A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of DAPs in the NILs
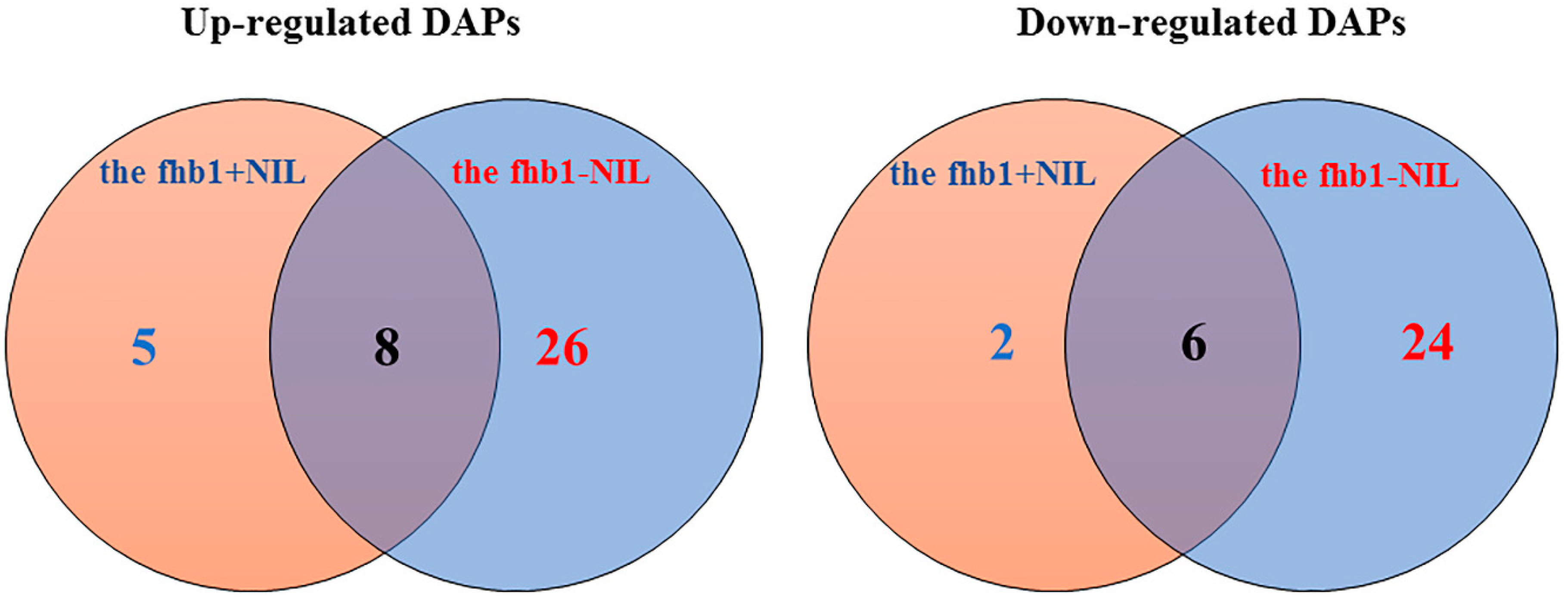
2.2. DAPs discovered in the fhb1+ NIL and the fhb1- NIL
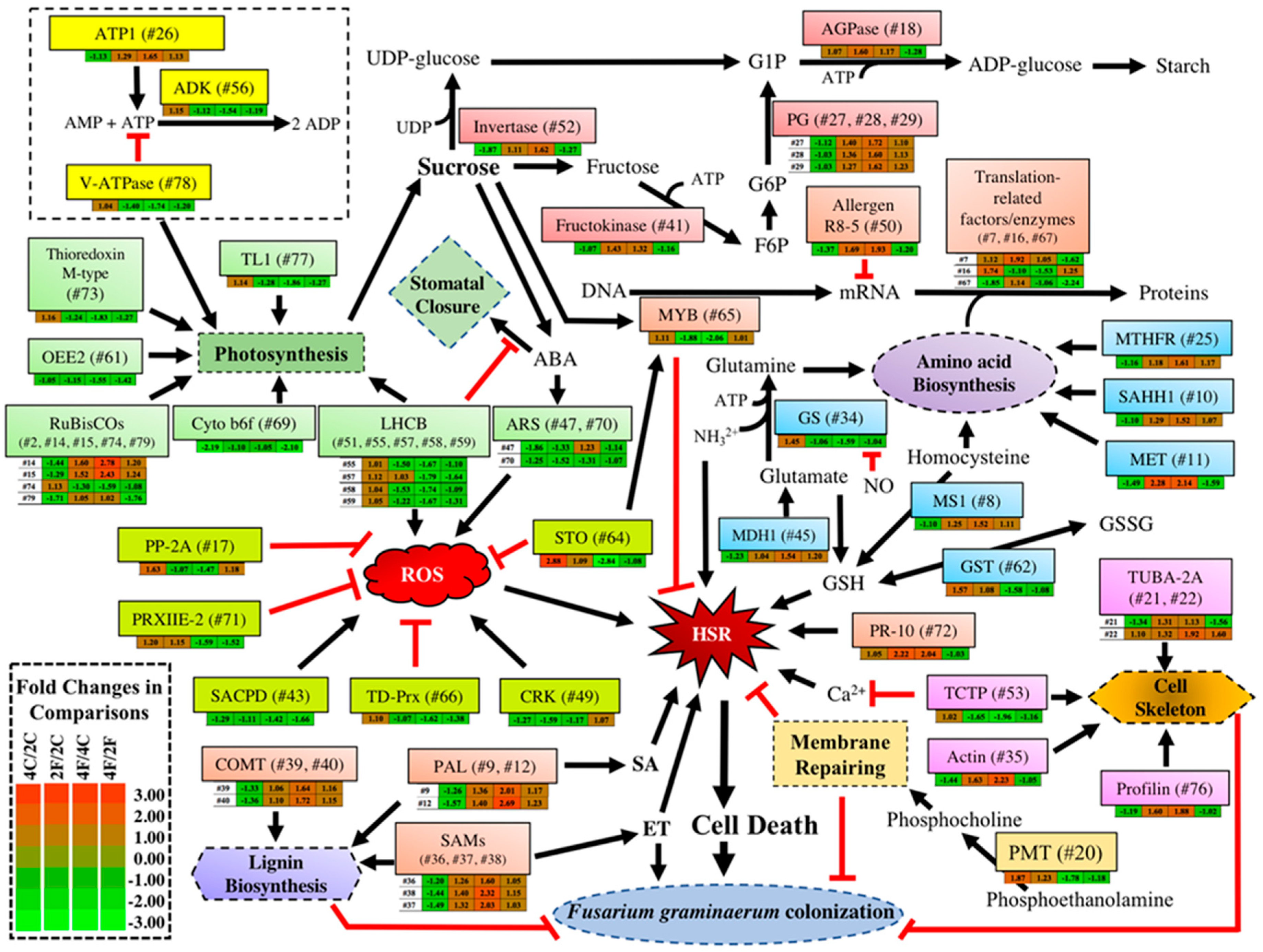
2.3. Constitutive DAPs in the Absence of Qfhb1
2.4. Fusarium-Induced DAPs Common in Both NILs
2.5. Fusarium-Induced DAPs That Were Specific to the fhb1+ NIL
2.6. Fusarium-Induced DAPs specific to the fhb1- NIL
3. Materials and Methods
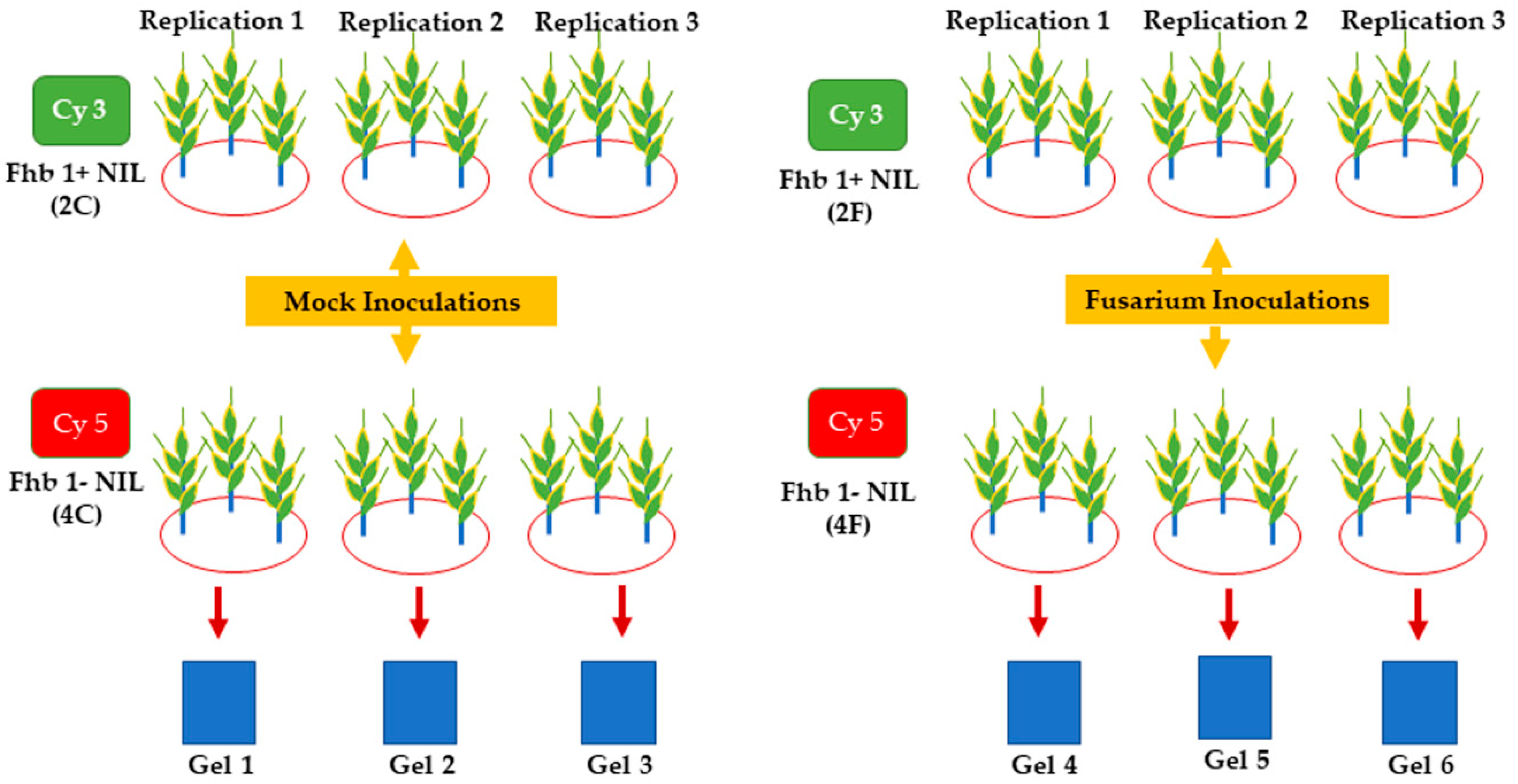
3.1. Plant Materials and Fusarium Inoculation
3.2. Proteomic Analyses and Identification of Differential Abundant Proteins
3.3. Real-Time RT-PCR Validation of Proteomic Data
3.4. Computational Analysis and Protein–Protein Interaction Predictions
3.5. Clustering and Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Desjardins, A.E.; Hohn, T.M. Mycotoxins in plant pathogenesis. Mol. Plant-Microbe Interact. 1997, 10, 147–152. [Google Scholar] [CrossRef]
- Snijders, C.H.A.; Perkowski, J. Effects of head blight caused by Fusarium culmorum on toxin content and weight of wheat kernels. Phytopathology 1990, 80, 566–570. [Google Scholar] [CrossRef]
- Parry, D.W.; Jenkinson, P.; Mcleod, L. Fusarium ear blight (Scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Christensen, J.J. Factors affecting resistance of wheat to scab caused by Gibberella Zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Foroud, N.A.; Ouellet, T.; Laroche, A.; Oosterveen, B.; Jordan, M.C.; Ellis, B.E.; Eudes, F. Differential transcriptome analyses of three wheat genotypes reveal different host response pathways associated with Fusarium head blight and trichothecene resistance. Plant Pathol. 2012, 61, 296–314. [Google Scholar] [CrossRef]
- Anderson, J.A.; Stack, R.W.; Liu, S.; Waldron, B.L.; Fjeld, A.D.; Coyne, C.; Moreno-Sevilla, B.; Fetch, J.M.; Song, Q.J.; Cregan, P.B.; et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 2001, 102, 1164–1168. [Google Scholar] [CrossRef]
- Bai, G.H.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology 1999, 89, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Basnet, B.R.; Glover, K.D.; Ibrahim, A.M.H.; Yen, Y.; Chao, S.M. A QTL on chromosome 2DS of ‘Sumai 3’ increases susceptibility to Fusarium head blight in wheat. Euphytica 2012, 186, 91–101. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Lemmens, M.; Hartl, L.; Doldi, L.; Steiner, B.; Stierschneider, M.; Ruckenbauer, P. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor. Appl. Genet. 2002, 104, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP mapping of QTL for fusarium head blight resistance in wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Zhou, W.C.; Kolb, F.L.; Bai, G.H.; Shaner, G.; Domier, L.L. Genetic analysis of scab resistance QTL in wheat with microsatellite and AFLP markers. Genome 2002, 45, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Ekramoddoullah, A.K.M. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 2006, 68, 3–13. [Google Scholar] [CrossRef]
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of wheat and barley: A re-emerging disease of devastating impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef]
- Wisniewska, H.; Kowalczyk, K. Resistance of cultivars and breeding lines of spring wheat to Fusarium culmorum and powdery mildew. J. Appl. Genet. 2005, 46, 35–40. [Google Scholar] [PubMed]
- Choulet, F.; Wicker, T.; Rustenholz, C.; Paux, E.; Salse, J.; Leroy, P.; Schlub, S.; Le Paslier, M.C.; Magdelenat, G.; Gonthier, C.; et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 2010, 22, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.X.; Pumphrey, M.O.; Gill, B.S.; Trick, H.N.; Zhang, J.X.; Dolezel, J.; Chalhoub, B.; Anderson, J.A. Toward positional cloning of Fhb1, a major QTL for Fusarium head blight resistance in wheat. Cereal Res. Commun. 2008, 36, 195–201. [Google Scholar] [CrossRef]
- Schweiger, W.; Steiner, B.; Vautrin, S.; Nussbaumer, T.; Siegwart, G.; Zamini, M.; Jungreithmeier, F.; Gratl, V.; Lemmens, M.; Mayer, K.F.X.; et al. Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 2016, 129, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Hofstad, A.N.; Nussbaumer, T.; Akhunov, E.; Shin, S.; Kugler, K.G.; Kistler, H.C.; Mayer, K.F.X.; Muehlbauer, G.J. Examining the transcriptional response in wheat Fhb1 near-isogenic lines to Fusarium graminearum infection and deoxynivalenol treatment. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Pumphrey, M.O. Towards Map-Based Cloning of Fusarium Head Blight Resistance QTL Fhb1 and Non-Additive Expression of Homoeologous Genes in Allohexaploid Wheat. Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2007. [Google Scholar]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jin, X.H.; Jia, X.P.; Wang, H.Y.; Cao, A.Z.; Zhao, W.P.; Pei, H.Y.; Xue, Z.K.; He, L.Q.; Chen, Q.G.; et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genom. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Gala, A.; Yen, Y. Identification of functional genic components of major fusarium head blight resistance quantitative trait loci in wheat cultivar Sumai 3. Mol. Plant-Microbe Interact. 2013, 26, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, M.; Scholz, U.; Berthiller, F.; Dall’Asta, C.; Koutnik, A.; Schuhmacher, R.; Adam, G.; Buerstmayr, H.; Mesterhazy, A.; Krska, R.; et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for fusarium head blight resistance in wheat. Mol. Plant-Microbe Interact. 2005, 18, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 2012, 7, e40695. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y. Genetic Dissection of Fusarium Head Blight in Wheat (Triticum aestivum L.). Ph.D. Thesis, South Dakota State University, Brookings, SD, USA, 2014. [Google Scholar]
- Li, G.; Yen, Y. Jasmonate and ethylene signaling pathway may mediate Fusarium head blight resistance in wheat. Crop Sci. 2008, 48, 1888–1896. [Google Scholar] [CrossRef]
- Kwon, S.J.; Jin, H.C.; Lee, S.; Nam, M.H.; Chung, J.H.; Il Kwon, S.; Ryu, C.M.; Park, O.K. GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 2009, 58, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Bhadauria, V.; Banniza, S.; Wang, L.X.; Wei, Y.D.; Peng, Y.L. Proteomic studies of phytopathogenic fungi, oomycetes and their interactions with hosts. Eur. J. Plant Pathol. 2010, 126, 81–95. [Google Scholar] [CrossRef]
- Mehta, A.; Brasileiro, A.C.M.; Souza, D.S.L.; Romano, E.; Campos, M.A.; Grossi-De-Sa, M.F.; Silva, M.S.; Franco, O.L.; Fragoso, R.R.; Bevitori, R.; et al. Plant-pathogen interactions: What is proteomics telling us? FEBS J. 2008, 275, 3731–3746. [Google Scholar] [CrossRef] [PubMed]
- Quirino, B.F.; Candido, E.S.; Campos, P.F.; Franco, O.L.; Kruger, R.H. Proteomic approaches to study plant-pathogen interactions. Phytochemistry 2010, 71, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jacobsen, S.; Jorgensen, H.J.L.; Collinge, D.B.; Svensson, B.; Finnie, C. Fusarium graminearum and its interactions with cereal heads: Studies in the proteomics era. Front. Plant Sci. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Finnie, C.; Melchior, S.; Roepstorff, P.; Svensson, B. Proteome analysis of grain filling and seed maturation in barley. Plant Physiol. 2002, 129, 1308–1319. [Google Scholar] [CrossRef] [PubMed]
- Thurston, G.; Regan, S.; Rampitsch, C.; Xing, T. Proteomic and phosphoproteomic approaches to understand plant-pathogen interactions. Physiol. Mol. Plant Pathol. 2005, 66, 3–11. [Google Scholar] [CrossRef]
- Das, A.; Paudel, B.; Rohila, J.S. Potentials of Proteomics in Crop Breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2015; pp. 513–537. [Google Scholar]
- Wang, Y.; Yang, L.M.; Xu, H.B.; Li, Q.F.; Ma, Z.Q.; Chu, C.G. Differential proteomic analysis of proteins in wheat spikes induced by Fusarium graminearum. Proteomics 2005, 5, 4496–4503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Fu, J.M.; Hiromasa, Y.; Pan, H.Y.; Bai, G.H. Differentially expressed proteins associated with fusarium head blight resistance in wheat. PLoS ONE 2013, 8, e82079. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.C.; Kolb, F.L.; Riechers, D.E. Identification of proteins induced or upregulated by fusarium head blight infection in the spikes of hexaploid wheat (Triticum aestivum). Genome 2005, 48, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Beranova-Giorgianni, S. Proteome analysis by two-dimensional gel electrophoresis and mass spectrometry: Strengths and limitations. Trends Anal. Chem. 2003, 22, 273–281. [Google Scholar] [CrossRef]
- Lilley, K.S.; Dupree, P. Methods of quantitative proteomics and their application to plant organelle characterization. J. Exp. Bot. 2006, 57, 1493–1499. [Google Scholar] [CrossRef] [PubMed]
- Lilley, K.S.; Friedman, D.B. All about DIGE: Quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteom. 2004, 1, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Schenkluhn, L.; Hohnjec, N.; Niehaus, K.; Schmitz, U.; Colditz, F. Differential gel electrophoresis (DIGE) to quantitatively monitor early symbiosis- and pathogenesis-induced changes of the Medicago truncatula root proteome. J. Proteom. 2010, 73, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Unlu, M.; Morgan, M.E.; Minden, J.S. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis 1997, 18, 2071–2077. [Google Scholar] [CrossRef] [PubMed]
- Amey, R.C.; Schleicher, T.; Slinn, J.; Lewis, M.; Macdonald, H.; Neill, S.J.; Spencer-Phillips, P.T.N. Proteomic analysis of a compatible interaction between Pisum sativum (pea) and the downy mildew pathogen Peronospora viciae. Eur. J. Plant Pathol. 2008, 122, 41–55. [Google Scholar] [CrossRef]
- Dornez, E.; Croes, E.; Gebruers, K.; Carpentier, S.; Swennen, R.; Laukens, K.; Witters, E.; Urban, M.; Delcour, J.A.; Courtin, C.M. 2-D DIGE reveals changes in wheat xylanase inhibitor protein families due to Fusarium graminearum deltaTri5 infection and grain development. Proteomics 2010, 10, 2303–2319. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, X.; Pumphrey, M.O.; Stack, R.W.; Gill, B.S.; Anderson, J.A. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genom. 2006, 6, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Yoo, S.D. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet. 2011, 7, e1001263. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wind, J.J.; Shi, X.L.; Zhang, H.L.; Hanson, J.; Smeekens, S.C.; Teng, S. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc. Natl. Acad. Sci. USA 2011, 108, 3436–3441. [Google Scholar] [CrossRef] [PubMed]
- Engelsdorf, T.; Horst, R.J.; Prols, R.; Proschel, M.; Dietz, F.; Huckelhoven, R.; Voll, L.M. Reduced carbohydrate availability enhances the susceptibility of Arabidopsis toward Colletotrichum higginsianum. Plant Physiol. 2013, 162, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.N.; Xu, H.B.; Yi, H.Y.; Yang, L.M.; Kong, Z.X.; Zhang, L.X.; Xue, S.L.; Jia, H.Y.; Ma, Z.Q. Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE 2011, 6, e19008. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Douglas, K.T. Mechanism of action of glutathione-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 1987, 59, 103–167. [Google Scholar] [PubMed]
- Gomez, L.D.; Noctor, G.; Knight, M.R.; Foyer, C.H. Regulation of calcium signalling and gene expression by glutathione. J. Exp. Bot. 2004, 55, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Rentel, M.C.; Knight, M.R. Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol. 2004, 135, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Vanacker, H.; Carver, T.L.W.; Foyer, C.H. Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol. 2000, 123, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Durian, G.; Rahikainen, M.; Alegre, S.; Brosche, M.; Kangasjarvi, S. Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front. Plant Sci. 2016, 7, 812. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inze, D.; Kushnir, S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.J. Defense-related proteins in higher plants. Annu. Rev. Biochem. 1990, 59, 873–907. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Pierpoint, W.S.; Boller, T.; Conejero, V. Recommendations for naming plant pathogenesis-related proteins. Plant Mol. Biol. Rep. 1994, 12, 245–264. [Google Scholar] [CrossRef]
- Wu, J.; Kim, S.G.; Kang, K.Y.; Kim, J.G.; Park, S.R.; Gupta, R.; Kim, Y.H.; Wang, Y.; Kim, S.T. Overexpression of a pathogenesis-related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol. J. 2016, 32, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Gachet, Y.; Tournier, S.; Lee, M.; Lazaris-Karatzas, A.; Poulton, T.; Bommer, U.A. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 1999, 112, 1257–1271. [Google Scholar] [PubMed]
- Nagano-Ito, M.; Ichikawa, S. Biological effects of mammalian translationally controlled tumor protein (TCTP) on cell death, proliferation, and tumorigenesis. Biochem. Res. Int. 2012, 2012, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Jin, S.; Bernardo, A.; Amand, P.S.; Bai, G. Development of High-Throughput Diagnostic Marker for Fhb1, a Major Gene for FHB Resistance in Wheat; 2016 National Fusarium Head Blight Forum: St. Louis, MO, USA, 2016. [Google Scholar]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Bufe, A.; Schramm, G.; Keown, M.B.; Schlaak, M.; Becker, W.M. Major allergen Phl p Vb in timothy grass is a novel pollen RNase. FEBS Lett. 1995, 363, 6–12. [Google Scholar] [CrossRef]
- Pontis, H.G. On the scent of the riddle of sucrose. Trends Biochem. Sci. 1978, 3, 137–139. [Google Scholar] [CrossRef]
- Tognetti, J.A.; Pontis, H.G.; Martinez-Noel, G.M. Sucrose signaling in plants: A world yet to be explored. Plant Signal. Behav. 2013, 8, e23316. [Google Scholar] [PubMed]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef] [PubMed]
- Grierson, C.; Du, J.S.; Zabala, M.D.; Beggs, K.; Smith, C.; Holdsworth, M.; Bevan, M. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato-tuber storage protein gene. Plant J. 1994, 5, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. The nuclear factor SP8BF binds to the 5′-upstream regions of three different genes coding for major proteins of sweet potato tuberous roots. Plant Mol. Biol. 1992, 18, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, K.A.; Elmore, J.M.; Creer, A.Y.; Feng, B.M.; Franco, J.Y.; Rufian, J.S.; He, P.; Phinney, B.; Coaker, G. A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 2017, 173, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Idanheimo, N.; Gauthier, A.; Salojarvi, J.; Siligato, R.; Brosche, M.; Kollist, H.; Mahonen, A.P.; Kangasjarvi, J.; Wrzaczek, M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Joazeiro, C.A.P.; Weissman, A.M. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 2000, 102, 549–552. [Google Scholar] [CrossRef]
- Lorick, K.L.; Jensen, J.P.; Fang, S.Y.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef] [PubMed]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Amitai-Zeigerson, H.; Scolnik, P.A.; Bar-Zvi, D. Tomato Asr1 mRNA and protein are transiently expressed following salt stress, osmotic stress and treatment with abscisic acid. Plant Sci. 1995, 110, 205–213. [Google Scholar] [CrossRef]
- Cakir, B.; Agasse, A.; Gaillard, C.; Saumonneau, A.; Delrot, S.; Atanassova, R. A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 2003, 15, 2165–2180. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Huang, C.; Deng, X.M.; Zhou, S.Y.; Chen, L.H.; Li, Y.; Wang, C.; Ma, Z.B.; Yuan, Q.Q.; Wang, Y.; et al. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 2013, 36, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Kim, Y.S.; Yoon, H.S. Rice ASR1 protein with reactive oxygen species scavenging and chaperone-like activities enhances acquired tolerance to abiotic stresses in Saccharomyces cerevisiae. Mol. Cells 2012, 33, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Klinkenberg, J.; Faist, H.; Saupe, S.; Lambertz, S.; Krischke, M.; Stingl, N.; Fekete, A.; Mueller, M.J.; Feussner, I.; Hedrich, R.; et al. Two fatty acid desaturases, stearoyl-acyl carrier protein Δ9-desaturase6 and fatty acid desaturase3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol. 2014, 164, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Goyer, A.; Haslekas, C.; Miginiac-Maslow, M.; Klein, U.; Le Marechal, P.; Jacquot, J.P.; Decottignies, P. Isolation and characterization of a thioredoxin-dependent peroxidase from Chlamydomonas reinhardtii. Eur. J. Biochem. 2002, 269, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta 1993, 1140, 215–250. [Google Scholar] [CrossRef]
- Kitagawa, N.; Mazon, H.; Heck, A.J.; Wilkens, S. Stoichiometry of the peripheral stalk subunits E and G of yeast V1-ATPase determined by mass spectrometry. J. Biol. Chem. 2008, 283, 3329–3337. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Tameling, W.I.L.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.C.; Takken, F.L.W. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006, 140, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.M.; Coschigano, K.T.; Oliveira, I.C.; MeloOliveira, R.; Coruzzi, G.M. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef] [PubMed]
- De Gara, L.; de Pinto, M.C.; Tommasi, F. The antioxidant systems vis-a-vis reactive oxygen species during plant-pathogen interaction. Plant Physiol. Biochem. 2003, 41, 863–870. [Google Scholar] [CrossRef]
- Seifi, H.S.; Van Bockhaven, J.; Angenon, G.; Hofte, M. Glutamate metabolism in plant disease and defense: Friend or foe? Mol. Plant-Microbe Interact. 2013, 26, 475–485. [Google Scholar] [CrossRef] [PubMed]
- von Gonner, M.; Schlosser, E. Oxidative stress in interactions between Avena sativa L. and Drechslera spp. Physiol. Mol. Plant Pathol. 1993, 42, 221–234. [Google Scholar] [CrossRef]
- Swamy, M.; Sirajudeen, K.N.S.; Chandran, G. Nitric oxide (NO), citrulline-NO cycle enzymes, glutamine synthetase, and oxidative status in kainic acid-mediated excitotoxicity in rat brain. Drug Chem. Toxicol. 2009, 32, 326–331. [Google Scholar] [CrossRef] [PubMed]
- De Stefano, M.; Ferrarini, A.; Delledonne, M. Nitric oxide functions in the plant hypersensitive disease resistance response. BMC Plant Biol. 2005, 5, S10. [Google Scholar] [CrossRef]
- Melo, P.M.; Silva, L.S.; Ribeiro, I.; Seabra, A.R.; Carvalho, H.G. Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 2011, 157, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Odanaka, S.; Bennett, A.B.; Kanayama, Y. Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of Frk1 and Frk2 expression in tomato. Plant Physiol. 2002, 129, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Hu, Z.R.; Li, Y.H.; Li, C.; Peng, F.X.; Yao, Y.Y.; Peng, H.R.; Ni, Z.F.; Xie, C.J.; Sun, Q.X. Overexpression of a wheat stearoyl-ACP desaturase (SACPD) gene TaSSI2 in Arabidopsis ssi2 mutant compromise its resistance to powdery mildew. Gene 2013, 524, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Hurkman, W.J.; Tanaka, C.K. Solubilization of plant membrane-proteins for analysis by two-dimensional gel-electrophoresis. Plant Physiol. 1986, 81, 802–806. [Google Scholar] [CrossRef] [PubMed]
- Damerval, C.; de Vienne, D.; Michel, Z.; Hervé, T. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 1986, 7, 52–54. [Google Scholar] [CrossRef]
- Amme, S.; Matros, A.; Schlesier, B.; Mock, H.P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006, 57, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Eldakak, M.; Paudel, B.; Kim, D.W.; Hemmati, H.; Basu, C.; Rohila, J.S. Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed Res. Int. 2016, 6021047. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Eldakak, M.; Rohila, J.S.; Basu, C. Biochemical analysis of ‘kerosene tree’ Hymenaea courbaril L. under heat stress. Plant Signal. Behav. 2014, 9, e972851. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, G.; Moro, C.F.; Rohila, J.S.; Shibato, J.; Kubo, A.; Imanaka, T.; Kimura, S.; Ozawa, S.; Fukutani, S.; Endo, S.; et al. 2D-DIGE-based proteome expression changes in leaves of rice seedlings exposed to low-level gamma radiation at Iitate village, Fukushima. Plant Signal. Behav. 2015, 10, e1103406. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.L.; Roy, A.; Wang, P.H.; Gaffoor, I.; Sekhon, R.S.; Buanafina, M.M.D.; Rohila, J.S.; Chopra, S. Comparative proteomics analysis by DIGE and iTRAQ provides insight into the regulation of phenylpropanoids in maize. J. Proteom. 2013, 93, 254–275. [Google Scholar] [CrossRef] [PubMed]
- Paudel, B.; Das, A.; Tran, M.; Boe, A.; Palmer, N.A.; Sarath, G.; Gonzalez-Hernandez, J.L.; Rushton, P.J.; Rohila, J.S. Proteomic responses of switchgrass and prairie cordgrass to senescence. Front. Plant Sci. 2016, 7, 293. [Google Scholar] [CrossRef] [PubMed]
- Poschmann, G.; Sitek, B.; Sipos, B.; Ulrich, A.; Wiese, S.; Stephan, C.; Warscheid, B.; Kloppel, G.; Vander Borght, A.; Ramaekers, F.C.S.; et al. Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteom. 2009, 8, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Skeeles, L.E.; Fleming, J.L.; Mahler, K.L.; Toland, A.E. The impact of 3′ UTR variants on differential expression of candidate cancer susceptibility genes. PLoS ONE 2013, 8, e58609. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. In Current Protocols in Bioinformatics; Wiley Online Library: Hoboken, NJ, USA, 2003; pp. 2.3.1–2.3.22. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Salavati, A.; Khatoon, A.; Nanjo, Y.; Komatsu, S. Analysis of proteomic changes in roots of soybean seedlings during recovery after flooding. J. Proteom. 2012, 75, 878–893. [Google Scholar] [CrossRef] [PubMed]
- Camon, E.; Magrane, M.; Barrell, D.; Lee, V.; Dimmer, E.; Maslen, J.; Binns, D.; Harte, N.; Lopez, R.; Apweiler, R. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004, 32, D262–D266. [Google Scholar] [CrossRef] [PubMed]
- De Hoon, M.J.L.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.M.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
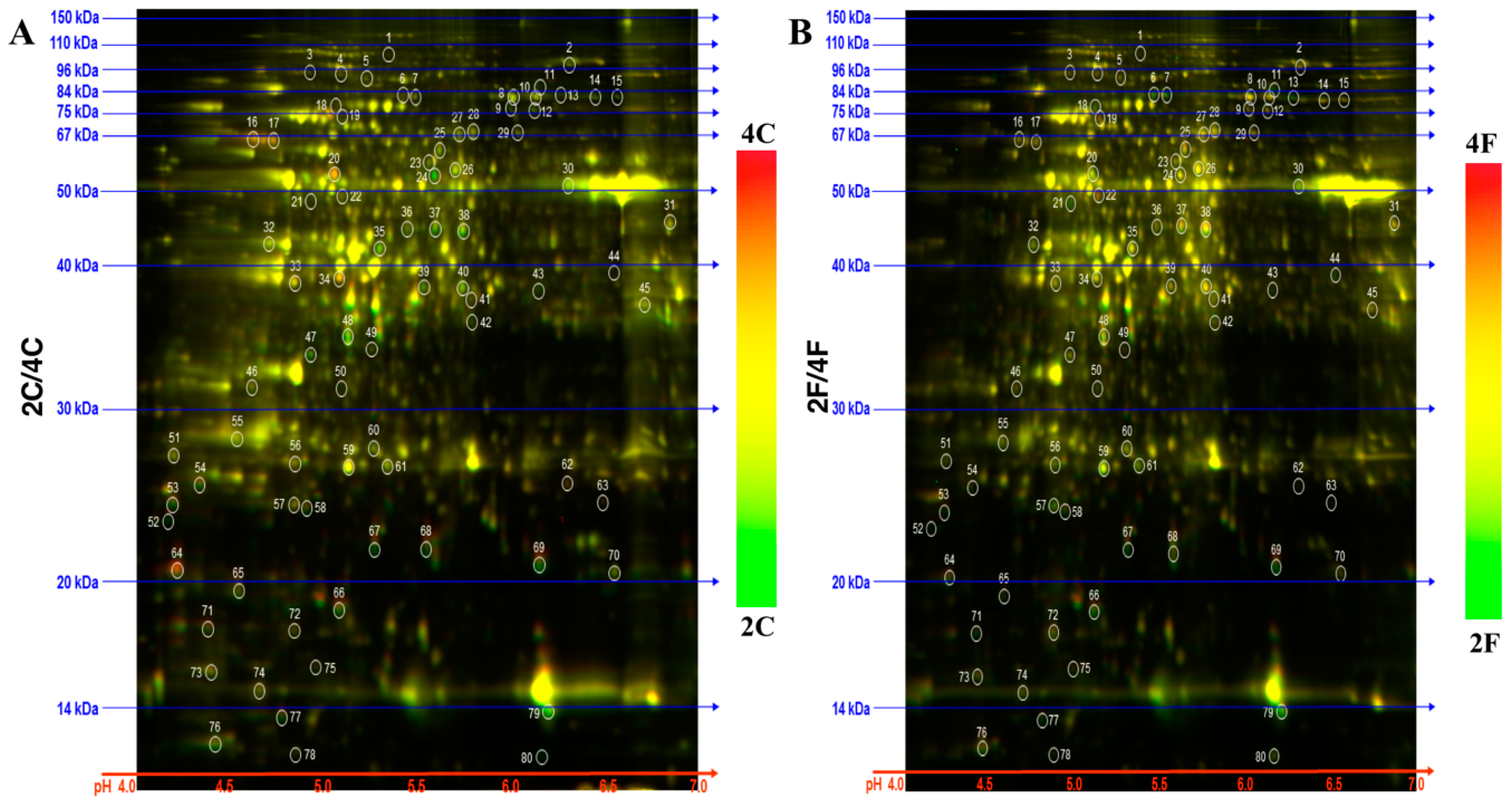
| Category | ID | Annotation | GenBank ID | kDa | pI | 4C/2C | 2F/2C | 4F/4C | 4F/2F |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 80 ** | 60S ribosomal protein L5 | gi|360042516 | 34.44 | 9.3 | −6.87 | 3.53 | 19.54 | −1.24 |
| 24 | Myo-inositol-1-phosphate synthase | gi|90289596 | 49.68 | 4.9 | −2.51 | 1.20 | 3.43 | 1.14 | |
| 12 | Phenylalanine ammonia-lyase | gi|3024363 | 75.89 | 6.1 | −1.57 | 1.40 | 2.69 | 1.23 | |
| 52 | Vacuolar invertase | gi|3219509 | 56.4 | 5.8 | −1.87 | 1.11 | 1.62 | −1.27 | |
| 1 ** | Glycosyltransferase | gi|56409844 | 65.08 | 6.0 | −1.71 | −1.18 | 1.19 | −1.21 | |
| 47 | Abscisic stress ripening protein | gi|357163453 | 25.95 | 5.1 | −1.86 | −1.33 | 1.23 | −1.14 | |
| 69 | Cytochrome b6-f complex iron-sulfur subunit, chloroplastic | gi|68566191 | 23.71 | 8.5 | −2.19 | −1.10 | −1.05 | −2.10 | |
| 67 | Eukaryotic translation initiation factor 5A1 | gi|74048999 | 17.35 | 5.7 | −1.85 | 1.14 | −1.06 | −2.24 | |
| 79 | Ribulose bisphosphate carboxylase small chain clone 512 | gi|132107 | 13.05 | 5.8 | −1.71 | 1.05 | 1.02 | −1.76 | |
| 2 | 17 | Protein phosphatase 2A structural subunit | gi|328775741 | 65.46 | 5.0 | 1.63 | −1.07 | −1.47 | 1.18 |
| 16 | Protein disulfide isomerase | gi|222446340 | 56.64 | 5.0 | 1.74 | −1.10 | −1.53 | 1.25 | |
| 20 | Phosphoethanolamine methyltransferase | gi|17887465 | 56.82 | 5.2 | 1.87 | 1.23 | −1.78 | −1.18 | |
| 62 | Glutathione transferase F5 | gi|23504745 | 23.42 | 5.8 | 1.57 | 1.08 | −1.58 | −1.08 | |
| 64 | Salt tolerant protein | gi|63021412 | 17.05 | 4.7 | 2.88 | 1.09 | −2.84 | −1.08 | |
| 3 | 34 | Glutamine synthetase isoform GSr1 | gi|40317416 | 38.71 | 5.4 | 1.45 | −1.06 | −1.59 | −1.04 |
| 51 | Chlorophyll a-b binding protein of LHCII type III, chloroplastic-like, predicted | gi|357122389 | 28.56 | 5.0 | 1.01 | −1.39 | −1.51 | −1.07 | |
| 56 | Adenylate kinase, chloroplastic-like | gi|357139457 | 31.43 | 7.1 | 1.15 | −1.12 | −1.54 | −1.19 | |
| 59 | Light-harvesting complex I chlorophyll a/b binding protein 3 | gi|79320443 | 23.73 | 5.6 | 1.05 | −1.22 | −1.67 | −1.31 | |
| 61 | 20S proteasome beta-4 subunit | gi|52548238 | 23.31 | 5.6 | −1.05 | −1.15 | −1.55 | −1.42 | |
| 66 | Thioredoxin-dependent peroxidase | gi|256708473 | 17.39 | 5.2 | 1.10 | −1.07 | −1.62 | −1.38 | |
| 68 | Eukaryotic translation initiation factor 5A1 | gi|74048999 | 17.35 | 5.7 | 1.29 | −1.05 | −1.66 | −1.22 | |
| 73 | Thioredoxin M-type, chloroplastic; Precursor | gi|11135474 | 19.12 | 8.7 | 1.16 | −1.24 | −1.83 | −1.27 | |
| 74 | Ribulose bisphosphate carboxylase small chain clone 512 (Fragment) | RBS3_WHEAT | 13.05 | 5.8 | 1.13 | −1.30 | −1.59 | −1.08 | |
| 77 | Thylakoid lumenal 15 kDa protein 1, chloroplastic, predicted | gi|225449424 | 23.42 | 6.5 | 1.14 | −1.28 | −1.86 | −1.27 | |
| 78 | Vacuolar ATPase subunit F | gi|94537548 | 14.37 | 5.3 | 1.04 | −1.40 | −1.74 | −1.20 | |
| 57 | Chlorophyll a-b binding protein | gi|225690794 | 26.96 | 5.4 | 1.12 | 1.03 | −1.79 | −1.64 | |
| 71 | Peroxiredoxin-2E-2, chloroplastic-like, predicted | gi|357139104 | 23.21 | 6.7 | 1.20 | 1.15 | −1.59 | −1.52 | |
| 4 | 22 | Alpha tubulin-2A | gi|90289596 | 49.68 | 4.9 | 1.10 | 1.32 | 1.92 | 1.60 |
| 3 ** | Glycosyltransferase | gi|56409844 | 65.08 | 6.0 | 1.01 | 1.49 | 1.68 | 1.14 | |
| 2 | Ribulose bisphosphate carboxylase small chain PW9, chloroplastic | RBS2_WHEAT | 19.44 | 8.5 | −1.20 | 1.35 | 1.80 | 1.11 | |
| 4 * | COP9 signalosome subunit | gi|159463648 | 50.44 | 6.0 | −1.05 | 1.12 | 1.52 | 1.29 | |
| 5 | Predicted barley protein | gi|326503600 | 99.43 | 5.2 | −1.04 | 1.34 | 1.56 | 1.13 | |
| 8 | Methionine synthase 1 enzyme | gi|68655495 | 84.51 | 5.7 | −1.10 | 1.25 | 1.52 | 1.11 | |
| 9 | Phenylalanine ammonia-lyase | gi|3024363 | 75.89 | 6.1 | −1.26 | 1.36 | 2.01 | 1.17 | |
| 10 | Adenosylhomocysteinase | SAHH_WHEAT | 53.4 | 5.7 | −1.10 | 1.29 | 1.52 | 1.07 | |
| 23 | Mitochondrial-processing peptidase subunit beta-like, predicted | gi|357113428 | 58.38 | 5.6 | −1.36 | 1.24 | 1.66 | −1.01 | |
| 25 | 5,10-methylene-tetrahydrofolate reductase | gi|115589742 | 64.83 | 5.9 | −1.16 | 1.18 | 1.61 | 1.17 | |
| 26 | ATPase subunit 1 | gi|81176509 | 55.27 | 5.7 | −1.13 | 1.29 | 1.65 | 1.13 | |
| 27 | Phosphoglucomutase | gi|18076790 | 62.75 | 5.7 | −1.12 | 1.40 | 1.72 | 1.10 | |
| 28 | Phosphoglucomutase | gi|18076790 | 62.75 | 5.7 | −1.03 | 1.36 | 1.60 | 1.13 | |
| 29 | Phosphoglucomutase | gi|18076790 | 62.75 | 5.7 | −1.03 | 1.27 | 1.62 | 1.23 | |
| 36 | S-adenosylmethionine synthase | gi|223635315 | 43.15 | 5.6 | −1.20 | 1.26 | 1.60 | 1.05 | |
| 38 | S-adenosylmethionine synthase | gi|223635315 | 43.15 | 5.6 | −1.44 | 1.40 | 2.32 | 1.15 | |
| 37 | Acyl transferase 5, putative | gi|151175359 | 46.83 | 5.5 | −1.49 | 1.32 | 2.03 | 1.03 | |
| 39 | Caffeic acid 3-O-methyltransferase | gi|145321007 | 38.57 | 5.7 | −1.33 | 1.06 | 1.64 | 1.16 | |
| 40 | Caffeic acid 3-O-methyltransferase | gi|145321007 | 38.57 | 5.7 | −1.36 | 1.10 | 1.72 | 1.15 | |
| 42 | 60S acidic ribosomal protein (Os08g0130500) | gi|115474653 | 34.36 | 5.4 | −1.36 | 1.46 | 1.76 | −1.13 | |
| 45 | Malate dehydrogenase 1, mitochondrial-like, predicted | gi|357132456 | 35.37 | 8.5 | −1.23 | 1.04 | 1.54 | 1.20 | |
| 5 | 11 | 5-methyltetrahydro-pteroyltriglutamate/hom-ocysteine methyl-transferase-like, predicted | gi|357155679 | 84.64 | 6.0 | −1.49 | 2.28 | 2.14 | −1.59 |
| 72 | Pathogenesis related protein 10 | gi|196051131 | 17.05 | 5.2 | 1.05 | 2.22 | 2.04 | −1.03 | |
| 50 | Group 5/9 grass pollen allergen R8-5 | gi|365769201 | 24.91 | 5.6 | −1.37 | 1.69 | 1.93 | −1.20 | |
| 35 | Actin-97-like, predicted | gi|357135037 | 41.7 | 5.2 | −1.44 | 1.63 | 2.23 | −1.05 | |
| 76 | Profilin | gi|300807845 | 14.18 | 4.9 | −1.19 | 1.60 | 1.88 | −1.02 | |
| 14 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|14017580 | 52.82 | 6.2 | −1.44 | 1.60 | 2.78 | 1.20 | |
| 15 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|14017580 | 52.82 | 6.2 | −1.29 | 1.52 | 2.43 | 1.24 | |
| 6 | 33 | Predicted barley protein | gi|326512860 | 36.77 | 5.0 | 1.22 | −1.76 | −1.99 | 1.08 |
| 53 | Translationally-controlled tumor protein | gi|146285306 | 18.77 | 4.6 | 1.02 | −1.65 | −1.96 | −1.16 | |
| 54 | Predicted barley protein | gi|326520557 | 16.18 | 4.8 | −1.03 | −1.55 | −1.66 | −1.11 | |
| 55 | Chlorophyll a/b binding protein, predicted | gi|302566696 | 28.2 | 5.1 | 1.01 | −1.50 | −1.67 | −1.10 | |
| 58 | Chlorophyll a-b binding protein CP24 10A, predicted | gi|356525886 | 27.67 | 6.2 | 1.04 | −1.53 | −1.74 | −1.09 | |
| 65 | MYB-related protein | gi|359952782 | 48.05 | 8.9 | 1.11 | −1.88 | −2.06 | 1.01 | |
| 7 | 13 ** | Mechanosensitive ion channel | gi|86439721 | 34.71 | 8.5 | −1.21 | 2.51 | 1.48 | −2.05 |
| 6 | Predicted Micromona protein | gi|255083775 | 44.95 | 10.4 | 1.07 | 2.27 | 1.24 | −1.72 | |
| 7 | Eukaryotic translation initiation factor 3 subunit D-like, predicted | gi|357128487 | 78.27 | 5.7 | 1.12 | 1.92 | 1.05 | −1.62 | |
| 44 | Uncharacterized maize protein LOC100273160 | gi|226528643 | 39.94 | 6.1 | −1.03 | 1.65 | 1.33 | −1.27 | |
| 18 | Glucose-1-phosphate adenylyltransferase large subunit, chloroplastic/amyloplastic (Fragment) | GLGL3_WHEAT | 55.52 | 6.6 | 1.07 | 1.60 | 1.17 | −1.28 | |
| 8 | 49 ** | Root phototropism 2, putative, expressed | gi|108862102 | 28.47 | 9.7 | −1.27 | −1.59 | −1.17 | 1.07 |
| 70 | Abscisic acid stress ripening 1 protein | gi|321155395 | 10.45 | 6.6 | −1.25 | −1.52 | −1.31 | −1.07 | |
| 9 | 19 * | Starch synthase IIa-3 | gi|8953573 | 86.74 | 6.3 | 1.43 | 1.18 | 1.40 | 1.70 |
| 21 | Alpha tubulin-2A | gi|90289596 | 49.68 | 4.9 | −1.34 | 1.31 | 1.13 | −1.56 | |
| 43 | Stearoyl-ACP desaturase | gi|319739540 | 44.43 | 8.2 | −1.29 | −1.11 | −1.42 | −1.66 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eldakak, M.; Das, A.; Zhuang, Y.; Rohila, J.S.; Glover, K.; Yen, Y. A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat. Pathogens 2018, 7, 58. https://doi.org/10.3390/pathogens7030058
Eldakak M, Das A, Zhuang Y, Rohila JS, Glover K, Yen Y. A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat. Pathogens. 2018; 7(3):58. https://doi.org/10.3390/pathogens7030058
Chicago/Turabian StyleEldakak, Moustafa, Aayudh Das, Yongbin Zhuang, Jai S. Rohila, Karl Glover, and Yang Yen. 2018. "A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat" Pathogens 7, no. 3: 58. https://doi.org/10.3390/pathogens7030058
APA StyleEldakak, M., Das, A., Zhuang, Y., Rohila, J. S., Glover, K., & Yen, Y. (2018). A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat. Pathogens, 7(3), 58. https://doi.org/10.3390/pathogens7030058





