The Influenza NS1 Protein: What Do We Know in Equine Influenza Virus Pathogenesis?
Abstract
:1. Introduction
2. Influenza NS1 Protein Inhibition of Host Antiviral Response
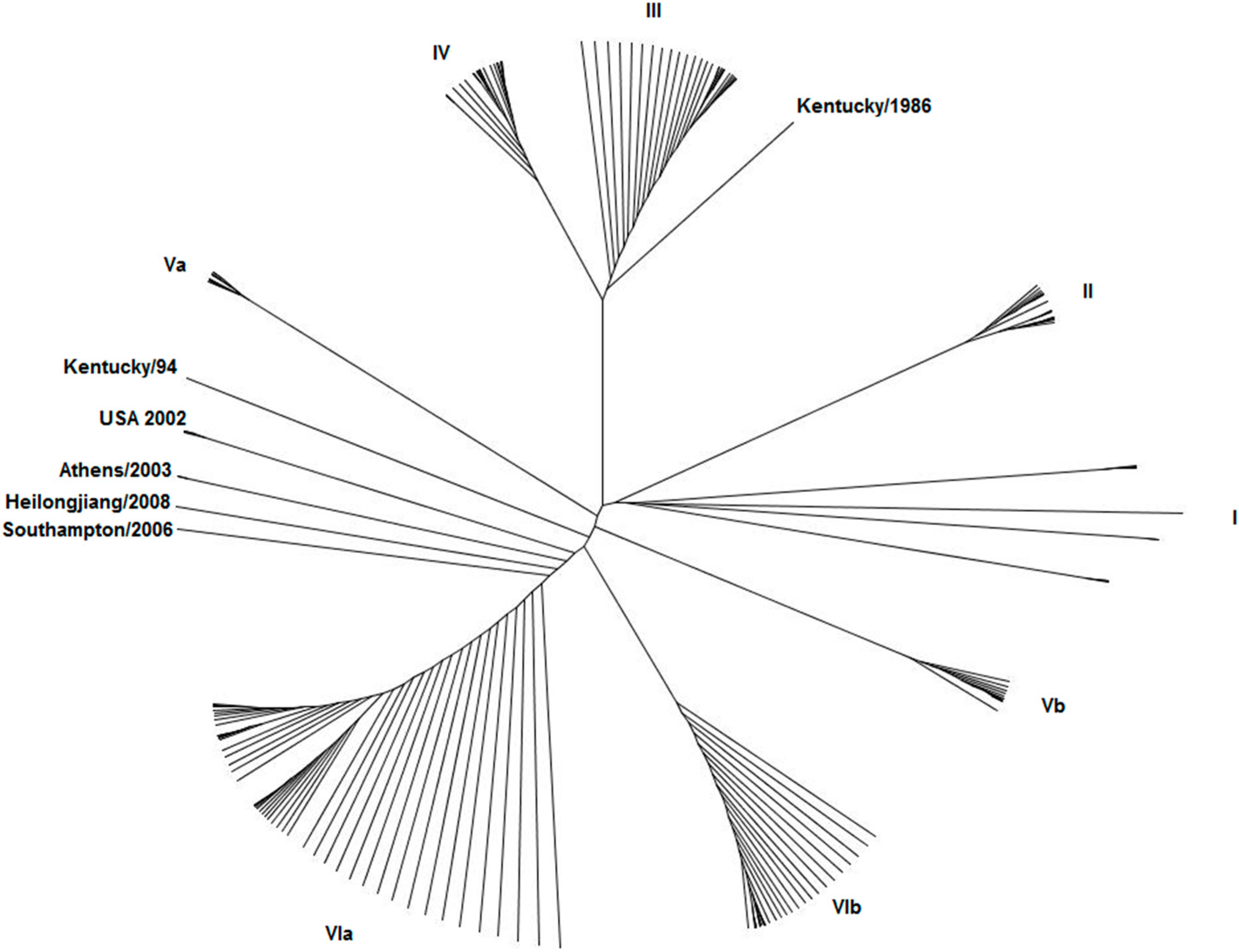
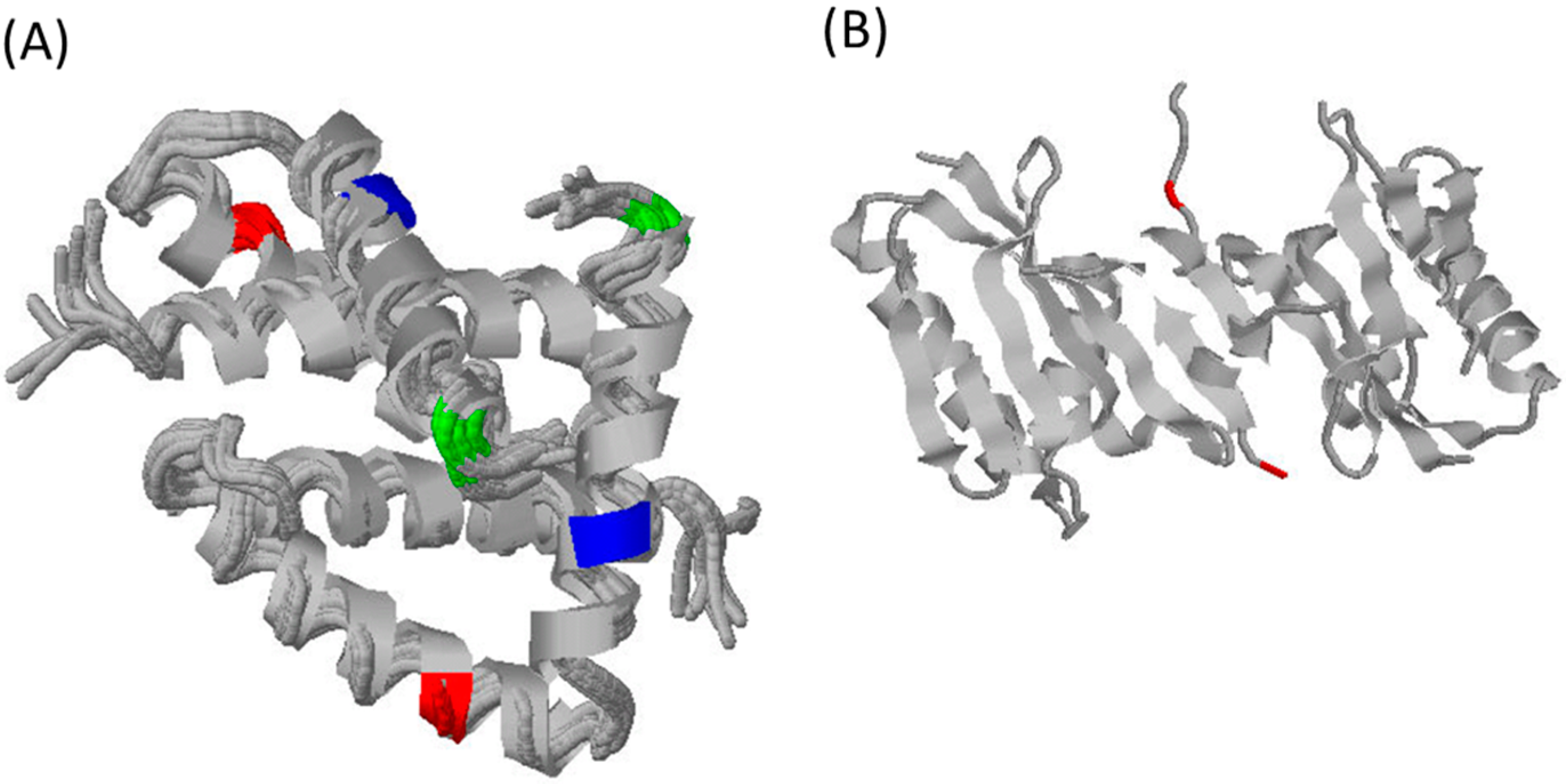
3. What Do We Know about Equine Influenza NS1?
4. Potential Role of NS1 in Equine Pathology and Inter-Species Transmission
5. Potential to Apply Knowledge about NS1 for Prevention and Control of Influenza
6. Concluding Remarks
Author Contributions
Conflicts of Interest
Abbreviations
| CPSF | Cleavage and Polyadenylation Specific Factor |
| eIF4G | eukaryotic translation initiation factor 4GI |
| EIV | equine influenza virus |
| HA | hemagglutinin |
| IFN | interferon |
| IL | interleukin |
| NA | neuraminidase |
| NES | nuclear export signal |
| NLS | nuclear localization signal |
| NS1 | non-structural protein 1 |
| OAS | 2′,5′-oligoadenylate synthetase |
| PABPII | poly-A binding protein II. |
| PAMPs | pathogen-associated molecular patterns |
| PI3K | phosphoinoside-3-kinase |
| PKR | protein kinase R |
| RIG-I | retinoic acid-inducible gene product I |
| TNF | tumor necrosis factor |
| TRIM | tripartite motif |
References
- Watson, J.; Halpin, K.; Selleck, P.; Axell, A.; Bruce, K.; Hansson, E.; Hammond, J.; Daniels, P.; Jeggo, M. Isolation and characterisation of an H3N8 equine influenza virus in australia, 2007. Aust. Vet. J. 2011, 89 (Suppl. 1), 35–37. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Niwa, H.; Tsujimura, K.; Kondo, T.; Matsumura, T. Epidemic of equine influenza among vaccinated racehorses in japan in 2007. J. Vet. Med. Sci. 2008, 70, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Andino, R.; Domingo, E. Viral quasispecies. Virology 2015, 479–480, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Murano, K.; Ohniwa, R.L.; Kawaguchi, A.; Nagata, K. Oseltamivir expands quasispecies of influenza virus through cell-to-cell transmission. Sci. Rep. 2015, 5, 9163. [Google Scholar] [CrossRef]
- Daly, J.M.; Lai, A.C.; Binns, M.M.; Chambers, T.M.; Barrandeguy, M.; Mumford, J.A. Antigenic and genetic evolution of equine H3N8 influenza a viruses. J. Gen. Virol. 1996, 77, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.C.; Chambers, T.M.; Holland, R.E., Jr.; Morley, P.S.; Haines, D.M.; Townsend, H.G.; Barrandeguy, M. Diverged evolution of recent equine-2 influenza (H3N8) viruses in the western hemisphere. Arch. Virol. 2001, 146, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Bryant, N.A.; Rash, A.S.; Russell, C.A.; Ross, J.; Cooke, A.; Bowman, S.; MacRae, S.; Lewis, N.S.; Paillot, R.; Zanoni, R.; et al. Antigenic and genetic variations in european and north american equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 2009, 138, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Resa-Infante, P.; Jorba, N.; Coloma, R.; Ortín, J. The influenza virus RNA synthesis machine: Advances in its structure and function. RNA Biol. 2011, 8, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.J.; Lomniczi, B.; Bellamy, A.R.; Skehel, J.J. Transcription of the influenza virus genome. Virology 1977, 83, 337–355. [Google Scholar] [CrossRef]
- Geiss, G.K.; Salvatore, M.; Tumpey, T.M.; Carter, V.S.; Wang, X.; Basler, C.F.; Taubenberger, J.K.; Bumgarner, R.E.; Palese, P.; Katze, M.G.; et al. Cellular transcriptional profiling in influenza a virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 2002, 99, 10736–10741. [Google Scholar] [CrossRef] [PubMed]
- Solorzano, A.; Webby, R.J.; Lager, K.M.; Janke, B.H.; Garcia-Sastre, A.; Richt, J.A. Mutations in the ns1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005, 79, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Aragon, T.; de la Luna, S.; Novoa, I.; Carrasco, L.; Ortin, J.; Nieto, A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 2000, 20, 6259–6268. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortin, J.; Jackson, D. The multifunctional NS1 protein of influenza a viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.Y.; Xu, Y.; Xiao, R.; Aramini, J.M.; Sahasrabudhe, P.V.; Krug, R.M.; Montelione, G.T. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza a virus: Evidence for a novel RNA-binding mode. Biochemistry 2004, 43, 1950–1962. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Basler, C.F.; Williams, B.R.; Silverman, R.H.; Palese, P.; Garcia-Sastre, A. Functional replacement of the carboxy-terminal two-thirds of the influenza a virus ns1 protein with short heterologous dimerization domains. J. Virol. 2002, 76, 12951–12962. [Google Scholar] [CrossRef] [PubMed]
- Satterly, N.; Tsai, P.L.; van Deursen, J.; Nussenzveig, D.R.; Wang, Y.; Faria, P.A.; Levay, A.; Levy, D.E.; Fontoura, B.M. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2007, 104, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wambach, M.; Katze, M.G.; Krug, R.M. Binding of the influenza virus ns1 protein to double-stranded rna inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 1995, 214, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Chen, L.M.; Zeng, H.; Gomez, J.A.; Plowden, J.; Fujita, T.; Katz, J.M.; Donis, R.O.; Sambhara, S. Ns1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 2007, 36, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Melen, K.; Kinnunen, L.; Fagerlund, R.; Ikonen, N.; Twu, K.Y.; Krug, R.M.; Julkunen, I. Nuclear and nucleolar targeting of influenza A virus NS1 protein: Striking differences between different virus subtypes. J. Virol. 2007, 81, 5995–6006. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Min, J.Y.; Krug, R.M.; Sen, G.C. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either pact or double-stranded RNA. Virology 2006, 349, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yamakita, Y.; Krug, R.M. Regulation of a nuclear export signal by an adjacent inhibitory sequence: The effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 1998, 95, 4864–4869. [Google Scholar] [CrossRef] [PubMed]
- Nemeroff, M.E.; Barabino, S.M.; Li, Y.; Keller, W.; Krug, R.M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Krug, R.M. Influenza A virus NS1 protein targets poly(a)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999, 18, 2273–2283. [Google Scholar] [CrossRef] [PubMed]
- Obenauer, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wang, J.; Ma, J.; Fan, Y.; et al. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Nemeroff, M.E.; Qian, X.Y.; Krug, R.M. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 1995, 212, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Krug, R.M. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 1996, 223, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Cheung, C.Y.; Chui, W.H.; Tsao, S.W.; Nicholls, J.M.; Chan, Y.O.; Chan, R.W.; Long, H.T.; Poon, L.L.; Guan, Y.; et al. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 2005, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Poon, L.L.; Lau, A.S.; Luk, W.; Lau, Y.L.; Shortridge, K.F.; Gordon, S.; Guan, Y.; Peiris, J.S. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: A mechanism for the unusual severity of human disease? Lancet 2002, 360, 1831–1837. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Zheng, H.; Muster, T.; Palese, P.; Beg, A.A.; Garcia-Sastre, A. Influenza A virus NS1 protein prevents activation of nf-kappab and induction of alpha/beta interferon. J. Virol. 2000, 74, 11566–11573. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.; Wang, X.; Ehrhardt, C.; Zheng, H.; Donelan, N.; Planz, O.; Pleschka, S.; Garcia-Sastre, A.; Heins, G.; Wolff, T. The influenza A virus NS1 protein inhibits activation of jun N-terminal kinase and AP-1 transcription factors. J. Virol. 2002, 76, 11166–11171. [Google Scholar] [CrossRef] [PubMed]
- Talon, J.; Horvath, C.M.; Polley, R.; Basler, C.F.; Muster, T.; Palese, P.; Garcia-Sastre, A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 2000, 74, 7989–7996. [Google Scholar] [CrossRef] [PubMed]
- Min, J.Y.; Krug, R.M. The primary function of rna binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (a) synthetase/rnase l pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 2007, 81, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Hoffmann, E.; Webster, R.G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 2002, 8, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Suarez, D.L.; Perdue, M.L. Multiple alignment comparison of the non-structural genes of influenza a viruses. Virus Res. 1998, 54, 59–69. [Google Scholar] [PubMed]
- Xu, J.; Zhong, H.A.; Madrahimov, A.; Helikar, T.; Lu, G. Molecular phylogeny and evolutionary dynamics of influenza A nonstructural (NS) gene. Infect. Genet. Evol. 2014, 22, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, M.L.; Wise, H.M.; Nicol, M.Q.; Smith, N.; Dunfee, R.L.; Beard, P.M.; Jagger, B.W.; Ligertwood, Y.; Hardisty, G.R.; Xiao, H.; et al. The role of the B-allele of the influenza A virus segment 8 in setting mammalian host range and pathogenicity. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, M.; Kawaoka, Y.; Gorman, O.; Ito, T.; Saito, T.; Webster, R.G. Characterization of a new avian-like influenza A virus from horses in china. Virology 1992, 188, 245–255. [Google Scholar] [CrossRef]
- Kwasnik, M.; Gora, I.M.; Rola, J.; Zmudzinski, J.F.; Rozek, W. NS-gene based phylogenetic analysis of equine influenza viruses isolated in poland. Vet. Microbiol. 2016, 182, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.Fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Martinez-Sobrido, L.; Lienenklaus, S.; Weiss, S.; Garcia-Sastre, A.; Staeheli, P. Strong interferon-inducing capacity of a highly virulent variant of influenza a virus strain PR8 with deletions in the NS1 gene. J. Gen. Virol. 2009, 90, 2990–2994. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Guo, J.; Zhang, A.; Zhang, W.; Zhao, Z.; Zhou, H.; Liu, C.; Chen, H.; Jin, M. Effects of the C-terminal truncation in NS1 protein of the 2009 pandemic H1N1 influenza virus on host gene expression. PLoS ONE 2011, 6, e26175. [Google Scholar] [CrossRef] [PubMed]
- Anastasina, M.; Schepens, B.; Soderholm, S.; Nyman, T.A.; Matikainen, S.; Saksela, K.; Saelens, X.; Kainov, D.E. The C terminus of NS1 protein of influenza A/WSN/1933(H1N1) virus modulates antiviral responses in infected human macrophages and mice. J. Gen. Virol. 2015, 96, 2086–2091. [Google Scholar] [CrossRef] [PubMed]
- Golebiewski, L.; Liu, H.; Javier, R.T.; Rice, A.P. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J. Virol. 2011, 85, 10639–10648. [Google Scholar] [CrossRef] [PubMed]
- Soubies, S.M.; Volmer, C.; Croville, G.; Loupias, J.; Peralta, B.; Costes, P.; Lacroux, C.; Guerin, J.L.; Volmer, R. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J. Virol. 2010, 84, 6733–6747. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.H.; Min, J.Y. A naturally truncated NS1 protein of influenza A virus impairs its interferon-antagonizing activity and thereby confers attenuation in vitro. Arch. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Crawford, P.C.; Dubovi, E.J.; Castleman, W.L.; Stephenson, I.; Gibbs, E.P.; Chen, L.; Smith, C.; Hill, R.C.; Ferro, P.; Pompey, J.; et al. Transmission of equine influenza virus to dogs. Science 2005, 310, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Crispe, E.; Finlaison, D.S.; Hurt, A.C.; Kirkland, P.D. Infection of dogs with equine influenza virus: Evidence for transmission from horses during the australian outbreak. Aust. Vet. J. 2011, 89 (Suppl. 1), 27–28. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.M.; Blunden, A.S.; Macrae, S.; Miller, J.; Bowman, S.J.; Kolodziejek, J.; Nowotny, N.; Smith, K.C. Transmission of equine influenza virus to english foxhounds. Emerg. Infect. Dis. 2008, 14, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, P.D.; Finlaison, D.S.; Crispe, E.; Hurt, A.C. Influenza virus transmission from horses to dogs, australia. Emerg. Infect. Dis. 2010, 16, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.M.; Macrae, S.; Newton, J.R.; Wattrang, E.; Elton, D.M. Equine influenza: A review of an unpredictable virus. Vet. J. 2011, 189, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Dundon, W.G. Variability among the neuraminidase, non-structural 1 and PB1-F2 proteins in the influenza a virus genome. Virus Genes 2012, 44, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Rivailler, P.; Perry, I.A.; Jang, Y.; Davis, C.T.; Chen, L.M.; Dubovi, E.J.; Donis, R.O. Evolution of canine and equine influenza (H3N8) viruses co-circulating between 2005 and 2008. Virology 2010, 408, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Begg, A.P.; Reece, R.L.; Hum, S.; Townsend, W.; Gordon, A.; Carrick, J. Pathological changes in horses dying with equine influenza in australia, 2007. Aust. Vet. J. 2011, 89 (Suppl. 1), 19–22. [Google Scholar] [CrossRef] [PubMed]
- Rynda-Apple, A.; Robinson, K.M.; Alcorn, J.F. Influenza and bacterial superinfection: Illuminating the immunologic mechanisms of disease. Infect. Immun. 2015, 83, 3764–3770. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014, 12, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Maamary, J.; Pica, N.; Belicha-Villanueva, A.; Chou, Y.Y.; Krammer, F.; Gao, Q.; Garcia-Sastre, A.; Palese, P. Attenuated influenza virus construct with enhanced hemagglutinin protein expression. J. Virol. 2012, 86, 5782–5790. [Google Scholar] [CrossRef] [PubMed]
- Quinlivan, M.; Zamarin, D.; Garcia-Sastre, A.; Cullinane, A.; Chambers, T.; Palese, P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 2005, 79, 8431–8439. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.M.; Quinlivan, M.; Sturgill, T.; Cullinane, A.; Horohov, D.W.; Zamarin, D.; Arkins, S.; Garcia-Sastre, A.; Palese, P. Influenza A viruses with truncated NS1 as modified live virus vaccines: Pilot studies of safety and efficacy in horses. Equine Vet. J. 2009, 41, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Engel, D.A. The influenza virus NS1 protein as a therapeutic target. Antiviral Res. 2013, 99, 409–416. [Google Scholar] [CrossRef] [PubMed]
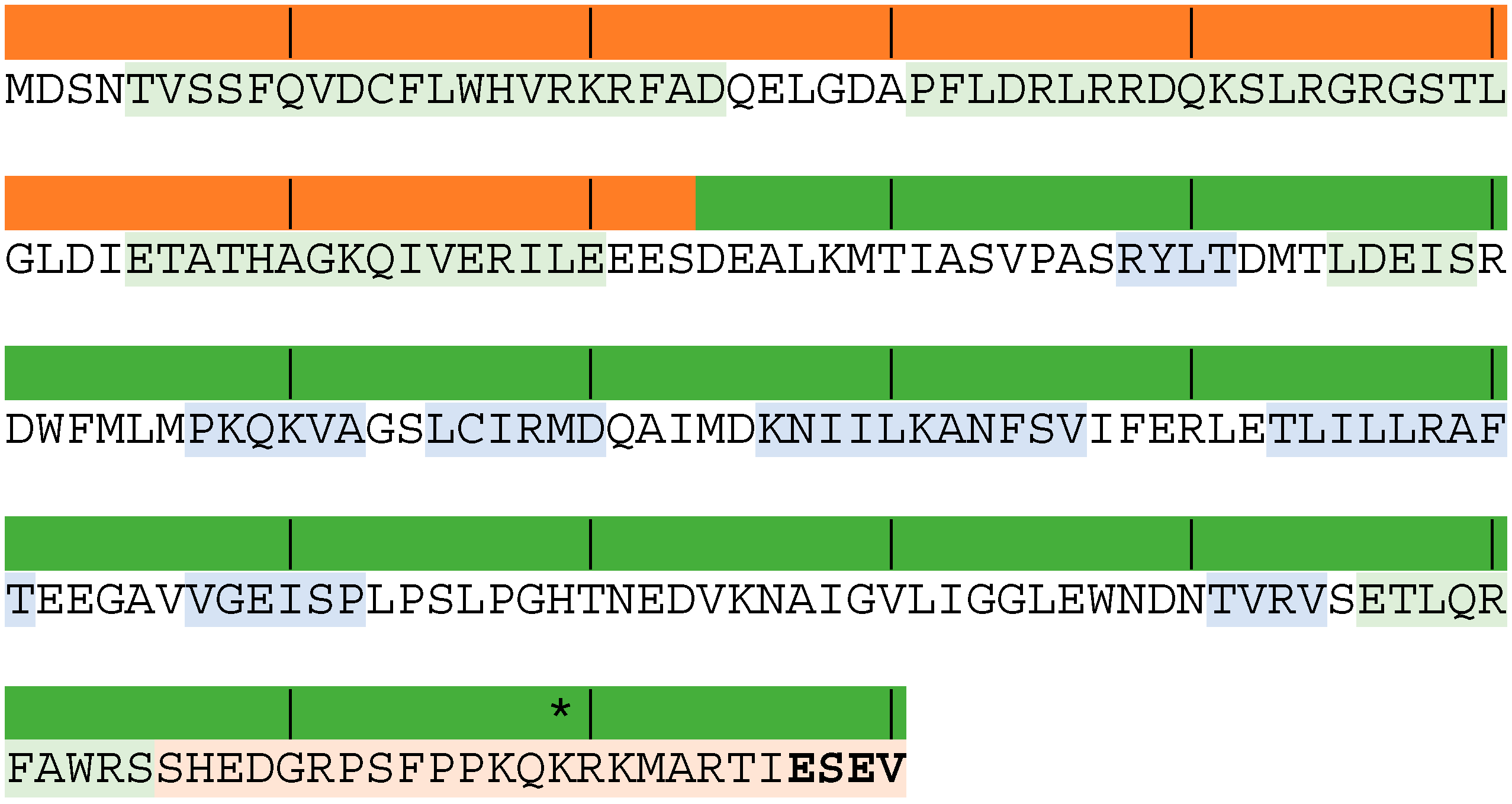
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barba, M.; Daly, J.M. The Influenza NS1 Protein: What Do We Know in Equine Influenza Virus Pathogenesis? Pathogens 2016, 5, 57. https://doi.org/10.3390/pathogens5030057
Barba M, Daly JM. The Influenza NS1 Protein: What Do We Know in Equine Influenza Virus Pathogenesis? Pathogens. 2016; 5(3):57. https://doi.org/10.3390/pathogens5030057
Chicago/Turabian StyleBarba, Marta, and Janet M. Daly. 2016. "The Influenza NS1 Protein: What Do We Know in Equine Influenza Virus Pathogenesis?" Pathogens 5, no. 3: 57. https://doi.org/10.3390/pathogens5030057
APA StyleBarba, M., & Daly, J. M. (2016). The Influenza NS1 Protein: What Do We Know in Equine Influenza Virus Pathogenesis? Pathogens, 5(3), 57. https://doi.org/10.3390/pathogens5030057






