Microfluidics-Based Approaches to the Isolation of African Trypanosomes
Abstract
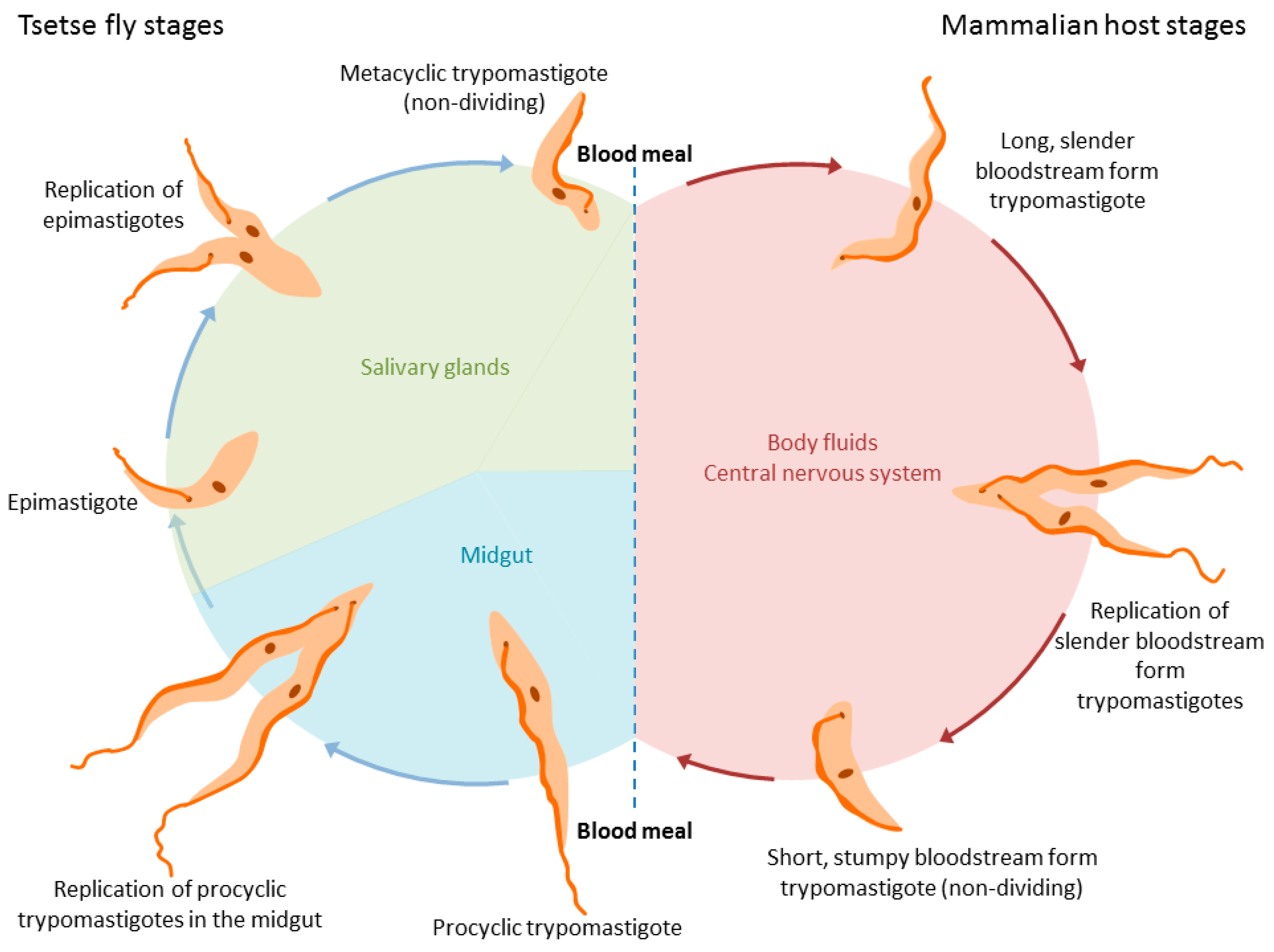
1. Introduction
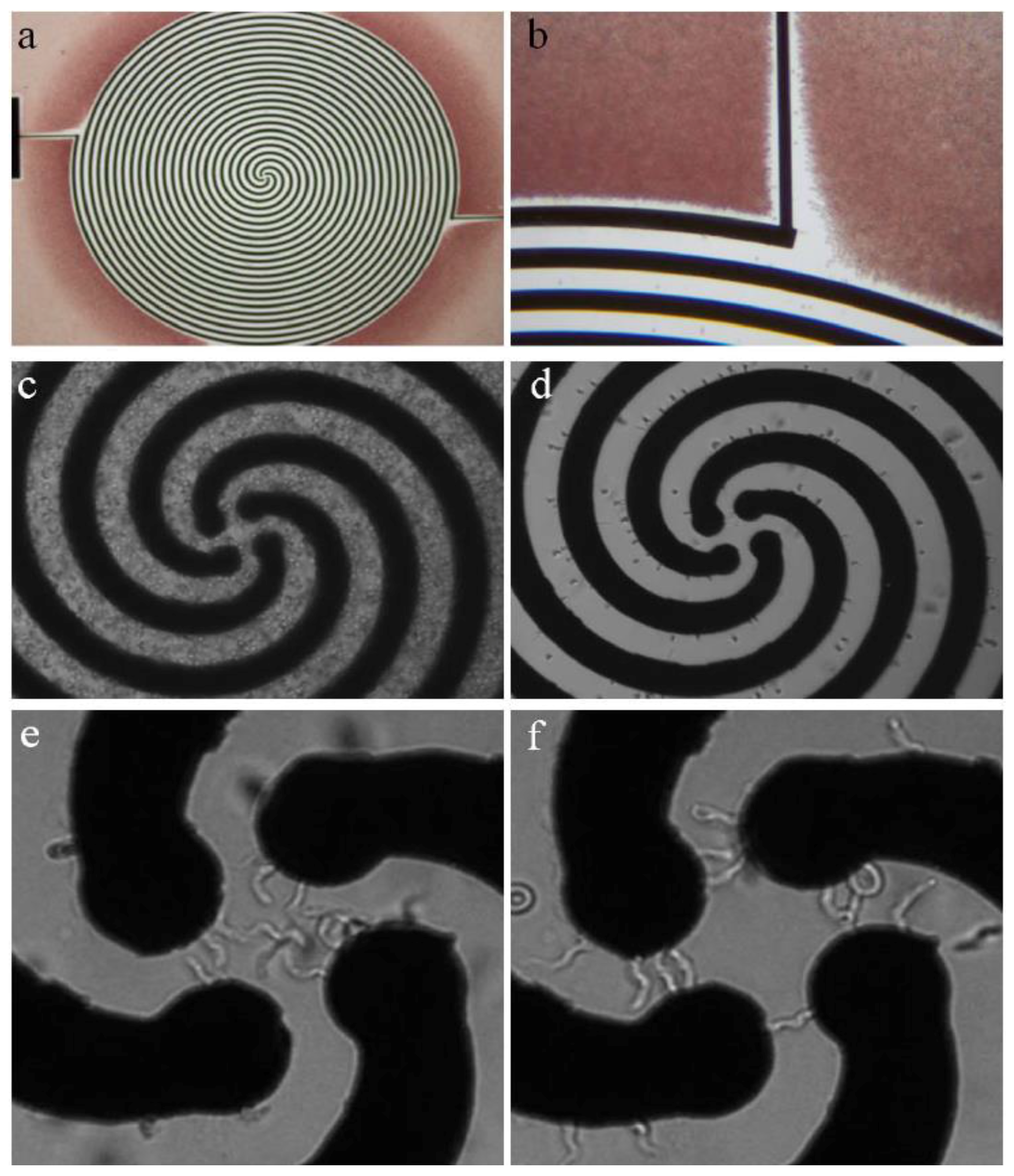
2. Separation by Dielectrophoresis
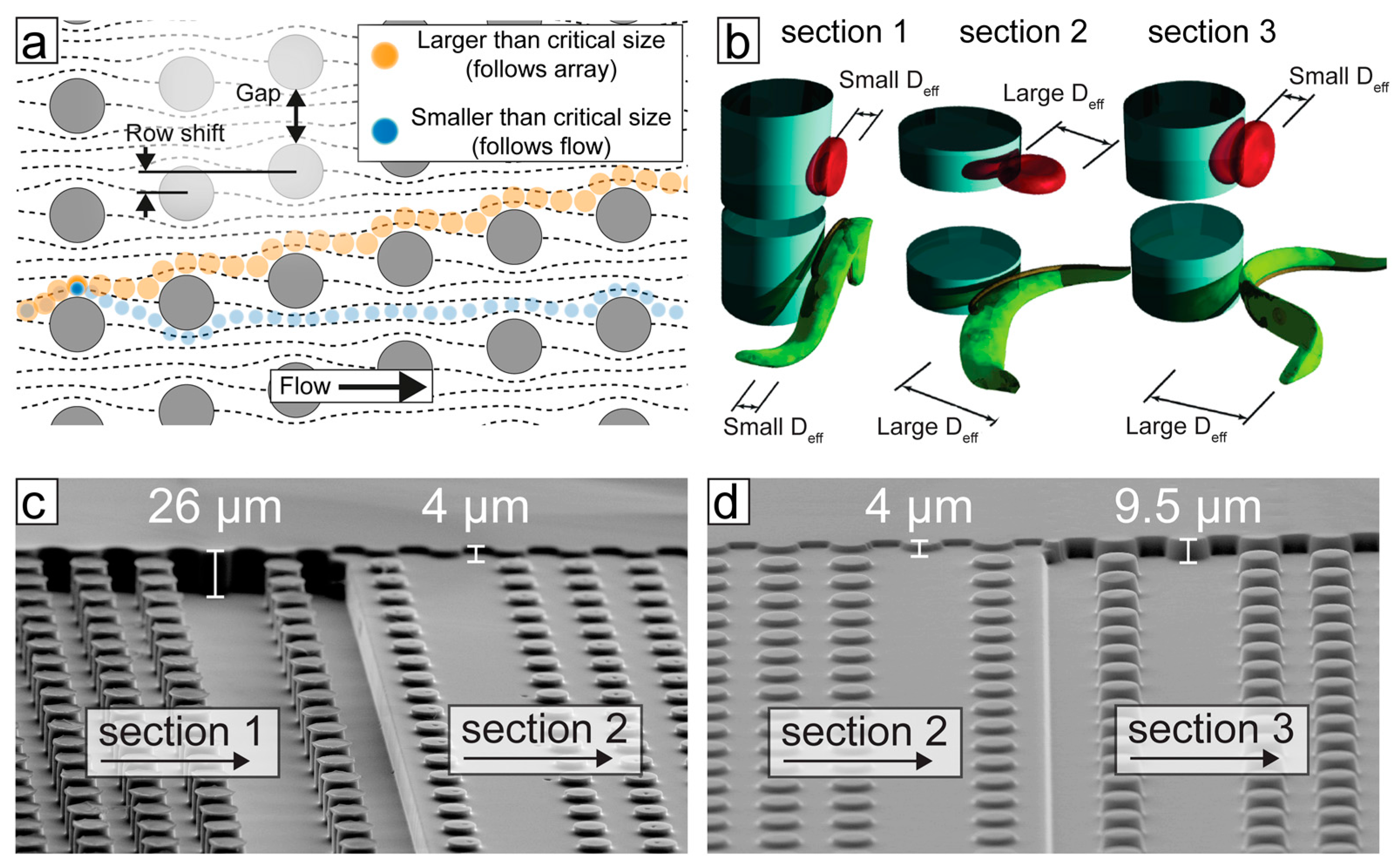
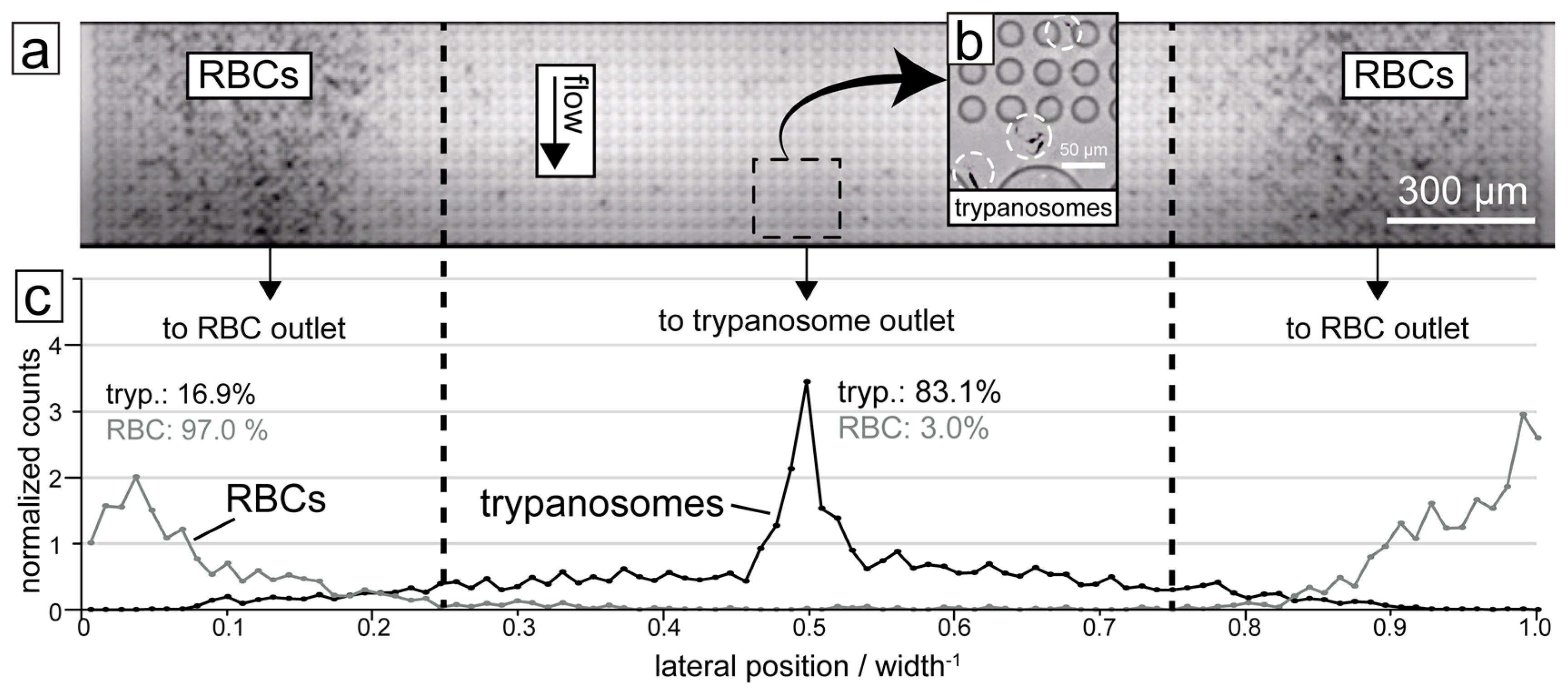
3. Separation by Deterministic Lateral Displacement (DLD)
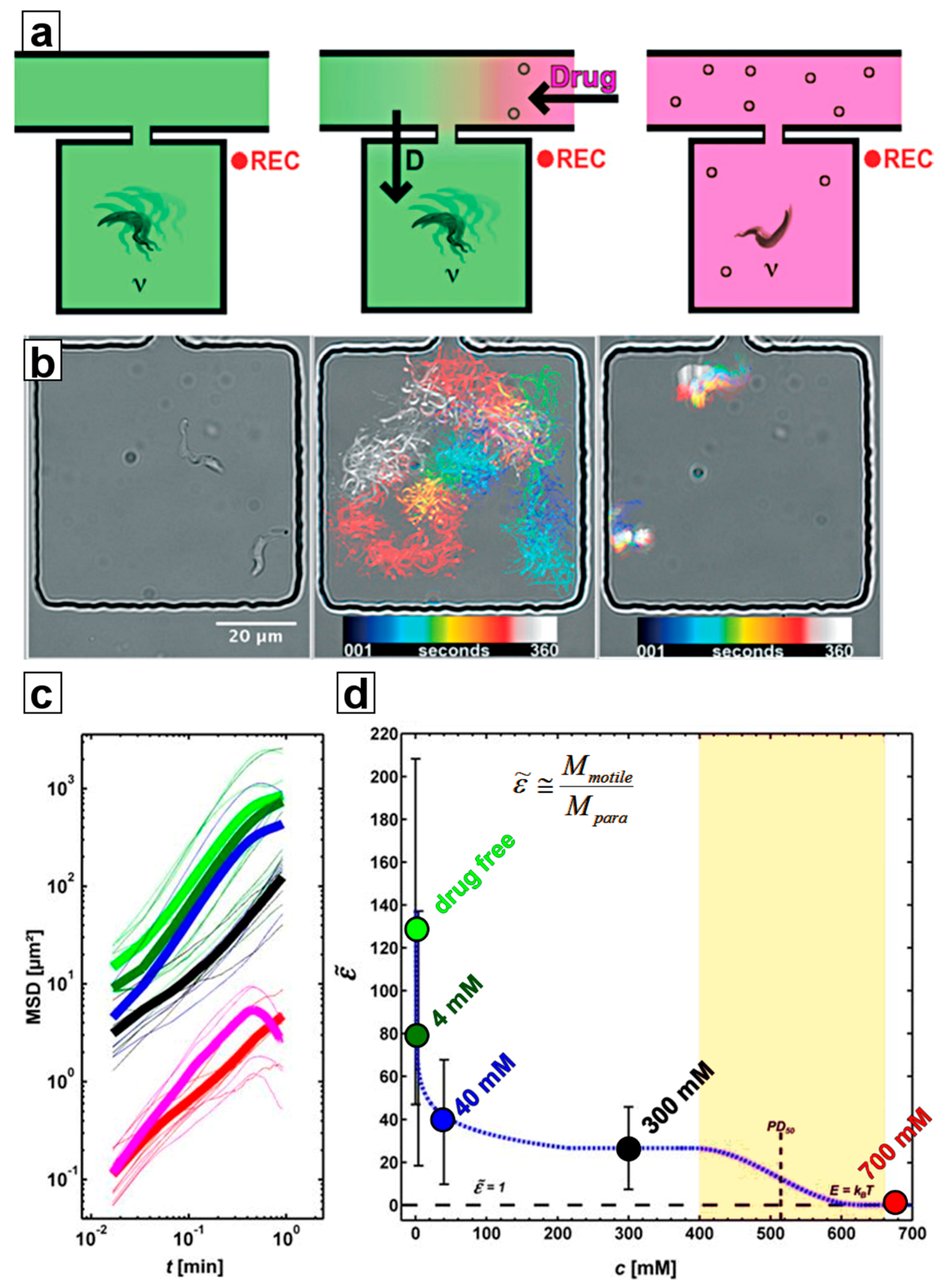
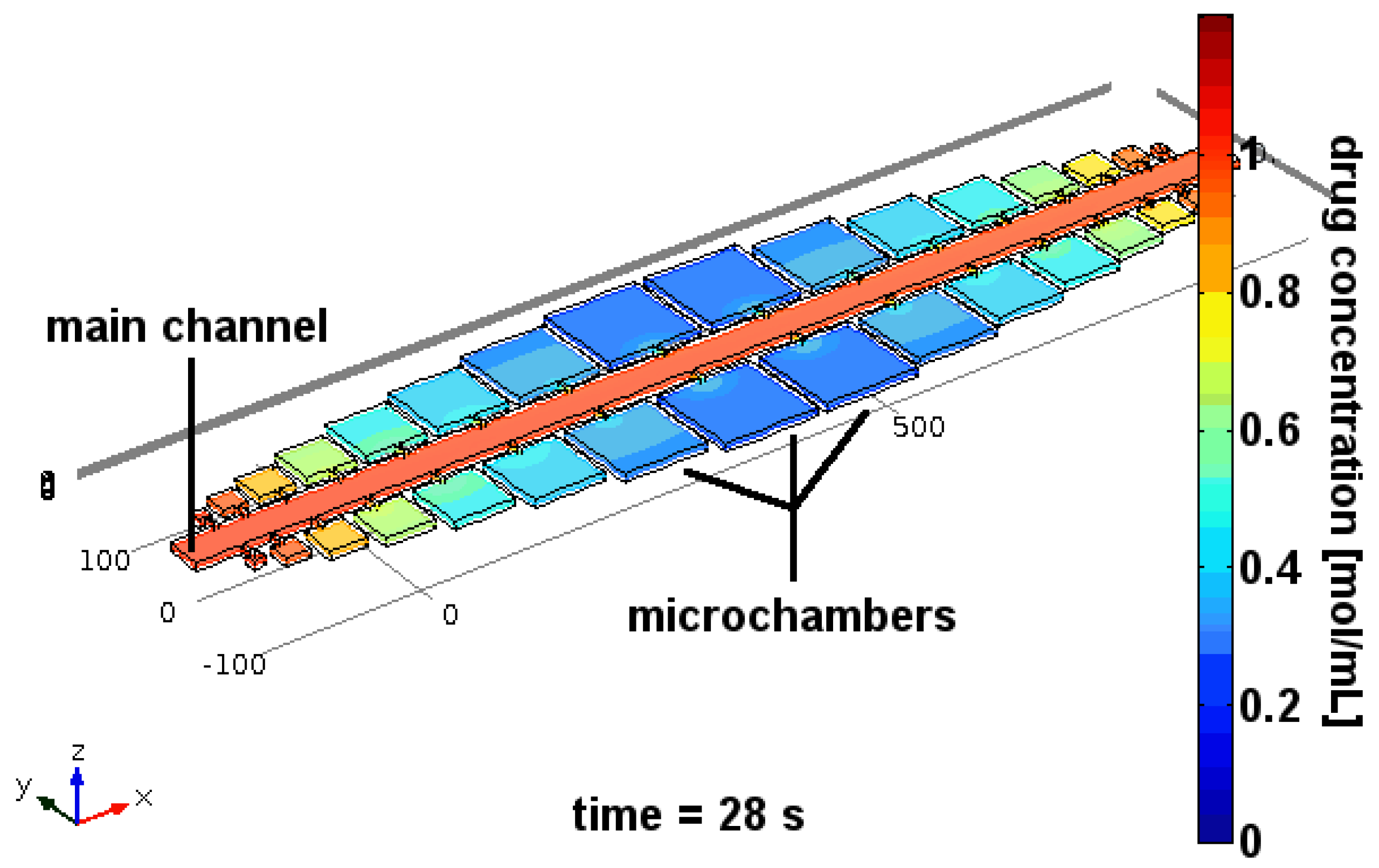
4. Separation of Trypanosomes Using Optical Tweezers and Drug Screening
4.1. Ramping
4.2. Constant Exposure
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Langousis, G.; Hill, K.L. Motility and more: The flagellum of Trypanosoma brucei. Nat. Rev. Microbiol. 2014, 12, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Mogk, S.; Boßelmann, C.M.; Mudogo, C.N.; Stein, J.; Wolburg, H.; Duszenko, M. African trypanosomes and brain infection—The unsolved question. Biol. Rev. 2017, 92, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Harrington, J.M.; Scelsi, C.; Hartel, A.; Jones, N.G.; Engstler, M.; Capewell, P.; MacLeod, A.; Hajduk, S. Novel African Trypanocidal Agents: Membrane Rigidifying Peptides. PLoS ONE 2012, 7, e44384. [Google Scholar] [CrossRef] [PubMed]
- Capewell, P.; Cren-Travaillé, C.; Marchesi, F.; Johnston, P.; Clucas, C.; Benson, R.A.; Gorman, T.-A.; Calvo-Alvarez, E.; Crouzols, A.; Jouvion, G.; et al. The skin is a significant but overlooked anatomical reservoir for vector-borne African trypanosomes. Elife 2016, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Beverley, S.M. African Trypanosomes Find a Fat Haven. Cell Host Microbe 2016, 19, 748–749. [Google Scholar] [CrossRef] [PubMed]
- Trindade, S.; Rijo-Ferreira, F.; Carvalho, T.; Pinto-Neves, D.; Guegan, F.; Aresta-Branco, F.; Bento, F.; Young, S.A.; Pinto, A.; Van Den Abbeele, J.; et al. Trypanosoma brucei Parasites Occupy and Functionally Adapt to the Adipose Tissue in Mice. Cell Host Microbe 2016, 19, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.A.; Zellweger, M.J.; Burri, C.; Hatz, C. Cardiac involvement in African and American trypanosomiasis. Lancet Infect. Dis. 2008, 8, 631–641. [Google Scholar] [CrossRef]
- McCarroll, C.S.; Rossor, C.L.; Morrison, L.R.; Morrison, L.J.; Loughrey, C.M. A Pre-clinical Animal Model of Trypanosoma brucei Infection Demonstrating Cardiac Dysfunction. PLoS Negl. Trop. Dis. 2015, 9, e0003811. [Google Scholar] [CrossRef] [PubMed]
- Claes, F.; Vodnala, S.K.; Van Reet, N.; Boucher, N.; Lunden-Miguel, H.; Baltz, T.; Goddeeris, B.M.; Büscher, P.; Rottenberg, M.E. Bioluminescent imaging of Trypanosoma brucei shows preferential testis dissemination which may hamper drug efficacy in sleeping sickness. PLoS Negl. Trop. Dis. 2009, 3, e486. [Google Scholar] [CrossRef] [PubMed]
- Engstler, M.; Pfohl, T.; Herminghaus, S.; Boshart, M.; Wiegertjes, G.; Heddergott, N.; Overath, P. Hydrodynamic Flow-Mediated Protein Sorting on the Cell Surface of Trypanosomes. Cell 2007, 131, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Stellamanns, E.; Uppaluri, S.; Hochstetter, A.; Heddergott, N.; Engstler, M.; Pfohl, T. Optical trapping reveals propulsion forces, power generation and motility efficiency of the unicellular parasites Trypanosoma brucei brucei. Sci. Rep. 2015, 4, 6515. [Google Scholar] [CrossRef] [PubMed]
- Mugnier, M.R.; Stebbins, C.E.; Papavasiliou, F.N. Masters of Disguise: Antigenic Variation and the VSG Coat in Trypanosoma brucei. PLoS Pathog. 2016, 12, e1005784. [Google Scholar] [CrossRef] [PubMed]
- Hirumi, H.; Hirumi, K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989, 75, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Schuster, F.L.; Sullivan, J.J. Cultivation of Clinically Significant Hemoflagellates. Clin. Microbiol. Rev. 2002, 15, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.T. Evaluation of the haematocrit centrifuge and other techniques for the field diagnosis of human trypanosomiasis and filariasis Evaluation of the Haematocrit Centrifuge and Other Techniques for the Field Diagnosis. Acta Trop. 1971, 28, 298–303. [Google Scholar] [PubMed]
- Lanham, S.M.; Godfrey, D.G. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 1970, 28, 521–534. [Google Scholar] [CrossRef]
- Lumsden, W.H.R.; Kimber, C.D.; Evans, D.A.; Doig, S.J. Trypanosoma brucei: Miniature anion-exchange centrifugation technique for detection of low parasitaemias: Adaptation for field use. Trans. R. Soc. Trop. Med. Hyg. 1979, 73, 312–317. [Google Scholar] [CrossRef]
- Camara, M.; Camara, O.; Ilboudo, H.; Sakande, H.; Kaboré, J.; N’Dri, L.; Jamonneau, V.; Bucheton, B. Sleeping sickness diagnosis: Use of buffy coats improves the sensitivity of the mini anion exchange centrifugation test. Trop. Med. Int. Health 2010, 15, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Mumba Ngoyi, D.; Ali Ekangu, R.; Mumvemba Kodi, M.F.; Pyana, P.P.; Balharbi, F.; Decq, M.; Kande Betu, V.; Van der Veken, W.; Sese, C.; Menten, J.; et al. Performance of Parasitological and Molecular Techniques for the Diagnosis and Surveillance of Gambiense Sleeping Sickness. PLoS Negl. Trop. Dis. 2014, 8, e2954. [Google Scholar] [CrossRef] [PubMed]
- Menachery, A.; Kremer, C.; Wong, P.E.; Carlsson, A.; Neale, S.L.; Barrett, M.P.; Cooper, J.M. Counterflow dielectrophoresis for trypanosome enrichment and detection in blood. Sci. Rep. 2012, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.R.; Cox, E.C.; Austin, R.H.; Sturm, J.C. Continuous particle separation through deterministic lateral displacement. Science 2004, 304, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Inglis, D.W.; Davis, J.A.; Austin, H.; Sturm, J.C. Critical particle size for fractionation by deterministic lateral displacement. Lab Chip 2006, 6, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.A. Microfluidic Separation of Blood Components through Deterministic Lateral Displacement. Ph.D. Thesis, Princeton University, Princeton, NJ, USA, 2008. [Google Scholar]
- Beech, J.P.; Holm, S.H.; Adolfsson, K.; Tegenfeldt, J.O. Sorting cells by size, shape and deformability. Lab Chip 2012, 12, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Henry, E.; Holm, S.H.; Zhang, Z.; Beech, J.P.; Tegenfeldt, J.O.; Fedosov, D.A.; Gompper, G. Sorting cells by their dynamical properties. Sci. Rep. 2016, 6, 34375. [Google Scholar] [CrossRef] [PubMed]
- Holm, S.H.; Beech, J.P.; Barrett, M.P.; Tegenfeldt, J.O. Separation of parasites from human blood using deterministic lateral displacement. Lab Chip 2011, 11, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Holm, S.H.; Beech, J.P.; Barrett, M.P.; Tegenfeldt, J.O. Simplifying microfluidic separation devices towards field-detection of blood parasites. Anal. Methods 2016, 8, 3291–3300. [Google Scholar] [CrossRef]
- De Thomaz, A.A.; Fontes, A.; Stahl, C.V.; Pozzo, L.Y.; Ayres, D.C.; Almeida, D.B.; Farias, P.M.A.; Santos, B.S.; Santos-Mallet, J.; Gomes, S.A.O.; et al. Optical tweezers for studying taxis in parasites. J. Opt. 2011, 13, 044015. [Google Scholar] [CrossRef]
- Hochstetter, A.; Stellamanns, E.; Deshpande, S.; Uppaluri, S.; Engstler, M.; Pfohl, T. Microfluidics-based single cell analysis reveals drug-dependent motility changes in trypanosomes. Lab Chip 2015, 15, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Tropea, C.; Eds, H.B. Nature-Inspired Fluid Mechanics; Tropea, C., Bleckmann, H., Eds.; Notes on Numerical Fluid Mechanics and Multidisciplinary Design; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2012; Volume 119. [Google Scholar]
- Kremer, C.; Neale, S.; Menachery, A.; Barrett, M.; Cooper, J.M. Optoelectronic tweezers for medical diagnostics. Proc SPIE Int. Soc. Opt. Eng. 2012, 8212, 1–7. [Google Scholar]
- Hochstetter, A.; Stellamanns, E.; Uppaluri, S.; Heddergott, N.; Engstler, M.; Pfohl, T. Tracing the microscopic motility of unicellular parasites. Eur. Biophys. J. 2013, 42 (Suppl. 1), S35–S235. [Google Scholar]
- Li, G.; Tang, J.X. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys. J. 2006, 91, 2726–2734. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.J. Use of chiral cell shape to ensure highly directional swimming in trypanosomes. PLoS Comput. Biol. 2017, 13, e1005353. [Google Scholar] [CrossRef] [PubMed]
- Bastin, P.; Sherwin, T.; Gull, K. Paraflagellar rod is vital for trypanosome motility. Nature 1998, 391, 548. [Google Scholar] [CrossRef] [PubMed]
- Rotureau, B.; Ooi, C.-P.; Huet, D.; Perrot, S.; Bastin, P. Forward motility is essential for trypanosome infection in the tsetse fly. Cell. Microbiol. 2014, 16, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Heddergott, N.; Krüger, T.; Babu, S.B.; Wei, A.; Stellamanns, E.; Uppaluri, S.; Pfohl, T.; Stark, H.; Engstler, M. Trypanosome Motion Represents an Adaptation to the Crowded Environment of the Vertebrate Bloodstream. PLoS Pathog. 2012, 8, e1003023. [Google Scholar] [CrossRef] [PubMed]
- Bourquin, Y.; Syed, A.; Reboud, J.; Ranford-Cartwright, L.C.; Barrett, M.P.; Cooper, J.M. Rare-cell enrichment by a rapid, label-free, ultrasonic isopycnic technique for medical diagnostics. Angew. Chem. Int. Ed. 2014, 53, 5587–5590. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L. Discovery and Development of Diagnostic Biomarkers for Human African Trypanosomiasis. Ph.D. Thesis, University of Dundee, Dundee, UK, 2012. [Google Scholar]
- Kennedy, P.G.E. Diagnostic and neuropathogenesis issues in human African trypanosomiasis. Int. J. Parasitol. 2006, 36, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Radwanska, M. Emerging trends in the diagnosis of Human African Trypanosomiasis. Parasitology 2010, 137, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- LaBarre, P.; Boyle, D.; Hawkins, K.; Weigl, B. Instrument-free nucleic acid amplification assays for global health settings. In Proceedings of the Event: SPIE Defense, Security, and Sensing, Orlando, FL, USA, 25–29 April 2011; Volume 8029, p. 802902. [Google Scholar]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Brun, R.; Blum, J.; Chappuis, F.; Burri, C. Human African trypanosomiasis. Lancet 2010, 375, 148–159. [Google Scholar] [CrossRef]
- Njiru, Z.K.; Mikosza, A.S.J.; Armstrong, T.; Enyaru, J.C.; Ndung’u, J.M.; Thompson, A.R.C. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl. Trop. Dis. 2008, 2, e147. [Google Scholar] [CrossRef] [PubMed]
- Njiru, Z.K. Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn. Microbiol. Infect. Dis. 2011, 69, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Kremer, C.; Witte, C.; Neale, S.L.; Reboud, J.; Barrett, M.P.; Cooper, J.M. Shape-dependent optoelectronic cell lysis. Angew. Chem. Int. Ed. 2014, 53, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Neale, S.L.; Witte, C.; Clark, A.W.; Reboud, J.; Cooper, J.M. Optoelectronic Cell Lysis. In Proceeding of the Nano Science + Engineering, San Diego, CA, USA, 17–21 August 2014. [Google Scholar]
- Witte, C.; Kremer, C.; Cooper, J.M.; Neale, S.L. Continuous cell lysis in microfluidics through acoustic and optoelectronic tweezers. SPIE. Digit. Libr. 2014, 8615, 2–7. [Google Scholar]
- Biéler, S.; Matovu, E.; Mitashi, P.; Ssewannyana, E.; Bi Shamamba, S.K.; Bessell, P.R.; Ndung’u, J.M. Improved detection of Trypanosoma brucei by lysis of red blood cells, concentration and LED fluorescence microscopy. Acta Trop. 2012, 121, 135–140. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrett, M.P.; Cooper, J.M.; Regnault, C.; Holm, S.H.; Beech, J.P.; Tegenfeldt, J.O.; Hochstetter, A. Microfluidics-Based Approaches to the Isolation of African Trypanosomes. Pathogens 2017, 6, 47. https://doi.org/10.3390/pathogens6040047
Barrett MP, Cooper JM, Regnault C, Holm SH, Beech JP, Tegenfeldt JO, Hochstetter A. Microfluidics-Based Approaches to the Isolation of African Trypanosomes. Pathogens. 2017; 6(4):47. https://doi.org/10.3390/pathogens6040047
Chicago/Turabian StyleBarrett, Michael P., Jonathan M. Cooper, Clément Regnault, Stefan H. Holm, Jason P. Beech, Jonas O. Tegenfeldt, and Axel Hochstetter. 2017. "Microfluidics-Based Approaches to the Isolation of African Trypanosomes" Pathogens 6, no. 4: 47. https://doi.org/10.3390/pathogens6040047
APA StyleBarrett, M. P., Cooper, J. M., Regnault, C., Holm, S. H., Beech, J. P., Tegenfeldt, J. O., & Hochstetter, A. (2017). Microfluidics-Based Approaches to the Isolation of African Trypanosomes. Pathogens, 6(4), 47. https://doi.org/10.3390/pathogens6040047





