey—carcass | 2014 | Europe | France | SAMN07734901 | [8] |
| 2014LSAL05133 | ANSES | Poultry | Avian | Meat from turkey—carcass | 2014 | Europe | France | SAMN07734953 | [8] |
| 2015LSAL02005 | ANSES | Poultry | Avian | Meat from duck—fresh | 2015 | Europe | France | SAMN07734993 | [8] |
| 2014LSAL03694 | ANSES | Poultry | Avian | Meat from guinea fowl | 2014 | Europe | France | SAMN07734940 | [8] |
| 2014LSAL03350 | ANSES | Poultry | Avian | Meat from broilers—Gallus gallus—carcass | 2014 | Europe | France | SAMN07734914 | [8] |
| 2014LSAL01779 | ANSES | Poultry | Avian | Meat from broilers—Gallus gallus—carcass | 2014 | Europe | France | SAMN08470240 | This study |
| 201402459 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2014 | Europe | France | SAMN09080915 | This study |
| 201407219 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2014 | Europe | France | SAMN09080917 | This study |
| S00530-16 | APHA | Poultry | Avian | Meleagris | 2016 | Europe | UK | SAMEA104448822 | This study |
| S00766-16 | APHA | Poultry | Avian | Meleagris | 2016 | Europe | UK | SAMEA104448826 | This study |
| S02574-17 | APHA | Poultry | Avian | Meleagris | 2017 | Europe | UK | SAMEA104448833 | This study |
| S04219-16 | APHA | Poultry | Avian | Meleagris | 2016 | Europe | UK | SAMEA104448829 | This study |
| 201402501 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2014 | Europe | France | SAMN09080916 | This study |
| 201506934 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2015 | Europe | France | SAMN09080917 | This study |
| 201507632 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2015 | Europe | France | SAMN09080919 | This study |
| 201510930 | Institut Pasteur | Human | Human | Human; Homo sapiens | 2015 | Europe | France | SAMN09080920 | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sévellec, Y.; Felten, A.; Radomski, N.; Granier, S.A.; Le Hello, S.; Petrovska, L.; Mistou, M.-Y.; Cadel-Six, S. Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe. Pathogens 2019, 8, 46. https://doi.org/10.3390/pathogens8020046
Sévellec Y, Felten A, Radomski N, Granier SA, Le Hello S, Petrovska L, Mistou M-Y, Cadel-Six S. Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe. Pathogens. 2019; 8(2):46. https://doi.org/10.3390/pathogens8020046
Chicago/Turabian StyleSévellec, Yann, Arnaud Felten, Nicolas Radomski, Sophie A. Granier, Simon Le Hello, Liljana Petrovska, Michel-Yves Mistou, and Sabrina Cadel-Six. 2019. "Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe" Pathogens 8, no. 2: 46. https://doi.org/10.3390/pathogens8020046
APA StyleSévellec, Y., Felten, A., Radomski, N., Granier, S. A., Le Hello, S., Petrovska, L., Mistou, M.-Y., & Cadel-Six, S. (2019). Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe. Pathogens, 8(2), 46. https://doi.org/10.3390/pathogens8020046





