The Epidemiology of Coccidioidomycosis (Valley fever) and the Disease Ecology of Coccidioides spp. in New Mexico (2006–2023)
Abstract
1. Introduction
2. Materials and Methods
2.1. New Mexico Department of Health (NMDOH)’s Epidemiological Data
2.2. Molecular and Phylogenetic Analyses of Clinical Coccidioides
2.3. Molecular Identification of Coccidioides in Soils
3. Results
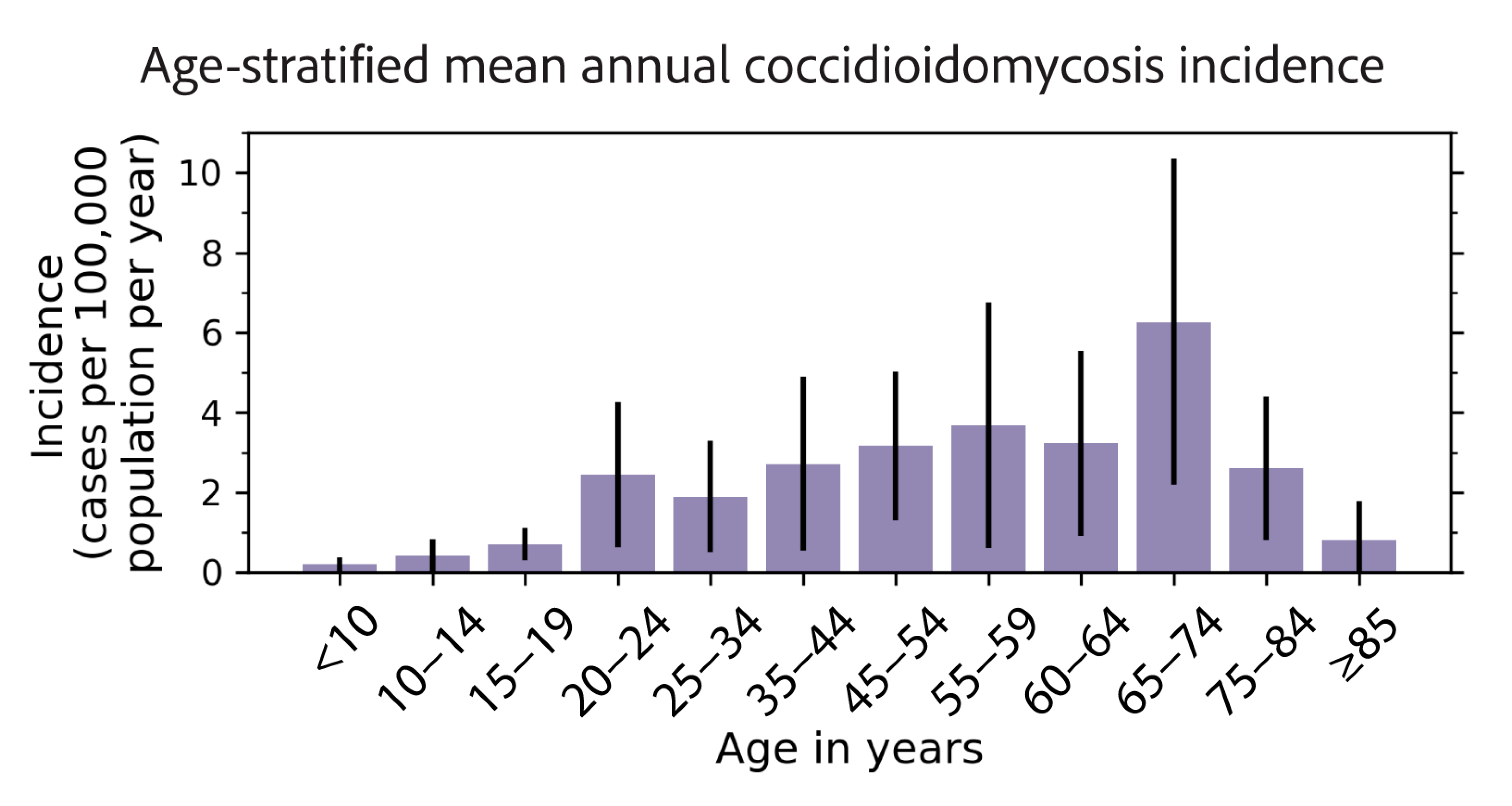
3.1. Epidemiology of Coccidioidomycosis in New Mexico
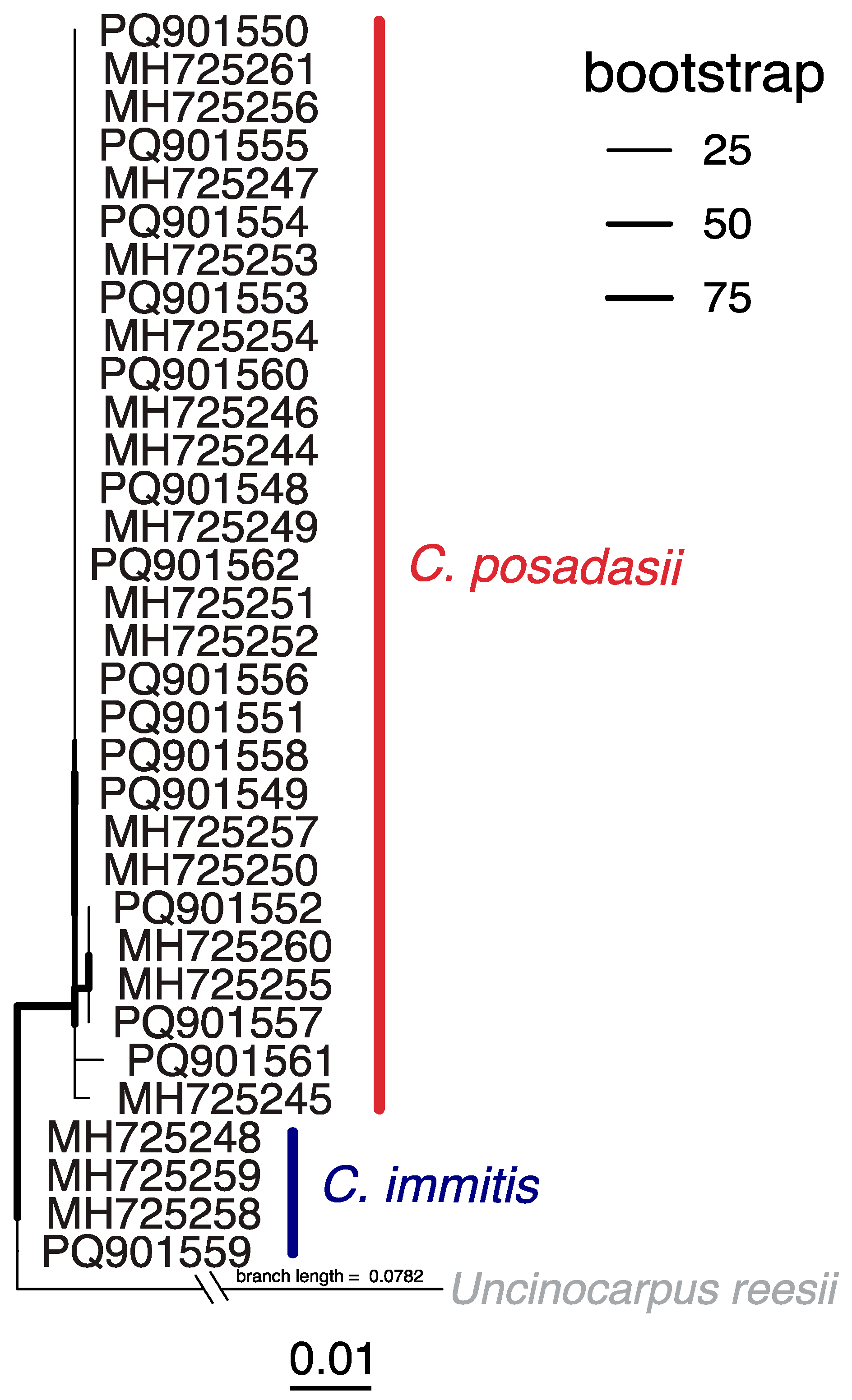
3.2. Clinical Coccidioidomycosis Sequence Analyses
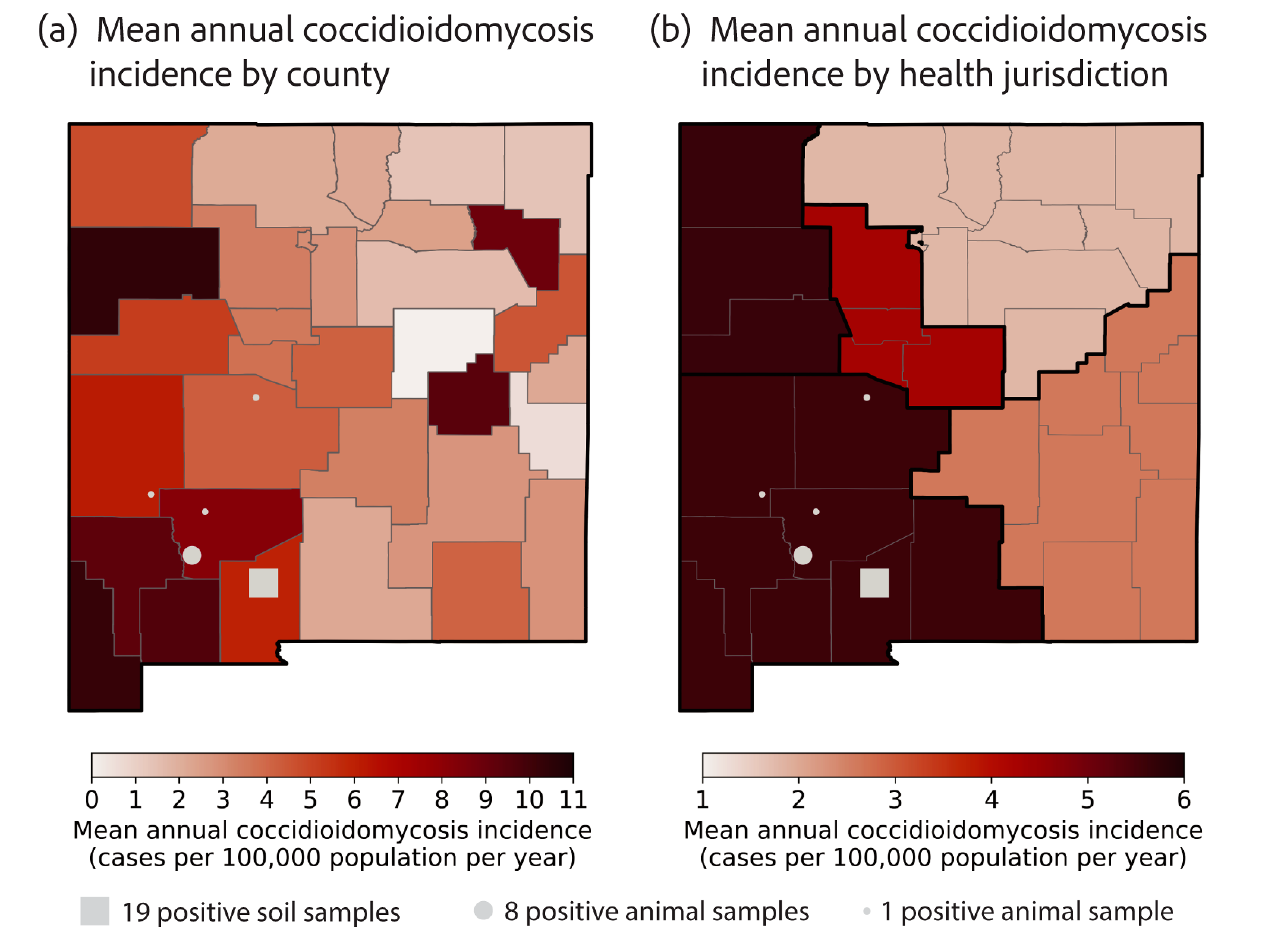
3.3. Coccidioides in New Mexico Soils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirschmann, J.V. The Early History of Coccidioidomycosis: 1892-1945. Clin. Infect. Dis. 2007, 44, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.L. An Epidemic of Coccidioidal Infection (Coccidioidomycosis). JAMA 1942, 118, 1182. [Google Scholar] [CrossRef]
- Edwards, P.Q.; Palmer, C.E. Prevalence of Sensitivity to Coccidioidin, with Special Reference to Specific and Nonspecific Reactions to Coccidioidin and to Histoplasmin. Dis. Chest 1957, 31, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Maddy, K. Ecological Factors Possibly Relating to the Geographic Distribution of Coccidioides immitis. In Proceedings of the Proceedings of the Symposium on Coccidioidomycosis, Phoenix, AZ, USA, 11–13 February 1957. [Google Scholar]
- Hamm, P.S.; Hutchison, M.I.; Leonard, P.; Melman, S.; Natvig, D.O. First Analysis of Human Coccidioides Isolates from New Mexico and the Southwest Four Corners Region: Implications for the Distributions of C. posadasii and C. immitis and Human Groups at Risk. J. Fungi 2019, 5, 74. [Google Scholar] [CrossRef]
- Salazar-Hamm, P.S.; Montoya, K.N.; Montoya, L.; Cook, K.; Liphardt, S.; Taylor, J.W.; Cook, J.A.; Natvig, D.O. Breathing Can Be Dangerous: Opportunistic Fungal Pathogens and the Diverse Community of the Small Mammal Lung Mycobiome. Front. Fungal Biol. 2022, 3, 996574. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Coccidioidomycosis/Valley Fever (Coccidioides spp.) 2023 Case Definition; 22-ID-07. Available online: https://ndc.services.cdc.gov/case-definitions/coccidioidomycosis-2023/ (accessed on 12 December 2024).
- Centers for Disease Control and Prevention. Coccidioidomycosis/Valley Fever (Coccidioides spp.) 2011 Case Definition; 10-ID-04. Available online: https://ndc.services.cdc.gov/case-definitions/coccidioidomycosis-2011/ (accessed on 12 December 2024).
- U.S. Census Bureau Annual Estimates of the Resident Population for Counties in New Mexico: April 1, 2010 to July 1. 2019. Available online: http://www.census.gov/data/datasets/time-series/demo/popest/2010s-counties-total.html (accessed on 18 April 2025).
- U.S. Census Bureau Intercensal Estimates of the Resident Population for Counties and States: April 1, 2000 to July 1. 2010. Available online: https://www.census.gov/data/datasets/time-series/demo/popest/2010s-counties-total.html (accessed on 18 April 2025).
- U.S. Census Bureau County Population Totals and Components of Change: 2020–2024. Available online: https://www.census.gov/data/tables/time-series/demo/popest/2020s-counties-total.html (accessed on 18 April 2025).
- U.S. Census Bureau American Community Survey, ACS 5-Year Estimates Data Profiles, Table DP05. Available online: https://data.census.gov/table?q=DP05 (accessed on 12 December 2024).
- Walker, K. Tidycensus: Load US Census Boundary and Attribute Data as “Tidyverse” and ’Sf’-Ready Data Frames; 2023. Available online: https://walkerke.r-universe.dev/tidycensus (accessed on 18 April 2025).
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.J.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols, a Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Sherry, S.T.; Yankie, L.; Karsch-Mizrachi, I. GenBank 2024 Update. Nucleic Acids Res. 2024, 52, D134–D137. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Bowers, J.R.; Parise, K.L.; Kelley, E.J.; Lemmer, D.; Schupp, J.M.; Driebe, E.M.; Engelthaler, D.M.; Keim, P.; Barker, B.M. Direct Detection of Coccidioides from Arizona Soils Using CocciENV, a Highly Sensitive and Specific Real-Time PCR Assay. Med. Mycol. 2019, 57, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Platt, A.R.; Woodhall, R.W.; George, A.L. Improved DNA Sequencing Quality and Efficiency Using an Optimized Fast Cycle Sequencing Protocol. BioTechniques 2007, 43, 58–62. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention Valley Fever (Coccidioidomycosis). Available online: https://www.cdc.gov/valley-fever/php/statistics/index.html (accessed on 16 January 2025).
- Benedict, K.; McCotter, O.Z.; Brady, S.; Komatsu, K.; Sondermeyer, G.L.; Cooksey; Nguyen, A.; Jain, S.; Vugia, D.J.; Jackson, B.R. Surveillance for Coccidioidomycosis—United States, 2011–2017. MMWR Surveill. Summ. 2019, 68, 1–15. [Google Scholar] [CrossRef]
- Mayfield, H.; Davila, V.; Penedo, E. Coccidioidomycosis-Related Hospital Visits, Texas, USA, 2016–2021. Emerg. Infect. Dis. 2024, 30, 231624. [Google Scholar] [CrossRef]
- Carey, A.; Gorris, M.E.; Chiller, T.; Jackson, B.; Beadles, W.; Webb, B.J. Epidemiology, Clinical Features, and Outcomes of Coccidioidomycosis, Utah, 2006–2015. Emerg. Infect. Dis. 2021, 27, 2269–2277. [Google Scholar] [CrossRef]
- Williams, S.L.; Smith, D.J.; Benedict, K.; Ahlers, J.R.; Austin, C.; Birn, R.; Carter, A.M.; Christophe, N.N.; Cibulskas, K.; Cieslak, P.R.; et al. Surveillance for Coccidioidomycosis, Histoplasmosis, and Blastomycosis during the COVID-19 Pandemic—United States, 2019–2021. MMWR Morb. Mortal. Wkly. Rep. 2024, 73, 239–244. [Google Scholar] [CrossRef]
- Van Deursen, B.; Hagenaars, M.; Meima, A.; Van Asten, L.; Richardus, J.H.; Fanoy, E.; Voeten, H. A Sharp Decrease in Reported Non-COVID-19 Notifiable Infectious Diseases during the First Wave of the COVID-19 Epidemic in the Rotterdam Region, the Netherlands: A Descriptive Study. BMC Infect. Dis. 2022, 22, 208. [Google Scholar] [CrossRef]
- Facciolà, A.; Laganà, A.; Genovese, G.; Romeo, B.; Sidoti, S.; D’Andrea, G.; Raco, C.; Visalli, G.; Di Pietro, A. Impact of the COVID-19 Pandemic on the Infectious Disease Epidemiology. J. Prev. Med. Hyg. 2023, 64, E274–E282. [Google Scholar] [CrossRef]
- Arizona Department of Health Services. Arizona Health Status and Vital Statistics; Arizona Department of Health Services: Phoenix, AZ, USA, 2021; pp. 203–206.
- Hepler, S.A.; Kaufeld, K.A.; Kline, D.; Greene, A.; Gorris, M.E. Estimating Coccidioidomycosis Endemicity While Accounting for Imperfect Detection Using Spatio—Temporal Occupancy Modeling. Am. J. Epidemiol. 2024, 194, kwae199. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.A.; Anderson, S.M.; Imholte, S.B.; Erhart, L.M.; Chen, S.; Park, B.J.; Christ, C.; Komatsu, K.K.; Chiller, T.; Sunenshine, R.H. Enhanced Surveillance of Coccidioidomycosis, Arizona, USA, 2007–2008. Emerg. Infect. Dis. 2010, 16, 1738–1744. [Google Scholar] [CrossRef]
- Wilken, J.A.; Sondermeyer, G.; Shusterman, D.; McNary, J.; Vugia, D.J.; McDowell, A.; Borenstein, P.; Gilliss, D.; Ancock, B.; Prudhomme, J.; et al. Coccidioidomycosis among Workers Constructing Solar Power Farms, California, USA, 2011–2014. Emerg. Infect. Dis. 2015, 21, 1997–2005. [Google Scholar] [CrossRef]
- Perez-Lockett, K. Coccidioidomycosis in New Mexico: An Epidemiological Summary. In Proceedings of the New Mexico Department of Health Training Presentation; 2013. [Google Scholar]
- Chen, S.; Erhart, L.M.; Anderson, S.; Komatsu, K.; Park, B.; Chiller, T.; Sunenshine, R. Coccidioidomycosis: Knowledge, Attitudes, and Practices among Healthcare Providers—Arizona, 2007. Med. Mycol. 2011, 49, 649–656. [Google Scholar] [CrossRef] [PubMed]
- James, A.E.; McCall, J.R.; Petersen, K.R.; Wohrle, R.D.; Oltean, H.N. A Survey of Veterinarians’ Knowledge, Attitudes and Practices Regarding an Emerging Disease: Coccidioidomycosis in Washington State. Zoonoses Public Health 2020, 67, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.E.; Ardon-Dryer, K.; Campuzano, A.; Castañón-Olivares, L.R.; Gill, T.E.; Greene, A.; Hung, C.-Y.; Kaufeld, K.A.; Lacy, M.; Sánchez-Paredes, E. Advocating for Coccidioidomycosis to Be a Reportable Disease Nationwide in the United States and Encouraging Disease Surveillance across North and South America. J. Fungi 2023, 9, 83. [Google Scholar] [CrossRef]
- Sievers, M. Coccidioidomycosis among Southwestern American Indians. Am. Rev. Respir. Dis. 1964, 90, 920–926. [Google Scholar]
- Mead, H.L.; Kollath, D.R.; Teixeira, M.D.M.; Roe, C.C.; Plude, C.; Nandurkar, N.; Donohoo, C.; O’Connor, B.L.W.; Terriquez, J.; Keim, P.; et al. Coccidioidomycosis in Northern Arizona: An Investigation of the Host, Pathogen, and Environment Using a Disease Triangle Approach. mSphere 2022, 7, e00352-22. [Google Scholar] [CrossRef]
- McCotter, O.; Kennedy, J.; McCollum, J.; Bartholomew, M.; Iralu, J.; Jackson, B.R.; Haberling, D.; Benedict, K. Coccidioidomycosis among American Indians and Alaska Natives, 2001–2014. Open Forum Infect. Dis. 2019, 6, ofz052. [Google Scholar] [CrossRef]
- Lucero-Obusan, C.; Deka, R.; Schirmer, P.; Oda, G.; Holodniy, M. Epidemiology of Coccidioidomycosis in the Veterans Health Administration, 2013-2022. J. Fungi Basel Switz. 2023, 9, 731. [Google Scholar] [CrossRef]
- Smith, C.E.; Beard, R.R. Varieties of Coccidioidal Infection in Relation to the Epidemiology and Control of the Diseases. Am. J. Public Health Nations Health 1946, 36, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Seitz, A.E.; Prevots, D.R.; Holland, S.M. Hospitalizations Associated with Disseminated Coccidioidomycosis, Arizona and California, USA. Emerg. Infect. Dis. 2012, 18, 1476–1479. [Google Scholar] [CrossRef] [PubMed]
- Cummings, K.C.; McDOWELL, A.; Wheeler, C.; McNARY, J.; Das, R.; Vugia, D.J.; Mohle-Boetani, J.C. Point-Source Outbreak of Coccidioidomycosis in Construction Workers. Epidemiol. Infect. 2010, 138, 507–511. [Google Scholar] [CrossRef] [PubMed]
- McCracken, B.E. Final Report of Coccidioidomycosis Research Project at Camp Roberts, California, 1 September 1952–15 October 1953; Surgeon’s Office 6th Army: San Francisco, CA, USA, 1953. [Google Scholar]
- Pappagianis, D.; Einstein, H. Tempest from Tehachapi Takes Toll or Coccidioides Conveyed Aloft and Afar. West J. Med. 1978, 129, 527–530. [Google Scholar]
- Pappagianis, D.; Lindsay, S.; Beall, S.; Williams, P. Ethnic Background and the Clinical Course of Coccidioidomycosis (Letter). Am. Rev. Respir. Dis. 1979, 120, 959–961. [Google Scholar]
- Hsu, A.P. The Known and Unknown “Knowns” of Human Susceptibility to Coccidioidomycosis. J. Fungi 2024, 10, 256. [Google Scholar] [CrossRef]
- El-Sayed, A. Complex Systems for a Complex Issue: Race in Health Research. Virtual Mentor 2014, 16, 450–454. [Google Scholar] [CrossRef]
- Holman, R.C.; Folkema, A.M.; Singleton, R.J.; Redd, J.T.; Christensen, K.Y.; Steiner, C.A.; Schonberger, L.B.; Hennessy, T.W.; Cheek, J.E. Disparities in Infectious Disease Hospitalizations for American Indian/Alaska Native People. Public Health Rep.® 2011, 126, 508–521. [Google Scholar] [CrossRef]
- Ehrenpreis, J.E.; Ehrenpreis, E.D. A Historical Perspective of Healthcare Disparity and Infectious Disease in the Native American Population. Am. J. Med. Sci. 2022, 363, 288–294. [Google Scholar] [CrossRef]
- Sondermeyer Cooksey, G.L.; Nguyen, A.; Vugia, D.; Jain, S. Regional Analysis of Coccidioidomycosis Incidence—California, 2000–2018. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1817–1821. [Google Scholar] [CrossRef]
- Ondo, A.L.; Zlotoff, B.J.; Mings, S.M.; Rochester, L.C.; Shanler, S.D. Primary Cutaneous Coccidioidomycosis: An Incidental Finding. Clin. Exp. Dermatol. 2010, 35, e42–e43. [Google Scholar] [CrossRef] [PubMed]
- Gumprecht, B. Las Cruces Sun-News Beware the Spore: Fungus in New Mexico Soils Causes Serious Illness. 2019. Available online: https://www.lcsun-news.com/story/life/wellness/2019/03/11/soil-fungus-serious-illness-new-mexico/2711787002/ (accessed on 18 April 2025).
- Lang, R.; Stokes, W.; Lemaire, J.; Johnson, A.; Conly, J. A Case Report of Coccidioides Posadasii Meningoencephalitis in an Immunocompetent Host. BMC Infect. Dis. 2019, 19, 722. [Google Scholar] [CrossRef]
- Gorris, M.E.; Cat, L.A.; Zender, C.S.; Treseder, K.K.; Randerson, J.T. Coccidioidomycosis Dynamics in Relation to Climate in the Southwestern United States. GeoHealth 2018, 2, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Head, J.R.; Sondermeyer-Cooksey, G.; Heaney, A.K.; Yu, A.T.; Jones, I.; Bhattachan, A.; Campo, S.K.; Wagner, R.; Mgbara, W.; Phillips, S.; et al. Effects of Precipitation, Heat, and Drought on Incidence and Expansion of Coccidioidomycosis in Western USA: A Longitudinal Surveillance Study. Lancet Planet. Health 2022, 6, e793–e803. [Google Scholar] [CrossRef] [PubMed]
- Kollath, D.R.; Mihaljevic, J.R.; Barker, B.M. PM10 and Other Climatic Variables Are Important Predictors of Seasonal Variability of Coccidioidomycosis in Arizona. Microbiol. Spectr. 2022, 10, e01483-21. [Google Scholar] [CrossRef]
- Heaney, A.K.; Camponuri, S.K.; Head, J.R.; Collender, P.; Weaver, A.; Sondermeyer Cooksey, G.; Yu, A.; Vugia, D.; Jain, S.; Bhattachan, A.; et al. Coccidioidomycosis Seasonality in California: A Longitudinal Surveillance Study of the Climate Determinants and Spatiotemporal Variability of Seasonal Dynamics, 2000–2021. Lancet Reg. Health-Am. 2024, 38, 100864. [Google Scholar] [CrossRef]
- Camponuri, S.K.; Head, J.R.; Collender, P.A.; Weaver, A.K.; Heaney, A.K.; Colvin, K.A.; Bhattachan, A.; Sondermeyer-Cooksey, G.; Vugia, D.J.; Jain, S.; et al. Prolonged Dry Seasons Lengthen Coccidioidomycosis Transmission Seasons: Implications for a Changing California. medRxiv 2024. [Google Scholar] [CrossRef]
- Sprigg, W.A.; Nickovic, S.; Galgiani, J.N.; Pejanovic, G.; Petkovic, S.; Vujadinovic, M.; Vukovic, A.; Dacic, M.; DiBiase, S.; Prasad, A.; et al. Regional Dust Storm Modeling for Health Services: The Case of Valley Fever. Aeolian Res. 2014, 14, 53–73. [Google Scholar] [CrossRef]
- Gorris, M.E.; Treseder, K.K.; Zender, C.S.; Randerson, J.T. Expansion of Coccidioidomycosis Endemic Regions in the United States in Response to Climate Change. GeoHealth 2019, 3, 308–327. [Google Scholar] [CrossRef]
- PRISM Climate Group, Oregon State University PRISM Climate Data 2014. Available online: https://prism.oregonstate.edu/ (accessed on 12 March 2025).
- Salazar-Hamm, P.; Torres-Cruz, T.J. The Impact of Climate Change on Human Fungal Pathogen Distribution and Disease Incidence. Curr. Clin. Microbiol. Rep. 2024, 11, 140–152. [Google Scholar] [CrossRef]
- Colella, J.P.; Cobos, M.E.; Salinas, I.; Cook, J.A. The PICANTE Consortium Advancing the Central Role of Non-Model Biorepositories in Predictive Modeling of Emerging Pathogens. PLOS Pathog. 2023, 19, e1011410. [Google Scholar] [CrossRef] [PubMed]
- Dunnum, J.L.; Yanagihara, R.; Johnson, K.M.; Armien, B.; Batsaikhan, N.; Morgan, L.; Cook, J.A. Biospecimen Repositories and Integrated Databases as Critical Infrastructure for Pathogen Discovery and Pathobiology Research. PLoS Negl. Trop. Dis. 2017, 11, e0005133. [Google Scholar] [CrossRef] [PubMed]
- Fisher, F.S.; Bultman, M.W.; Johnson, S.M.; Pappagianis, D.; Zaborsky, E. Coccidioides Niches and Habitat Parameters in the Southwestern United States: A Matter of Scale. Ann. N. Y. Acad. Sci. 2007, 1111, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Mead, H.L.; Hamm, P.S.; Shaffer, I.N.; Teixeira, M.D.M.; Wendel, C.S.; Wiederhold, N.P.; Thompson, G.R.; Muñiz-Salazar, R.; Castañón-Olivares, L.R.; Keim, P.; et al. Differential Thermotolerance Adaptation between Species of Coccidioides. J. Fungi 2020, 6, 366. [Google Scholar] [CrossRef]
- Fisher, M.C.; Koenig, G.L.; White, T.J.; Taylor, J.W. Molecular and Phenotypic Description of Coccidioides posadasii Sp. Nov., Previously Recognized as the Non-California Population of Coccidioides Immitis. Mycologia 2002, 94, 73–84. [Google Scholar] [CrossRef]
- Elconin, A.F.; Egeberg, R.O.; Lubarsky, R. Growth Pattern of Coccidioides Immitis in the Soil of an Endemic Area. In Proceedings of the Symposium on Coccidioidomycosis; Public Health Service: Washington, DC, USA, 1957; pp. 168–170. [Google Scholar]
- Greene, D.R.; Koenig, G.; Fisher, M.C.; Taylor, J.W. Soil Isolation and Molecular Identification of Coccidioides Immitis. Mycologia 2000, 92, 406–410. [Google Scholar] [CrossRef]
- Emmons, C.W. Isolation of Coccidioides from Soil and Rodents. Public Health Rep. 1942, 57, 109–111. [Google Scholar] [CrossRef]
- Kollath, D.R.; Teixeira, M.M.; Funke, A.; Miller, K.J.; Barker, B.M. Investigating the Role of Animal Burrows on the Ecology and Distribution of Coccidioides Spp. in Arizona Soils. Mycopathologia 2019, 185, 145–159. [Google Scholar] [CrossRef]
- Head, J.R.; Camponuri, S.K.; Weaver, A.K.; Montoya, L.; Lee, E.; Radosevich, M.; Jones, I.; Wagner, R.; Bhattachan, A.; Campbell, G.; et al. Small Mammals and Their Burrows Shape the Distribution of Coccidioides in Soils: A Long-Term Ecological Experiment. BioRxiv 2024. [Google Scholar] [CrossRef]
- Wagner, R.; Montoya, L.; Head, J.R.; Campo, S.; Remais, J.; Taylor, J.W. Coccidioides Undetected in Soils from Agricultural Land and Uncorrelated with Time or the Greater Soil Fungal Community on Undeveloped Land. PLoS Pathog. 2023, 19, e1011391. [Google Scholar] [CrossRef]
- Egeberg, R.O.; Ely, A.F. Coccidioides Immitis in the Soil of the Southern San Joaquin Valley. Am. J. Med. Sci. 1956, 231, 151–154. [Google Scholar] [CrossRef]
- Taylor, J.W.; Barker, B.M. The Endozoan, Small-Mammal Reservoir Hypothesis and the Life Cycle of Coccidioides Species. Med. Mycol. 2019, 57, S16–S20. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J.; Stajich, J.E.; Rounsley, S.D.; Gardner, M.J.; Wortman, J.R.; Jordar, V.S.; Maiti, R.; Kodira, C.D.; Neafsey, D.E.; Zeng, Q.; et al. Comparative Genomic Analyses of the Human Fungal Pathogens Coccidioides and Their Relatives. Genome Res. 2009, 19, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.L.; Bestelmeyer, B.T.; Campanella, A. Shrub Encroachment, Productivity Pulses, and Core-transient Dynamics of Chihuahuan Desert Rodents. Ecosphere 2018, 9, e02330. [Google Scholar] [CrossRef]
- Anderson, J. Jornada Basin LTER: Wireless Meteorological Station at NPP C-CALI Site: Daily Summary Data: 2013—Ongoing; 2024. Available online: https://portal.edirepository.org/nis/mapbrowse?packageid=knb-lter-jrn.210437046.47 (accessed on 12 March 2025).
- Bailey, V. Mammals of New Mexico; U.S. Government Printing Office: Washington, DC, USA, 1931.
- Bartlett, R.D.; Bartlett, P. New Mexico’s Reptiles and Amphibians: A Field Guide; University of New Mexico Press: Albuquerque, NM, USA, 2013; ISBN 978-0-8263-5207-1. [Google Scholar]
- Thompson, B.; Patricia, A.; Carold, F. Analyses of Burrowing Owl Populations in New Mexico. J. Raptor. Res. 2001, 35, 362–370. [Google Scholar]
- Barker, B.M.; Tabor, J.A.; Shubitz, L.F.; Perrill, R.; Orbach, M.J. Detection and Phylogenetic Analysis of Coccidioides Posadasii in Arizona Soil Samples. Fungal Ecol. 2012, 5, 163–176. [Google Scholar] [CrossRef]
- Catalán-Dibene, J.; Johnson, S.M.; Eaton, R.; Romero-Olivares, A.L.; Baptista-Rosas, R.C.; Pappagianis, D.; Riquelme, M. Detection of Coccidioidal Antibodies in Serum of a Small Rodent Community in Baja California, Mexico. Fungal Biol. 2014, 118, 330–339. [Google Scholar] [CrossRef]
- Baptista-Rosas, R.C.; Catalán-Dibene, J.; Romero-Olivares, A.L.; Hinojosa, A.; Cavazos, T.; Riquelme, M. Molecular Detection of Coccidioides Spp. from Environmental Samples in Baja California: Linking Valley Fever to Soil and Climate Conditions. Fungal Ecol. 2012, 5, 177–190. [Google Scholar] [CrossRef]
- Engelthaler, D.M.; Roe, C.C.; Hepp, C.M.; Teixeira, M.; Driebe, E.M.; Schupp, J.M.; Gade, L.; Waddell, V.; Komatsu, K.; Arathoon, E.; et al. Local Population Structure and Patterns of Western Hemisphere Dispersal for Coccidioides spp., the Fungal Cause of Valley Fever. mBio 2016, 7, e00550-16. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Barker, B.M. Use of Population Genetics to Assess the Ecology, Evolution, and Population Structure of Coccidioides. Emerg. Infect. Dis. 2016, 22, 1022–1030. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Lang, B.F.; Matute, D.R.; Stajich, J.E.; Barker, B.M. Mitochondrial Genomes of the Human Pathogens Coccidioides Immitis and Coccidioides Posadasii. G3 2021, 11, jkab132. [Google Scholar] [CrossRef] [PubMed]

| Variable | Number (%) | |
|---|---|---|
| Case classification | 1541 (100) | |
| Confirmed | 946 (61.4) | |
| Probable | 56 (3.6) | |
| Suspect | 539 (35.0) | |
| Age | 843 (54.7) | |
| Age, y, median (IQR) | 55 (40–66) | |
| Pediatric < 18 y | 32 (3.8) | |
| Geriatric ≥ 65 y | 241 (28.6) | |
| Sex | 1533 (99.5) | |
| Female | 653 (42.6) | |
| Male | 880 (57.4) | |
| Ethnicity | 581 (37.7) | |
| Hispanic or Latino | 237 (40.8) | |
| Non-Hispanic or -Latino | 344 (59.2) | |
| Race | 854 (55.4) | |
| American Indian or Alaska Native | 197 (23.1) | |
| Asian | 9 (1.1) | |
| Black or African American | 19 (2.2) | |
| White | 496 (58.1) | |
| Other race | 27 (3.2) | |
| Multi-race | 106 (12.4) | |
| Outcomes | 631 (40.9) | |
| Hospital admission | 438 (69.4) | |
| Hospital length of stay, d, median (IQR) | 6 (3–11) | |
| 312 (20.2) | ||
| Mortality | 20 (6.4) |
| Demographic | IRR (95% CI) | |
|---|---|---|
| Sex | ||
| Female | Referent | |
| Male | 1.6 (1.4–1.7) | |
| Ethnicity | ||
| Non-Hispanic or -Latino | Referent | |
| Hispanic or Latino | 1.3 (0.8–1.8) | |
| Race | ||
| White | Referent | |
| American Indian or Alaska Native | 1.9 (1.4–2.4) | |
| Asian | 0.1 (0.0–0.3) | |
| Black or African American | 1.0 (0.3–1.6) | |
| Other race | 0.6 (0.0–1.2) |
| Jurisdiction | Total Number | Mean Annual Cases Per Year | Mean Population | Mean Annual Incidence (Cases Per 100,000 Population Per Year) | |
|---|---|---|---|---|---|
| Metro | 696 | 39 | 897,656 | 4.3 | |
| Northeast | 95 | 5 | 294,730 | 1.8 | |
| Northwest | 228 | 13 | 224,919 | 5.6 | |
| Northwest Tribal | 27 | 2 | |||
| Southeast | 136 | 8 | 288,132 | 2.6 | |
| Southwest | 372 | 21 | 369,042 | 5.6 | |
| 1527 | |||||
| Tribal | 10 | 1 | |||
| Out of state | 2 | <1 | |||
| Total | 1539 |
| Site | Microsite | Positivity |
|---|---|---|
| Grassland | 12 (12%) | |
| Topsoil | 6 (12%) | |
| Burrow | 6 (12%) | |
| Ecotone | 4 (4%) | |
| Topsoil | 2 (4%) | |
| Burrow | 2 (4%) | |
| Shrubland | 3 (3%) | |
| Topsoil | 0 (0%) | |
| Burrow | 3 (3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salazar-Hamm, P.S.; Shrum Davis, S.; Catalán-Dibene, J.; Romero-Olivares, A.L.; Edge, K.; Bartlow, A.W.; Natvig, D.O.; Gorris, M.E. The Epidemiology of Coccidioidomycosis (Valley fever) and the Disease Ecology of Coccidioides spp. in New Mexico (2006–2023). Pathogens 2025, 14, 607. https://doi.org/10.3390/pathogens14060607
Salazar-Hamm PS, Shrum Davis S, Catalán-Dibene J, Romero-Olivares AL, Edge K, Bartlow AW, Natvig DO, Gorris ME. The Epidemiology of Coccidioidomycosis (Valley fever) and the Disease Ecology of Coccidioides spp. in New Mexico (2006–2023). Pathogens. 2025; 14(6):607. https://doi.org/10.3390/pathogens14060607
Chicago/Turabian StyleSalazar-Hamm, Paris S., Sarah Shrum Davis, Jovani Catalán-Dibene, Adriana L. Romero-Olivares, Karen Edge, Andrew W. Bartlow, Donald O. Natvig, and Morgan E. Gorris. 2025. "The Epidemiology of Coccidioidomycosis (Valley fever) and the Disease Ecology of Coccidioides spp. in New Mexico (2006–2023)" Pathogens 14, no. 6: 607. https://doi.org/10.3390/pathogens14060607
APA StyleSalazar-Hamm, P. S., Shrum Davis, S., Catalán-Dibene, J., Romero-Olivares, A. L., Edge, K., Bartlow, A. W., Natvig, D. O., & Gorris, M. E. (2025). The Epidemiology of Coccidioidomycosis (Valley fever) and the Disease Ecology of Coccidioides spp. in New Mexico (2006–2023). Pathogens, 14(6), 607. https://doi.org/10.3390/pathogens14060607








