Diversity in the Common Fold: Structural Insights into Class D β-Lactamases from Gram-Negative Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence and Structural Analysis
2.2. Molecular Dynamics Simulations
3. Results and Discussion
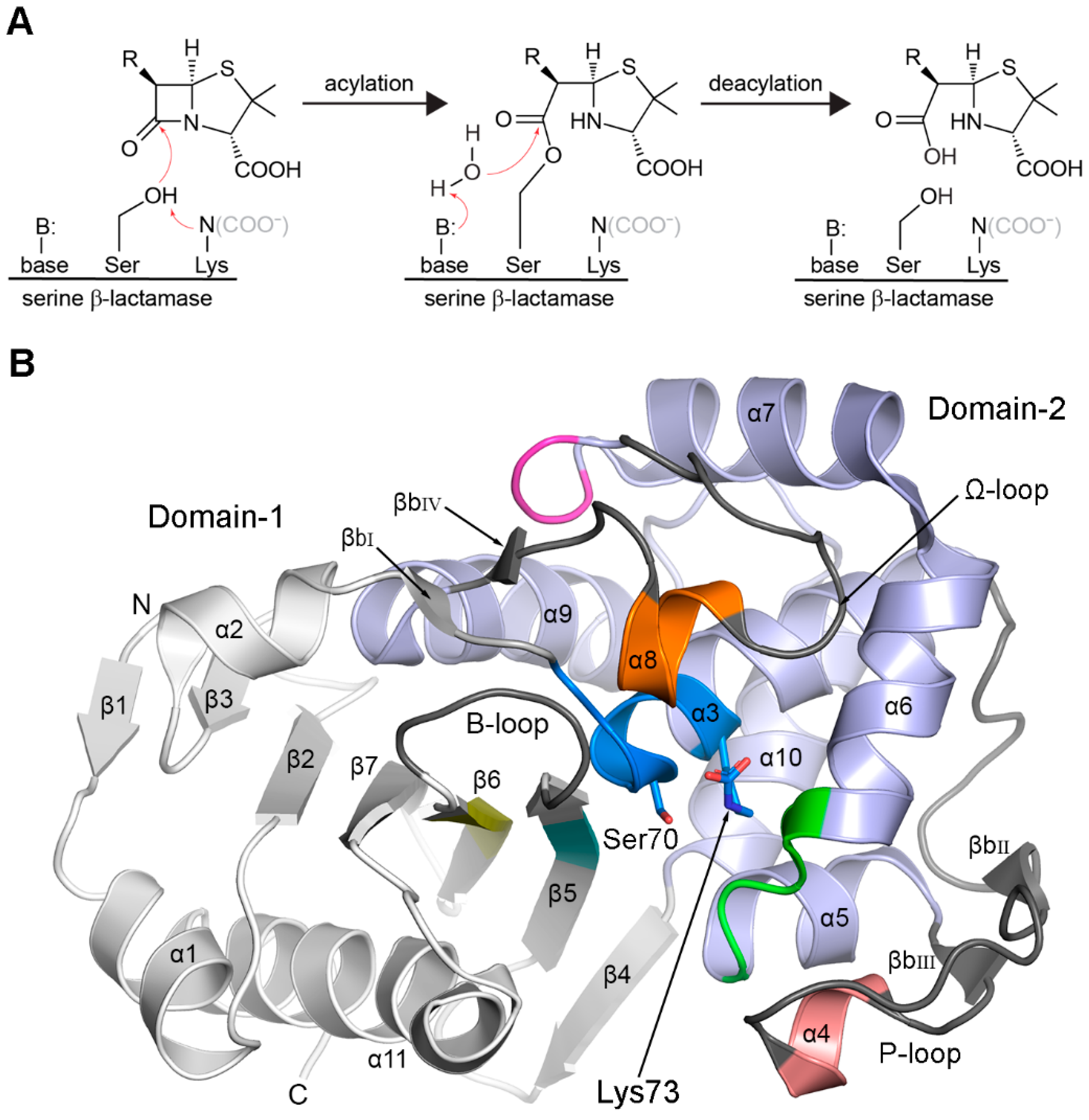
3.1. The Common DBL Structural Fold
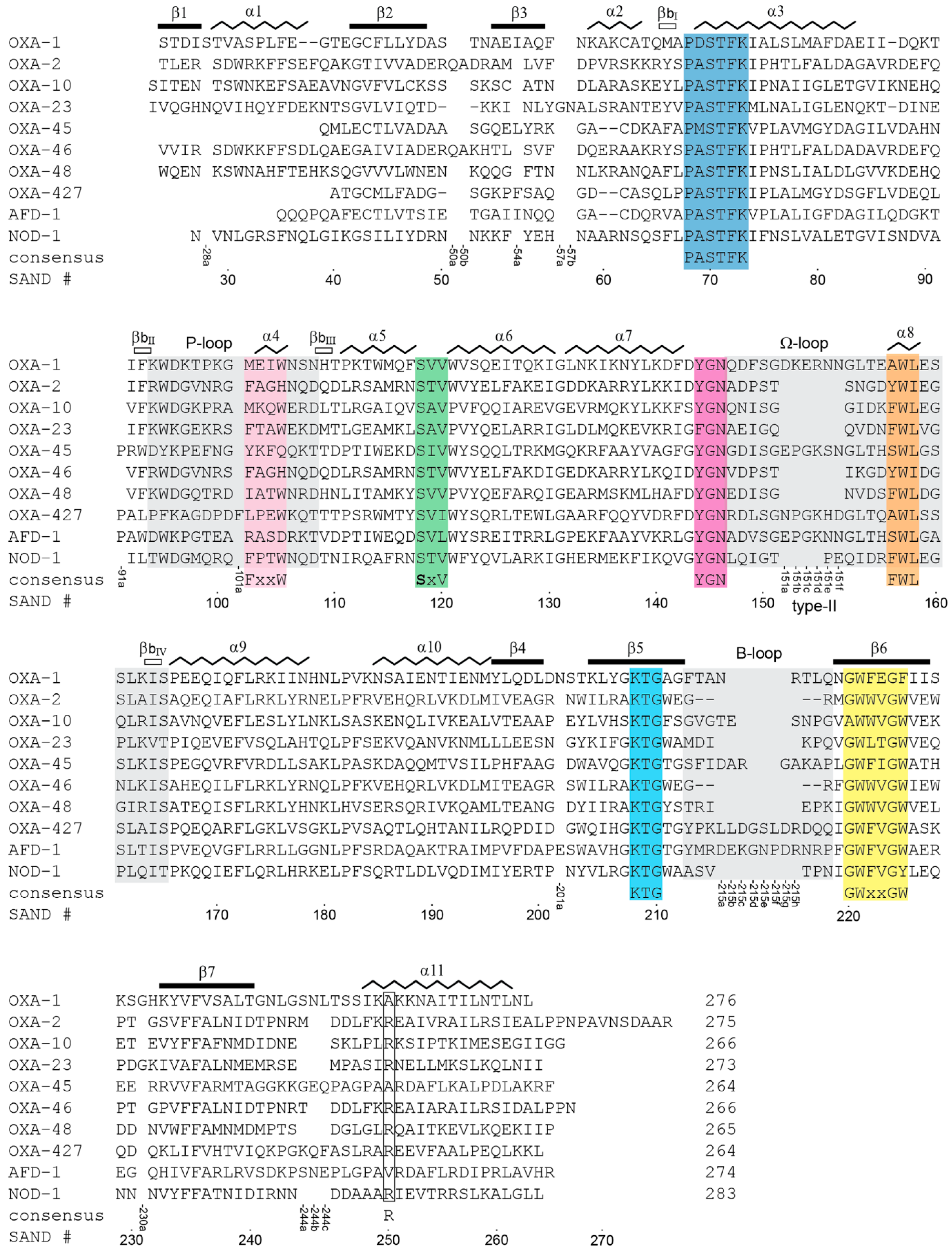
3.2. Class D Fingerprint Sequence Motifs
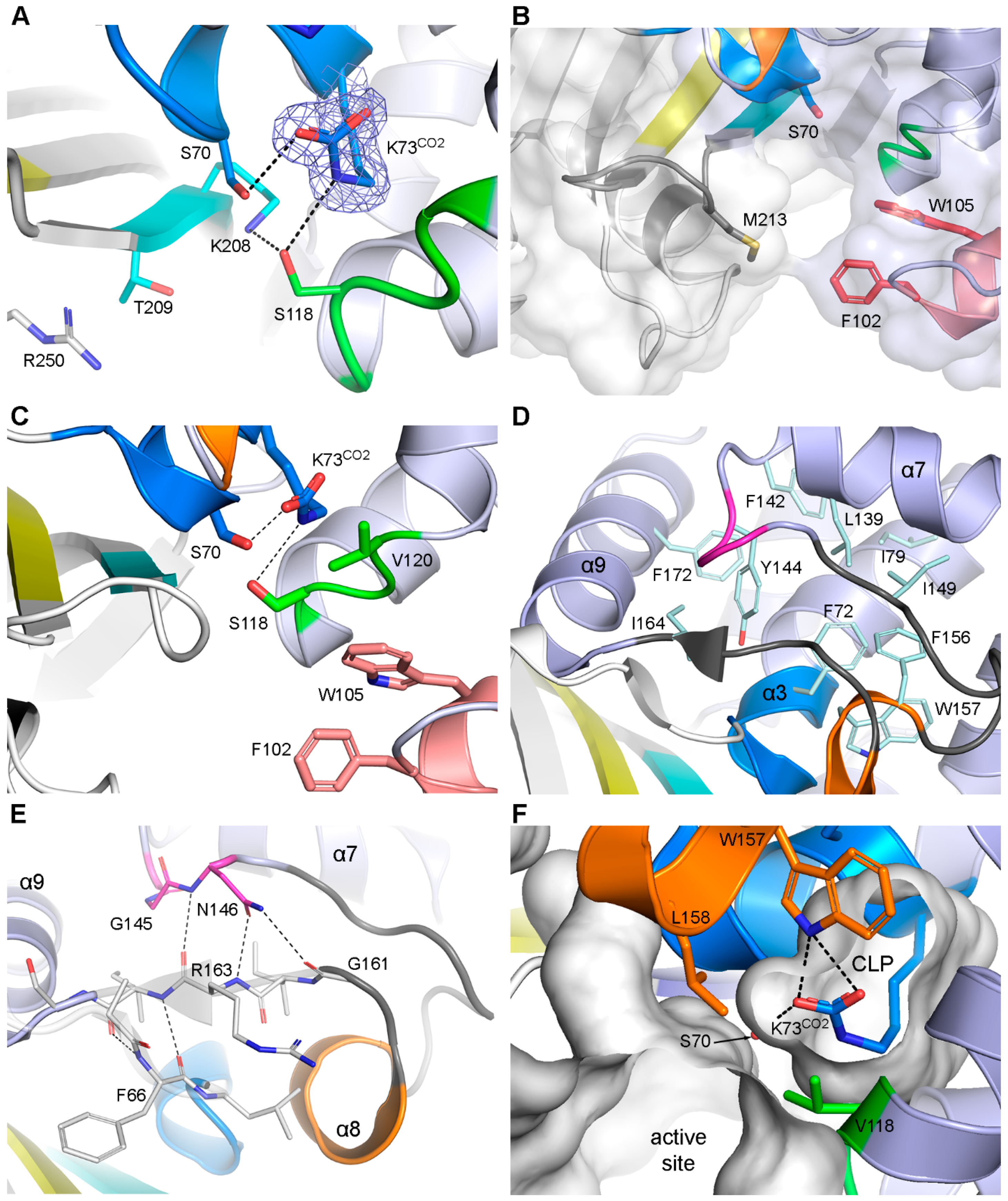
3.2.1. Motif-1 (PASTFK; Residues 68–73)
3.2.2. Motif-2 (FxxW; Residues 102–105)
3.2.3. Motif-3 (SAV; Residues 118–120)
3.2.4. Motif-4 (YGN; Residues 144–146)
3.2.5. Motif-5 (FWL; Residues 156–158)
3.2.6. Motif-6 (K(S/T)G; Residues 208–210)
3.2.7. Motif-7 (GWx(T/V)GW; Residues 220–225)
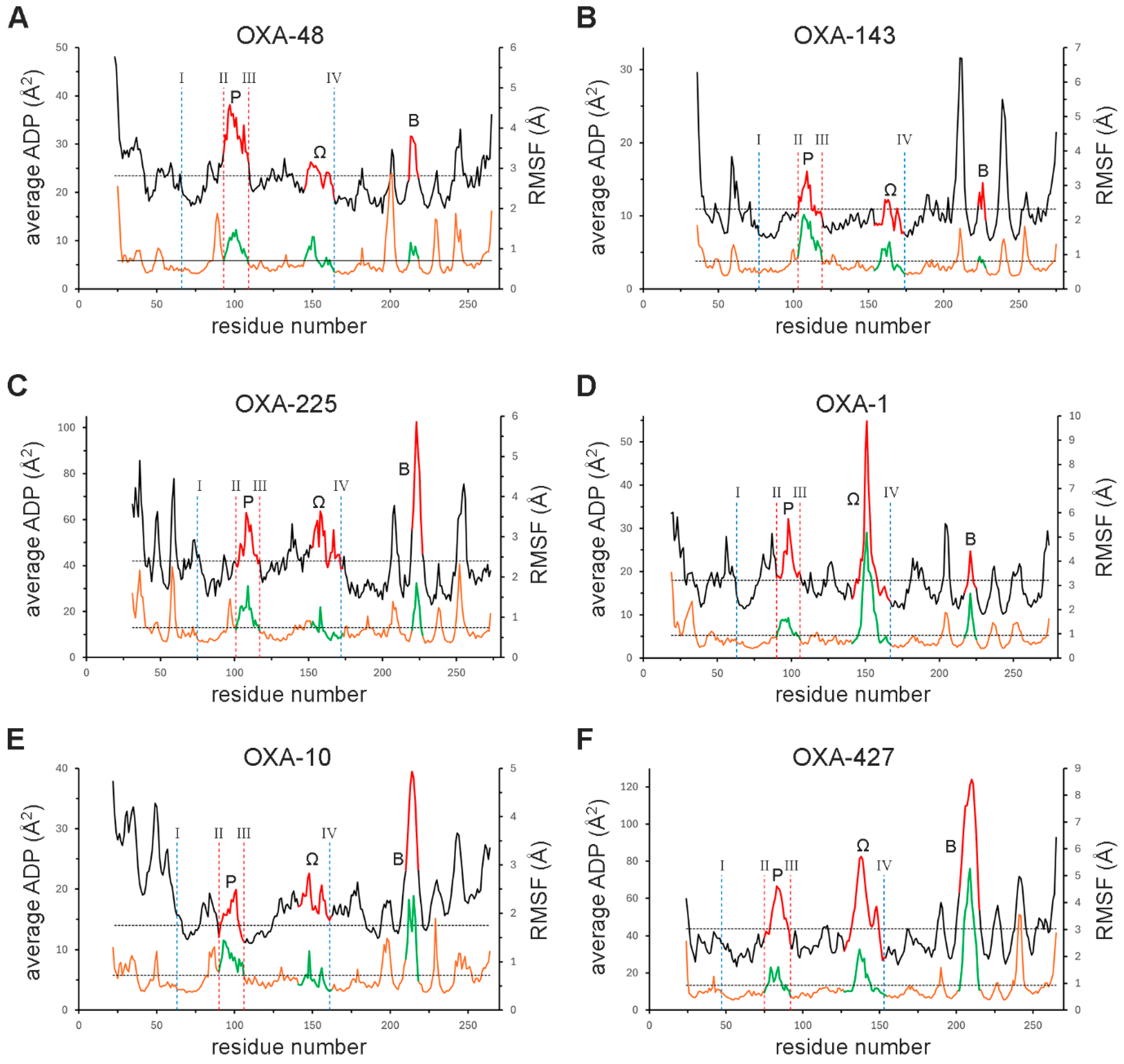
3.3. The P-Loop
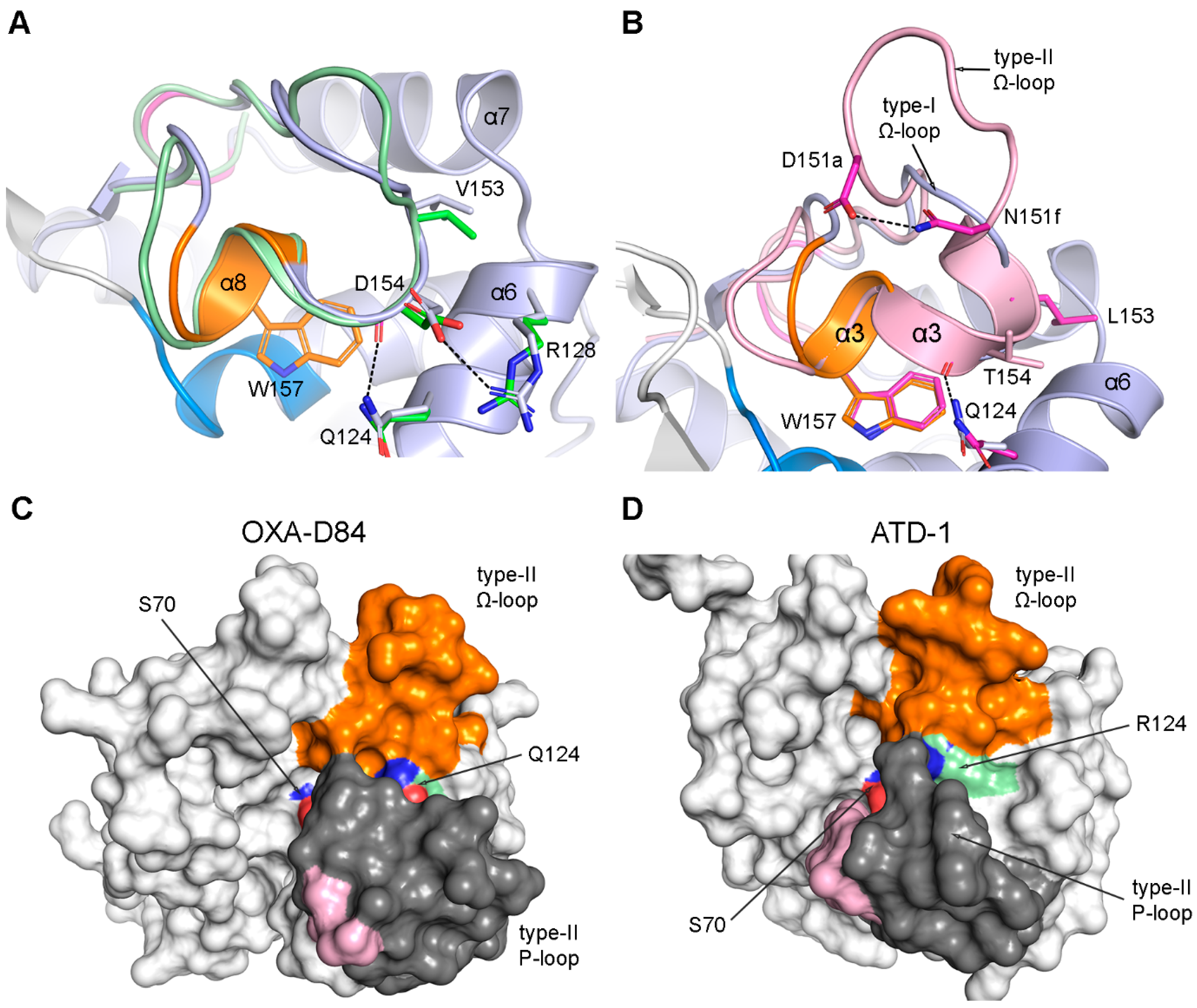
3.4. The Ω-Loop
3.5. The B-Loop
The Type-III B-Loop
3.6. Dynamics and Stability Within the DBL Fold
3.7. The β-Bridges
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
Abbreviations
References
- Poole, K. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 2004, 61, 2200–2223. [Google Scholar] [CrossRef]
- Livermore, D.M.; Woodford, N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006, 14, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Bench-to-bedside review: The role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit. Care 2010, 14, 224. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Proliferation and significance of clinically relevant β-lactamases. Ann. New York Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A. β-lactamases: A focus on current challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a025239. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-Lactamase DataBase (BLDB)—Structure and Function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of β-lactamases. Philos. Trans. R. Soc. London. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Ambler, R.P.; Coulson, A.F.W.; Frere, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A Standard Numbering Scheme for the Class-A β-Lactamases. Biochem. J. 1991, 276, 269–270. [Google Scholar] [CrossRef]
- Fisher, J.F.; Meroueh, S.O.; Mobashery, S. Bacterial Resistance to β-Lactam Antibiotics: Compelling Opportunism, Compelling Opportunity. Chem. Rev. 2005, 105, 395–424. [Google Scholar] [CrossRef]
- Leonard, D.A.; Bonomo, R.A.; Powers, R.A. Class D β-lactamases: A reappraisal after five decades. Acc. Chem. Res. 2013, 46, 2407–2425. [Google Scholar] [CrossRef]
- Golemi, D.; Maveyraud, L.; Vakulenko, S.; Samama, J.P.; Mobashery, S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. USA 2001, 98, 14280–14285. [Google Scholar] [CrossRef]
- Che, T.; Bethel, C.R.; Pusztai-Carey, M.; Bonomo, R.A.; Carey, P.R. The Different Inhibition Mechanisms of OXA-1 and OXA-24 β-Lactamases Are Determined by the Stability of Active Site Carboxylated Lysine. J. Biol. Chem. 2014, 289, 6152–6164. [Google Scholar] [CrossRef]
- Smith, C.A.; Stewart, N.K.; Toth, M.; Vakulenko, S.B. Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 β-Lactamase. Antimicrob. Agents Chemother. 2019, 63, e01202-19. [Google Scholar] [CrossRef]
- Stewart, N.K.; Toth, M.; Stasyuk, A.; Vakulenko, S.B.; Smith, C.A. In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D β-lactamases. ACS Infect. Dis. 2021, 7, 1765–1776. [Google Scholar] [CrossRef]
- Toth, M.; Stewart, N.K.; Maggiolo, A.O.; Quan, P.; Khan, M.M.K.; Buynak, J.D.; Smith, C.A.; Vakulenko, S.B. Decarboxylation of the Catalytic Lysine Residue by the C5α-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase. ACS Infect. Dis. 2024, 10, 4347–4359. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Hoiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Matagne, A.; Dubus, A.; Galleni, M.; Frère, J.-M. The β-lactamase cycle: A tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 1999, 16, 1–19. [Google Scholar] [CrossRef]
- Maveyraud, L.; Golemi, D.; Kotra, L.P.; Tranier, S.; Vakulenko, S.; Mobashery, S.; Samama, J.P. Insights Into Class D β-Lactamases are Revealed by the Crystal Structure of the OXA10 Enzyme from Pseudomonas Aeruginosa. Structure 2000, 8, 1289–1298. [Google Scholar] [CrossRef]
- Paetzel, M.; Danel, F.; de Castro, L.; Mosimann, S.C.; Page, M.G.P.; Strynadka, N.C.J. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 2000, 7, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.A.; Amyes, S.G. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Paton, R.; Miles, R.S.; Hood, J.; Amyes, S.G.B. ARI-I: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 1993, 2, 81–88. [Google Scholar] [CrossRef]
- Bou, G.; Oliver, A.; Martinez-Beltran, J. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 2000, 44, 1556–1561. [Google Scholar] [CrossRef]
- Poirel, L.; Heritier, C.; Toluen, V.; Nordmann, P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Toth, M.; Antunes, N.T.; Stewart, N.K.; Frase, H.; Bhattacharya, M.; Smith, C.A.; Vakulenko, S.B. Class D β-lactamases do exist in Gram-positive bacteria. Nat. Chem. Biol. 2016, 12, 9–14. [Google Scholar] [CrossRef]
- Toth, M.; Stewart, N.K.; Smith, C.; Vakulenko, S.B. Intrinsic Class D β-Lactamases of Clostridium difficile. mBio 2019, 9, e01803–e01818. [Google Scholar] [CrossRef]
- Sandhu, B.K.; Edwards, A.N.; Anderson, S.E.; Woods, E.C.; McBride, S.M. Regulation and anaerobic function of the Clostridioides difficile β-lactamase. Antimicrob. Agents Chemother. 2019, 64, e01496. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef]
- Stasyuk, A.; Smith, C.A. Standardized Residue Numbering and Secondary Structure Nomenclature in the Class D β-Lactamases. ACS Infect. Dis. 2025, 11, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Attana, F.; Kim, S.; Spencer, J.; Iorga, B.I.; Docquier, J.-D.; Rossolini, G.M.; Perilli, M.; Amicosante, G.; Vila, A.J.; Vakulenko, S.B.; et al. SAND: A comprehensive annotation of class D β-lactamases using structural alignment-based numbering. Antimicrob. Agents Chemother. 2025, 69, e00150-25. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview version 2: A Multiple Sequence Alignment and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004, D60, 2256–2268. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. 2010, D66, 486–501. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossváry, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar] [CrossRef]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Neria, E.; Fischer, S.; Karplus, M. Simulation of activation energies in molecular systems. J. Chem. Phys. 1996, 105, 1902–1921. [Google Scholar] [CrossRef]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé-Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Szarecka, A.; Lesnock, K.R.; Ramirez-Mondragon, C.A.; Nicholas, H.B.J.; Wymore, T. The Class D β-lactamase family: Residues governing the maintenance and diversity of function. Protein Eng. Des. Sel. 2011, 24, 801–809. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Jeong, S.-H. Class D β-lactamases. J. Antimicrob. Chemother. 2021, 76, 836–864. [Google Scholar] [CrossRef] [PubMed]
- Stewart, N.K.; Bhattacharya, M.; Toth, M.; Smith, C.A.; Vakulenko, S.B. A Surface Loop Modulates Activity of the Bacillus Class D β-Lactamases. J. Struct. Biol. 2020, 211, 107544. [Google Scholar] [CrossRef] [PubMed]
- Santillana, E.; Beceiro, A.; Bou, G.; Romero, A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. USA 2007, 104, 5354–5359. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.D.; Ortega, C.J.; Renck, N.A.; Bonomo, R.A.; Powers, R.A.; Leonard, D.A. Structures of the class D carbapenase OXA-24 from Acinetobacter baumanii in complex with doripenem. J. Mol. Biol. 2011, 406, 583–594. [Google Scholar] [CrossRef]
- Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Toth, M.; Kumarasiri, M.; Chang, M.; Mobashery, S.; Vakulenko, S.B. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem. Biol. 2013, 20, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Stewart, N.K.; Smith, C.A.; Antunes, N.T.; Toth, M.; Vakulenko, S.B. Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D β-Lactamases. Antimicrob. Agents Chemother. 2019, 63, e02191-18. [Google Scholar] [CrossRef]
- Stewart, N.K.; Toth, M.; Alqurafi, M.A.; Chai, W.; Nguyen, T.Q.; Quan, P.; Lee, M.; Buynak, J.D.; Smith, C.A.; Vakulenko, S.B. C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation. mBio 2022, 13, e00367-22. [Google Scholar] [CrossRef]
- Schneider, K.D.; Karpen, M.E.; Bonomo, R.A.; Leonard, D.A.; Powers, R.A. The 1.4 Å crystal structure of the class D β-lactamase OXA-1 complexed with doripenem. Biochemistry 2009, 48, 11840–11847. [Google Scholar] [CrossRef]
- Toth, M.; Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Maltz, L.; Vakulenko, S.B. The role of conserved surface hydrophobic residues in the carbapenemase activity of the class D β-lactamases. Acta Crystallogr. 2017, D73, 692–701. [Google Scholar] [CrossRef]
- Pincus, N.B.; Rosas-Lemus, M.; Gatesy, S.W.M.; Bertucci, H.K.; Brunzelle, J.S.; Minasov, G.; Shuvalova, L.A.; Lebrun-Corbin, M.; Satchell, K.J.F.; Ozer, E.A.; et al. Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family β-Lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2022, 66, e0098522. [Google Scholar] [CrossRef]
- King, D.T.; King, A.M.; Lal, S.M.; Wright, G.D.; Strynadka, N.C. Molecular Mechanism of Avibactam-Mediated β-Lactamase Inhibition. ACS Infect. Dis. 2015, 1, 175–184. [Google Scholar] [CrossRef]
- Leiros, H.-K.S.; Thomassen, A.M.; Samuelsen, O.; Flach, C.-F.; Kotsakis, S.D.; Larsson, D.G.J. Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10. FEBS Open Bio 2020, 10, 1821–1832. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.D.; Mangani, S.; Jahic, H.; Benvenuti, M.; Durand-Reville, T.F.; De Luca, F.; Ehmann, D.E.; Rossolini, G.M.; Alm, R.A.; Docquier, J.D. Molecular Basis of Selective Inhibition and Slow Reversibility of Avibactam against Class D Carbapenemases: A Structure-Guided Study of OXA-24 and OXA-48. ACS Chem. Biol. 2015, 10, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Stewart, N.K.; Smith, C.A.; Toth, M.; Stasyuk, A.; Vakulenko, S.B. The Crystal Structures of CDD-1, the Intrinsic Class D β-Lactamase from the Pathogenic Gram-Positive Bacterium Clostridioides difficile, and its Complex with Cefotaxime. J. Struct. Biol. 2019, 208, 1073913. [Google Scholar] [CrossRef]
- Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Frase, H.; Toth, M.; Kantardjieff, K.A.; Vakulenko, S. Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D β-Lactamases. ACS Chem. Biol. 2015, 10, 1791–1796. [Google Scholar] [CrossRef]
- Docquier, J.D.; Benvenuti, M.; Calderone, V.; Giuliani, F.; Kapetis, D.; De Luca, F.; Rossolini, G.M.; Mangani, S. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: Relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob. Agents Chemother. 2010, 54, 2167–2174. [Google Scholar] [CrossRef]
- June, C.M.; Muckenthaler, T.J.; Schroder, E.C.; Klamer, Z.L.; Wawrzak, Z.; Powers, R.A.; Szarecka, A.; Leonard, D.A. The structure of a doripenem-bound OXA-51 class D β-lactamase variant with enhanced carbapenemase activity. Protein Sci. 2016, 25, 2152–2163. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, epidemiology, and genetics of class D b-lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef]
- Bragginton, E.C.; Colenso, C.K.; Calvopiña, K.; Hinchliffe, P.; Shaw, J.M.; Tooke, C.L.; Seng, R.; Chantratita, N.; Mulholland, A.J.; Schofield, C.J.; et al. Structure and dynamics of Burkholderia pseudomallei OXA-57, a distinctive low efficiency class D β-lactamase with carbapenem-hydrolyzing activity. bioRxiv 2024. [Google Scholar] [CrossRef]
- Antunes, N.T.; Lamoureaux, T.L.; Toth, M.; Stewart, N.K.; Frase, H.; Vakulenko, S.B. Class D β-lactamases: Are they all carbapenemases? Antimicrob. Agents Chemother. 2014, 58, 2119–2125. [Google Scholar] [CrossRef] [PubMed]
- Antunes, V.U.; Llontop, E.E.; da Costa Vasconcelos, F.N.; Lopez de los Santos, Y.; Oliveira, R.J.; Lincopan, N.; Farah, C.S.; Doucet, N.; Mittermaier, A.; Favaro, D.C. Importance of the β5-β6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D β-Lactamase Subfamily OXA-143. Biochemistry 2019, 58, 3604–3616. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Retailleau, P.; Marchini, L.; Berthault, C.; Dortet, L.; Bonnin, R.A.; Iorga, B.I.; Naas, T. Role of Arginine 214 in the Substrate Specificity of OXA-48. Antimicrob. Agents Chemother. 2020, 64, e02329-19. [Google Scholar] [CrossRef]
- De Luca, F.; Benvenuti, M.; Carboni, F.; Pozzi, C.; Rossolini, G.M.; Mangani, S.; Docquier, J. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design. Proc. Natl. Acad. Sci. USA. 2011, 108, 18424–18429. [Google Scholar] [CrossRef]
- Dabos, L.; Zavala, A.; Bonnin, R.A.; Beckstein, O.; Retailleau, P.; Iorga, B.I.; Naas, T. Substrate Specificity of OXA-48 after β5-β6 Loop Replacement. ACS Infect. Dis. 2020, 6, 1032–1043. [Google Scholar] [CrossRef]
- Poirel, L.; Castanheira, M.; Carrër, A.; Rodriguez, C.P.; Jones, R.N.; Smayevsky, J.; Nordmann, P. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 2011, 55, 2546–2551. [Google Scholar] [CrossRef]
- Dortet, L.; Oueslati, S.; Jeannot, K.; Tandé, D.; Naas, T.; Nordmann, P. Genetic and Biochemical Characterization of OXA-405, an OXA-48-Type Extended-Spectrum β-Lactamase without Significant Carbapenemase Activity. Antimicrob. Agents Chemother. 2015, 59, 3823–3828. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.M.; Clasman, J.R.; June, C.M.; Kaitany, K.C.; LaFleur, J.R.; Taracila, M.A.; Klinger, N.V.; Bonomo, R.A.; Wymore, T.; Szarecka, A.; et al. Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D β-Lactamases from Acinetobacter baumannii. Biochemistry 2015, 54, 1976–1987. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Retailleau, P.; Marchini, L.; Dortet, L.; Bonnin, R.A.; Iorga, B.I.; Naas, T. Biochemical and Structural Characterization of OXA-405, an OXA-48 Variant with Extended-Spectrum β-Lactamase Activity. Microorganisms 2020, 8, 24. [Google Scholar] [CrossRef]
- Dabos, L.; Raczynska, J.E.; Bogaerts, P.; Zavala, A.; Girlich, D.; Bonnin, R.A.; Dortet, L.; Peyrat, A.; Retailleau, P.; Iorga, B.I.; et al. Structural and Biochemical Features of OXA-517: A Carbapenem and Expanded-Spectrum Cephalosporin Hydrolyzing OXA-48 Variant. Antimicrob. Agents Chemother. 2023, 67, e0109522. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, P.; Naas, T.; Saegeman, V.; Bonnin, R.A.; Schuermans, A.; Evrard, S.; Bouchahrouf, W.; Jove, T.; Tande, D.; de Bolle, X.; et al. OXA-427, a new plasmid-borne carbapenem-hydrolysing class D β-lactamase in Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2469–2477. [Google Scholar] [CrossRef]
- Kaitany, K.C.; Klinger, N.V.; June, C.M.; Ramey, M.E.; Bonomo, R.A.; Powers, R.A.; Leonard, D.A. Structures of the class D carbapenemases OXA-23 and OXA-146: Mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins and aztreonam. Antimicrob. Agents Chemother. 2013, 58, 4848–4855. [Google Scholar] [CrossRef]
- Hirvonen, V.H.A.; Spencer, J.; van der Kamp, M.W. Antimicrobial Resistance Conferred by OXA-48 β-Lactamases: Towards a Detailed Mechanistic Understanding. Antimicrob. Agents Chemother. 2021, 65, e00184-21. [Google Scholar] [CrossRef]
- Schroder, E.C.; Klamer, Z.L.; Saral, A.; Sugg, K.A.; June, C.M.; Wymore, T.; Szarecka, A.; Leonard, D.A. Clinical Variants of the Native Class D β-Lactamase of Acinetobacter baumannii Pose an Emerging Threat through Increased Hydrolytic Activity against Carbapenems. Antimicrob. Agents Chemother. 2016, 60, 6155–6164. [Google Scholar] [CrossRef] [PubMed]
- Saino, H.; Sugiyabu, T.; Ueno, G.; Yamamoto, M.; Ishii, Y.; Miyano, M. Crystal Structure of OXA-58 with the Substrate-Binding Cleft in a Closed State: Insights into the Mobility and Stability of the OXA-58 Structure. PLoS ONE 2015, 10, e0145869. [Google Scholar] [CrossRef] [PubMed]

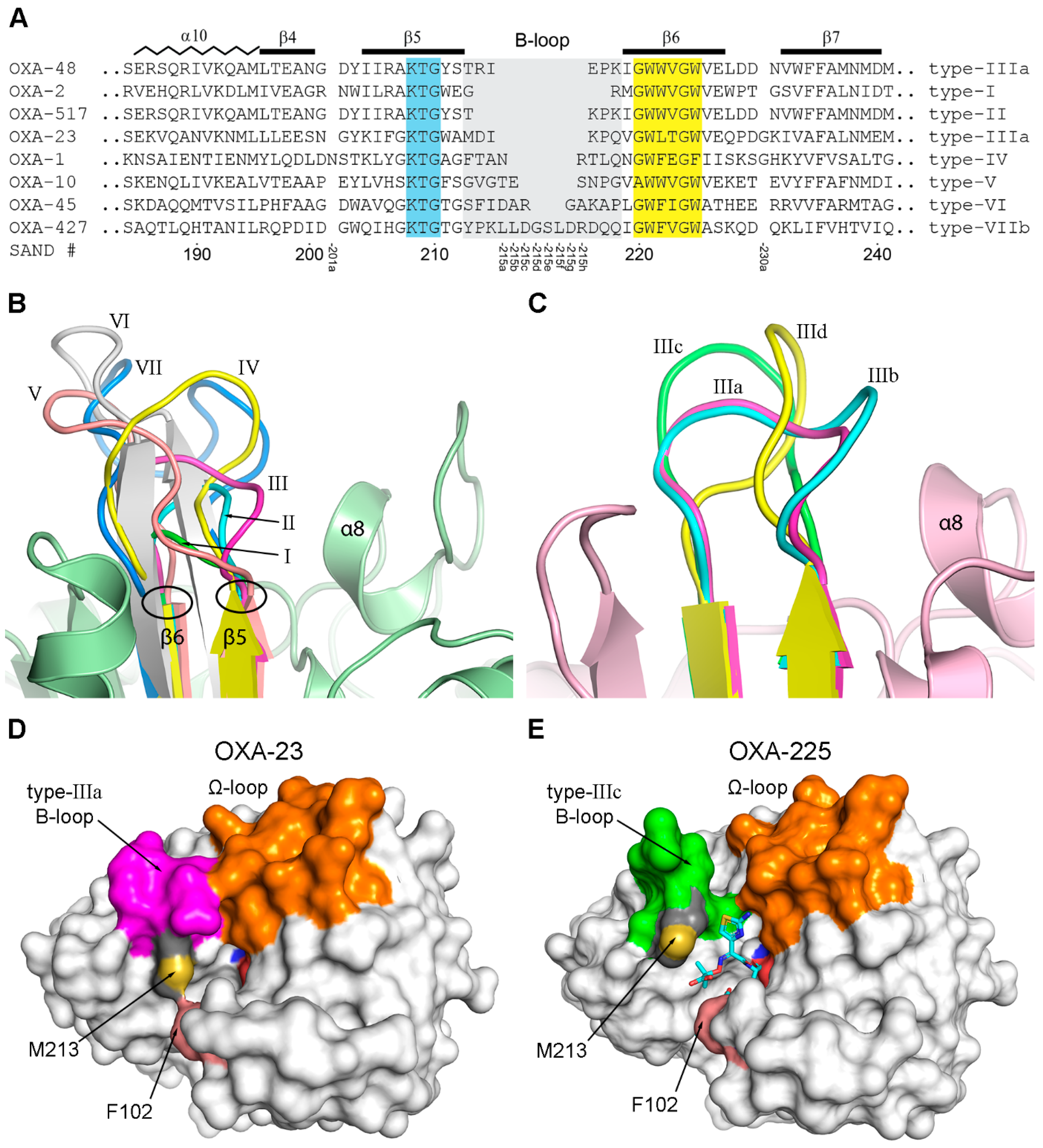
| Motif | Consensus Sequence a | Sequence Identity at Variable Positions b | Residue Numbering c |
|---|---|---|---|
| 1 | PASTFK | 1:P 99.3% (T, S, A and Q < 0.7%) 2:A 95.3%, E 1.2%, D 1%, C 0.8%, Q 0.8% (M, V, G, T and Y < 1%) 4:T 99.5%, (S and I < 0.5%) 5:F 93%, Y 6.9% | 68–73 |
| 2 | FxxW | 1:F 64.1%, L 9.5%, I 7.8%, M 6.6%, G 5.4%, Y 2.7%, (N, V, R, S, C, P and T < 3.9%) 4:W 96.2%, H 3.3% (T and C < 0.5%) | 102–105 |
| 3 | SAV | 1:S 99.9% (P and Y < 0.1%) 2:A 61.8%, V 16%, N 9.5%, Q 5.1%, T 4%, C 3.3%, I 0.3% 3:V 56.2%, I 40.1%, L 2.1%, F 0.9% (Y, M and A < 0.7%) | 118–120 |
| 4 | YGN | 1:Y 77.7%, F 22.2% 2:G 99.6% (S, D, K and R < 0.4%) 3:N 99% (E, K, A, S, D, G, V and Y < 1%) | 144–146 |
| 5 | FWL | 1:F 85.8%, A 8.1%, V 4.7%, S 1.2% 2:W 99% (G, C, L, R and D < 1%) 3:L 87.5%, I 8.9%, V 2.8% (H, Q, E and K < 0.7%) | 156–158 |
| 6 | K(S/T)G | 1:K 99.7% (R, E and Q < 0.3%) 2:S 52.3%, T 47.6% | 208–210 |
| 7 | GWx(T/V)GW | 1:G 87.5%, A 12.2% (S, C and T < 0.3%) 2:W 99.6% (Q, G and R < 0.4%) 4:T 55.2%, V 42.3%, E 1%, I 0.9%, A 0.5% (F and S < 0.1%) 6:W 87.1%, F 10.3%, Y 1.9% (M, H, I, L, S and V < 0.7% | 220–225 |
| B-Loop Type | Variants | Residues | Enzyme(s) | Activity |
|---|---|---|---|---|
| I | - | 2 | OXA-2 a | NSBL, ESBL |
| II | - | 4 | OXA-517 | CHDL, ESBL |
| III | a | 6 | OXA-23 b | CHDL |
| III | b | 7 c | OXA-146 | CHDL |
| III | c | 6 | OXA-160 d | CHDL |
| III | d | 6 | OXA-239 | CHDL |
| III | e | 7 | OXA-85 | NSBL |
| IV | - | 8 | OXA-1 | NSBL |
| V | - | 9 | OXA-10 e | ESBL |
| VI | - | 11 | OXA-45 | ESBL |
| VII | a | 14 | OXA-D84 f | Unknown |
| VII | b | 14 | OXA-427 | CHDL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, C.A.; Stasyuk, A. Diversity in the Common Fold: Structural Insights into Class D β-Lactamases from Gram-Negative Pathogens. Pathogens 2025, 14, 761. https://doi.org/10.3390/pathogens14080761
Smith CA, Stasyuk A. Diversity in the Common Fold: Structural Insights into Class D β-Lactamases from Gram-Negative Pathogens. Pathogens. 2025; 14(8):761. https://doi.org/10.3390/pathogens14080761
Chicago/Turabian StyleSmith, Clyde A., and Anastasiya Stasyuk. 2025. "Diversity in the Common Fold: Structural Insights into Class D β-Lactamases from Gram-Negative Pathogens" Pathogens 14, no. 8: 761. https://doi.org/10.3390/pathogens14080761
APA StyleSmith, C. A., & Stasyuk, A. (2025). Diversity in the Common Fold: Structural Insights into Class D β-Lactamases from Gram-Negative Pathogens. Pathogens, 14(8), 761. https://doi.org/10.3390/pathogens14080761






