From Microbial Ecology to Clinical Challenges: The Respiratory Microbiome’s Role in Antibiotic Resistance
Abstract
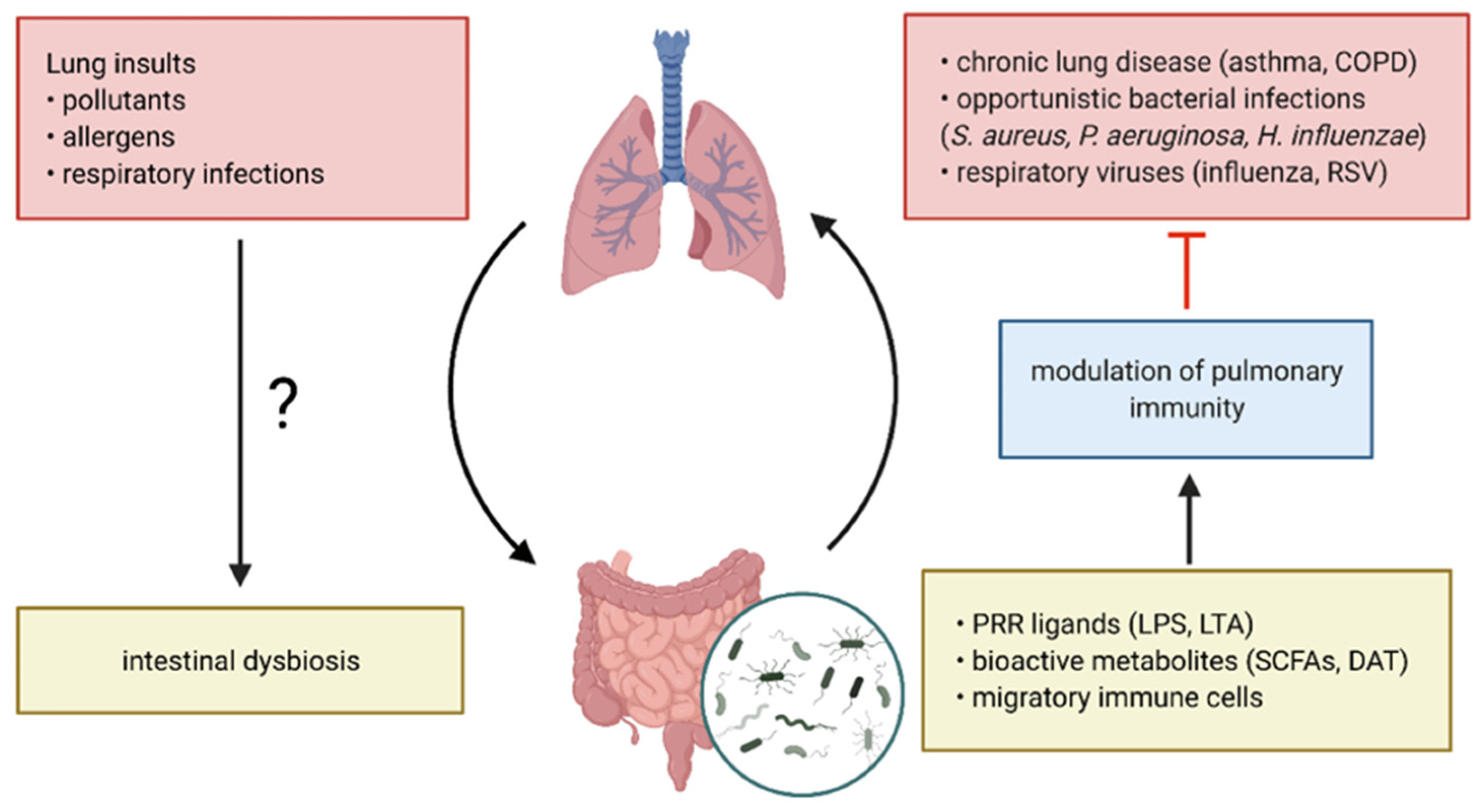
1. Introduction
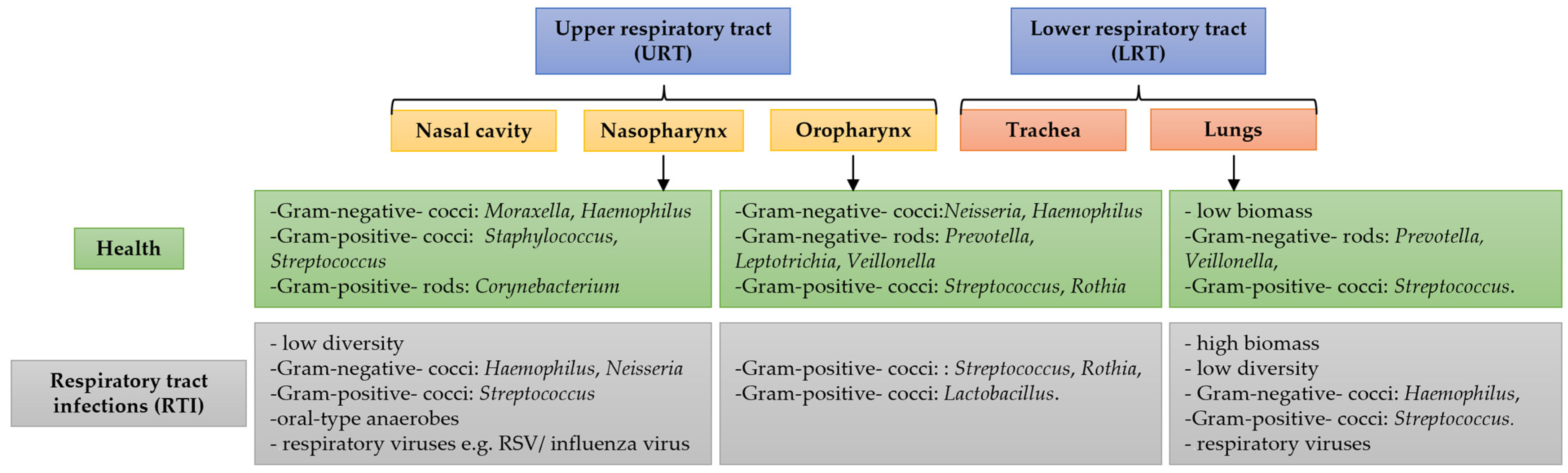
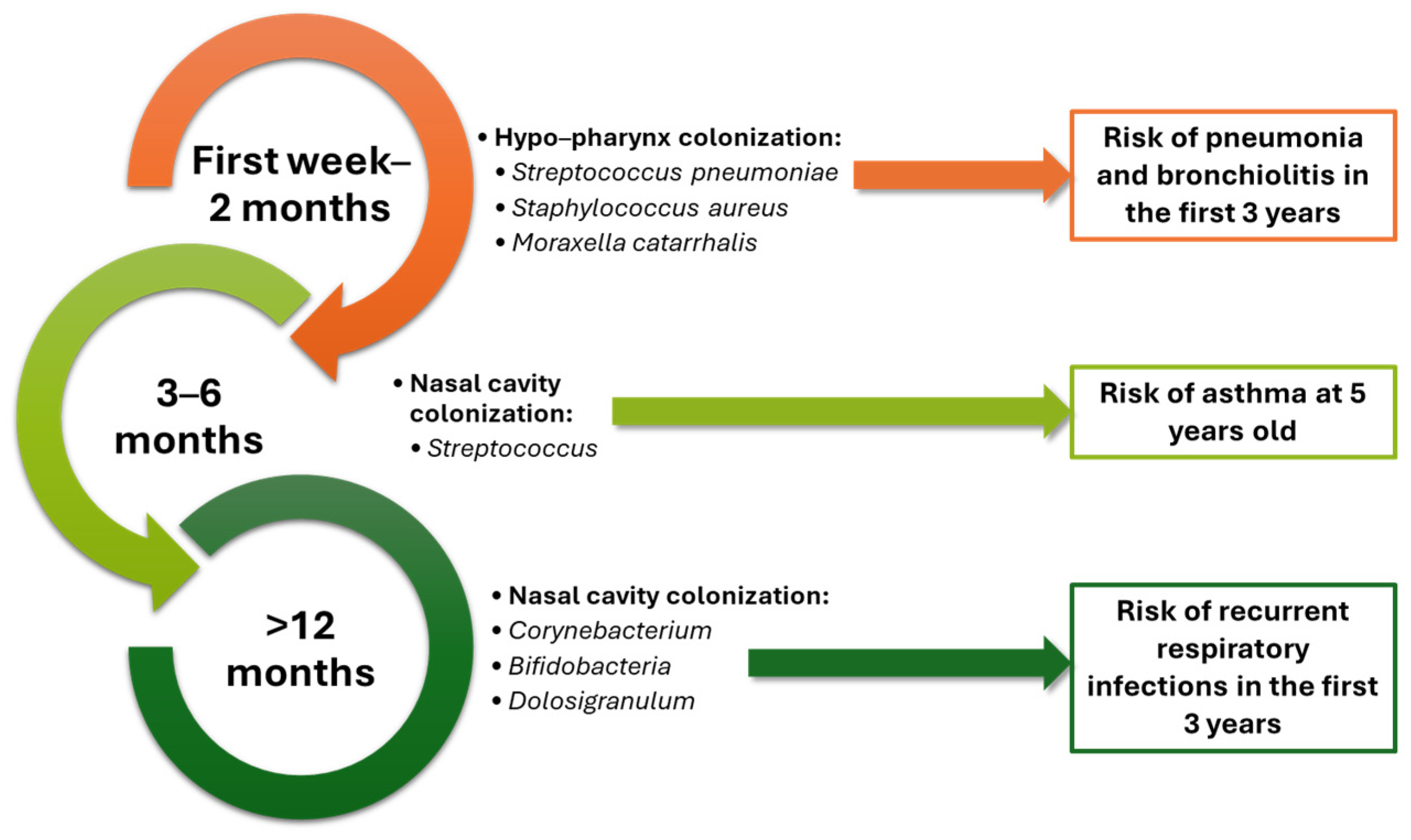
2. Respiratory Microbiome in Health and Disease
2.1. Impact of Antibiotics on the Respiratory Microbiome
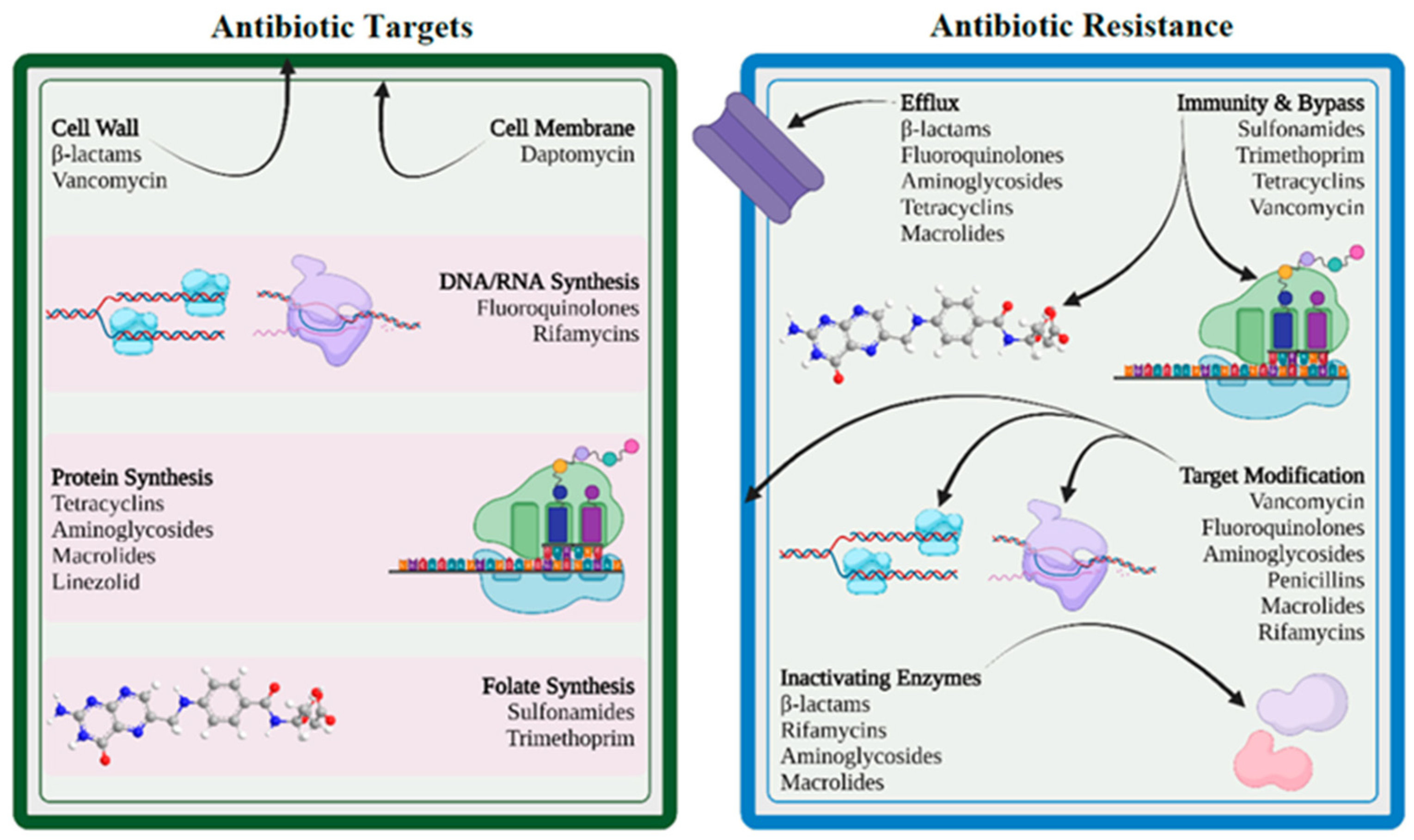
2.2. Antibiotic Resistance in Respiratory Pathogens
3. Therapeutic Strategies to Preserve Respiratory Microbiome Health and Combat Antibiotic Resistance
3.1. Antibiotic Stewardship Programs
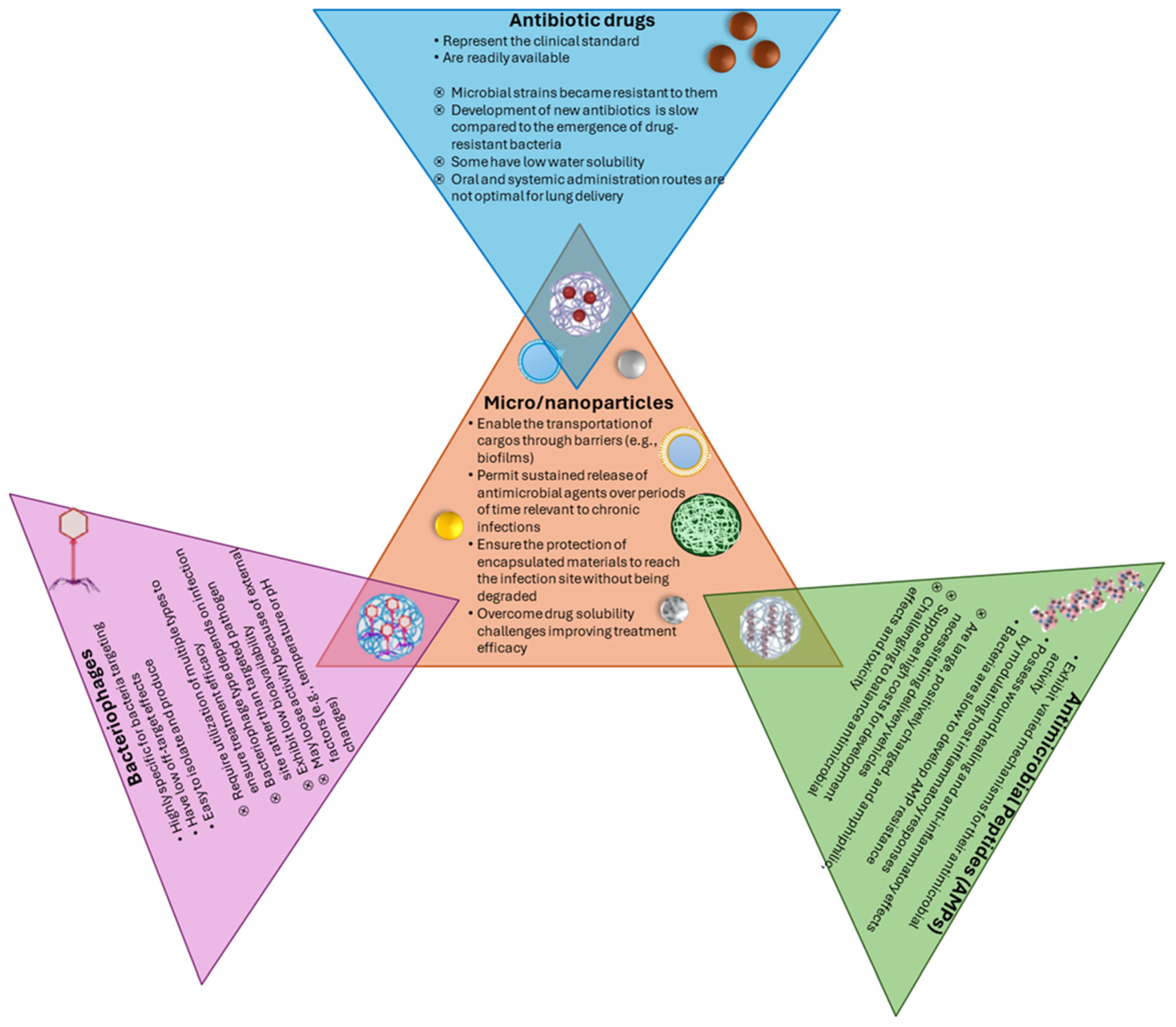
3.2. Alternatives to Conventional Antibiotics
3.3. Probiotics and Microbiome Restoration
4. Future Research Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, R.M.; Tsimring, L.; Hasty, J. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. eLife 2017, 6, e25950. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; He, L.-Y.; Gao, F.-Z.; Yao, K.-S.; Zhang, M.; Qiao, L.-K.; Chen, Z.-Y.; He, L.-X.; Liu, Y.-S.; Zhao, J.-L.; et al. Airborne antibiotic resistome and microbiome in pharmaceutical factories. Environ. Int. 2024, 186, 108639. [Google Scholar] [CrossRef] [PubMed]
- Guitor, A.K.; Wright, G.D. Antimicrobial Resistance and Respiratory Infections. Chest 2018, 154, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Forst, C.V.; Gordon, A.; Gussin, G.; Geber, A.B.; Fernandez, P.J.; Ding, T.; Lashua, L.; Wang, M.; Balmaseda, A.; et al. Characterization of antibiotic resistance and host-microbiome interactions in the human upper respiratory tract during influenza infection. Microbiome 2020, 8, 39. [Google Scholar] [CrossRef]
- Mindt, B.C.; DiGiandomenico, A. Microbiome Modulation as a Novel Strategy to Treat and Prevent Respiratory Infections. Antibiotics 2022, 11, 474. [Google Scholar] [CrossRef]
- Mac Aogain, M.; Lau, K.J.X.; Cai, Z.; Kumar Narayana, J.; Purbojati, R.W.; Drautz-Moses, D.I.; Gaultier, N.E.; Jaggi, T.K.; Tiew, P.Y.; Ong, T.H. Metagenomics reveals a core macrolide resistome related to microbiota in chronic respiratory disease. Am. J. Respir. Crit. Care Med. 2020, 202, 433–447. [Google Scholar] [CrossRef]
- Abreu, N.A.; Nagalingam, N.A.; Song, Y.; Roediger, F.C.; Pletcher, S.D.; Goldberg, A.N.; Lynch, S.V. Sinus Microbiome Diversity Depletion and Corynebacterium tuberculostearicum Enrichment Mediates Rhinosinusitis. Sci. Transl. Med. 2012, 4, ra124–ra151. [Google Scholar] [CrossRef]
- Hadian, S.A.; Rezayatmand, R. Economic impact of acute respiratory disease pandemics: A scoping review. J. Res. Med. Sci. 2022, 27, 88. [Google Scholar] [CrossRef]
- Houde, L.; Law, A.W.; Averin, A.; Weycker, D.; Cane, A.; Pugh, S.; Shea, K.M. Annual Clinical and Economic Burden of Medically Attended Lower Respiratory Tract Illnesses Due to Respiratory Syncytial Virus among US Infants Aged <12 Months. J. Infect Dis 2024, jiae544. [Google Scholar] [CrossRef]
- Carrico, J.; Hicks, K.A.; Wilson, E.; Panozzo, C.A.; Ghaswalla, P. The Annual Economic Burden of Respiratory Syncytial Virus in Adults in the United States. J. Infect. Dis. 2024, 230, e342–e352. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Baquero, F.; de Pablo, R.; Soriano, M.C.; Coque, T.M. Altered Ecology of the Respiratory Tract Microbiome and Nosocomial Pneumonia. Front. Microbiol. 2022, 12, 709421. [Google Scholar]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Faner, R.; Sibila, O.; Agustí, A.; Bernasconi, E.; Chalmers, J.D.; Huffnagle, G.B.; Manichanh, C.; Molyneaux, P.L.; Paredes, R.; Brocal, V.P. The microbiome in respiratory medicine: Current challenges and future perspectives. Eur. Respir. J. 2017, 49, 1602086. [Google Scholar] [CrossRef] [PubMed]
- Inglis, L.K.; Edwards, R.A. How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research. Microorganisms 2022, 10, 1671. [Google Scholar] [CrossRef]
- Schwartz, D.J.; Langdon, A.E.; Dantas, G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 2020, 12, 82. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, X. Gut Microecological Prescription: A Novel Approach to Regulating Intestinal Micro-Ecological Balance. Int. J. Gen. Med. 2025, 18, 603–626. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Kim, D.W.; Cha, C.J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef]
- Yoo, S.; Jung, S.C.; Kwak, K.; Kim, J.S. The Role of Prebiotics in Modulating Gut Microbiota: Implications for Human Health. Int. J. Mol. Sci. 2024, 25, 4834. [Google Scholar] [CrossRef]
- Petrariu, O.A.; Barbu, I.C.; Niculescu, A.G.; Constantin, M.; Grigore, G.A.; Cristian, R.E.; Mihaescu, G.; Vrancianu, C.O. Role of probiotics in managing various human diseases, from oral pathology to cancer and gastrointestinal diseases. Front. Microbiol. 2024, 14, 1296447. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Nelson, C.E.; Brodie, E.L.; Desantis, T.Z.; Baek, M.S.; Liu, J.; Woyke, T.; Allgaier, M.; Bristow, J.; Wiener-Kronish, J.P.; et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011, 127, e1–e3. [Google Scholar] [CrossRef]
- Yadav, M. Chapter 14—Microbiome therapeutics in respiratory illnesses. In Microbiome Therapeutics; Chauhan, N.S., Kumar, S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 401–419. [Google Scholar]
- Baughman, R.P.; Thorpe, J.E.; Staneck, J.; Rashkin, M.; Frame, P.T. Use of the protected specimen brush in patients with endotracheal or tracheostomy tubes. Chest 1987, 91, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, J.E.; Baughman, R.P.; Frame, P.T.; Wesseler, T.A.; Staneck, J.L. Bronchoalveolar lavage for diagnosing acute bacterial pneumonia. J. Infect. Dis. 1987, 155, 855–861. [Google Scholar] [CrossRef]
- Mitchell, A.B.; Glanville, A.R. The Human Respiratory Microbiome: Implications and Impact. Semin Respir Crit Care Med. 2018, 39, 199–212. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Gellatly, S.L.; Budden, K.F.; Mac Aogain, M.; Shukla, S.D.; Wood, D.L.A.; Hugenholtz, P.; Pethe, K.; Hansbro, P.M. Microbiomes in respiratory health and disease: An Asia-Pacific perspective. Respirology 2017, 22, 240–250. [Google Scholar]
- Drigot, Z.G.; Clark, S.E. Insights into the role of the respiratory tract microbiome in defense against bacterial pneumonia. Curr. Opin. Microbiol. 2024, 77, 102428. [Google Scholar] [CrossRef]
- Santacroce, L.; Charitos, I.A.; Ballini, A.; Inchingolo, F.; Luperto, P.; De Nitto, E.; Topi, S. The Human Respiratory System and its Microbiome at a Glimpse. Biology 2020, 9, 318. [Google Scholar] [CrossRef]
- Hélène, P.; Jean-Louis, H.; Lourdes, V.-S.; Claudie, L.; Clémence, B.; Pierre-Régis, B.; Geneviève, H.-A. Antibiotic resistance in chronic respiratory diseases: From susceptibility testing to the resistome. Eur. Respir. Rev. 2022, 31, 210259. [Google Scholar] [CrossRef]
- Le Noci, V.; Guglielmetti, S.; Arioli, S.; Camisaschi, C.; Bianchi, F.; Sommariva, M.; Storti, C.; Triulzi, T.; Castelli, C.; Balsari, A. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: A strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018, 24, 3528–3538. [Google Scholar]
- O’Connor, J.B.; Wagner, B.D.; Harris, J.K.; Frank, D.N.; Clabots, D.E.; Laguna, T.A. Detection and identification of fungi in the lower airway of children with and without cystic fibrosis. Front. Microbiol. 2023, 14, 1119703. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; He, Y.; Deng, L.; Chen, D.; Zhang, Y.; Luo, H.; Lei, W. Rapid detection of pathogenic fungi from coastal population with respiratory infections using microfluidic chip technology. BMC Infect. Dis. 2024, 24, 326. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Huang, X.; Gu, S.; Cai, H.; Wang, M.; Wang, H.; Wang, S.; Jiang, C.; Huang, L. Landscape of fungal detection in the lungs of patients with severe pneumonia in the ICU, a multicenter study based on clinical metagenomics. J. Infect. 2024, 89, 106195. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, K.; Pausan, M.R.; Perras, A.K.; Beck, M.; Bang, C.; Mora, M.; Schilhabel, A.; Schmitz, R.; Moissl-Eichinger, C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824-17. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Ginevra, C.; Rusniok, C.; Jarraud, S.; Buchrieser, C. The respiratory tract microbiome, the pathogen load, and clinical interventions define severity of bacterial pneumonia. Cell Rep. Med. 2023, 4, 101167. [Google Scholar] [CrossRef]
- Yasuda, I.; Suzuki, M.; Maeda, H.; Terada, M.; Sando, E.; Ng, C.F.S.; Otomaru, H.; Yoshida, L.M.; Morimoto, K. Respiratory virus detection in the upper respiratory tract of asymptomatic, community-dwelling older people. BMC Infect. Dis. 2022, 22, 411. [Google Scholar] [CrossRef]
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.H.; Adcock, I.M.; Chotirmall, S.H.; Chung, K.F.; et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019, 7, 907–920. [Google Scholar] [CrossRef]
- Guarnieri, G.; Olivieri, B.; Senna, G.; Vianello, A. Relative Humidity and Its Impact on the Immune System and Infections. Int. J. Mol. Sci. 2023, 24, 9456. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhou, Y.; Chang, Y.; Liang, X.; Zhang, H.; Lin, X.; Qing, K.; Zhou, X.; Luo, Z. The Effects of Ventilation, Humidity, and Temperature on Bacterial Growth and Bacterial Genera Distribution. Int. J. Environ. Res. Public. Health. 2022, 19, 15345. [Google Scholar] [CrossRef]
- Rasheed, A.; Parmar, K.; Jain, S.; Chakravortty, D.; Basu, S. Weather-related changes in the dehydration of respiratory droplets on surfaces bolster bacterial endurance. J. Colloid. Interface Sci. 2024, 674, 653–662. [Google Scholar] [CrossRef]
- Zhang, W.; Ruan, Y.; Ling, J.; Wang, L. A study of the correlation between meteorological factors and hospitalization for acute lower respiratory infections in children. BMC Public. Health. 2024, 24, 3135. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Jesudasen, S.; Bringhurst, L.; Sui, H.Y.; McIver, L.; Whiteson, K.; Hanselmann, K.; O’Toole, G.A.; Richards, C.J.; Sicilian, L.; et al. Supplemental Oxygen Alters the Airway Microbiome in Cystic Fibrosis. mSystems. 2022, 7, e0036422. [Google Scholar] [CrossRef]
- Sun, M.; Lu, F.; Yu, D.; Wang, Y.; Chen, P.; Liu, S. Respiratory diseases and gut microbiota: Relevance, pathogenesis, and treatment. Front. Microbiol. 2024, 15, 1358597. [Google Scholar] [CrossRef] [PubMed]
- Quinn-Bohmann, N.; Freixas-Coutin, J.A.; Seo, J.; Simmons, R.; Diener, C.; Gibbons, S.M. Meta-analysis of the human upper respiratory tract microbiome reveals robust taxonomic associations with health and disease. BMC Biol. 2024, 22, 93. [Google Scholar] [CrossRef] [PubMed]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef]
- Paulo, A.C.; Lança, J.; Almeida, S.T.; Hilty, M.; Sá-Leão, R. The upper respiratory tract microbiota of healthy adults is affected by Streptococcus pneumoniae carriage, smoking habits, and contact with children. Microbiome 2023, 11, 199. [Google Scholar] [CrossRef]
- Theodosiou, A.A.; Dorey, R.B.; Laver, J.R.; Cleary, D.W.; Read, R.C.; Jones, C.E. Manipulating the infant respiratory microbiomes to improve clinical outcomes: A review of the literature. J. Infect. 2021, 82, 247–252. [Google Scholar] [CrossRef]
- Natalini, J.G.; Singh, S.; Segal, L.N. The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 2023, 21, 222–235. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef]
- Chu, V.T.; Tsitsiklis, A.; Mick, E.; Ambroggio, L.; Kalantar, K.L.; Glascock, A.; Osborne, C.M.; Wagner, B.D.; Matthay, M.A.; DeRisi, J.L.; et al. The antibiotic resistance reservoir of the lung microbiome expands with age in a population of critically ill patients. Nat. Commun. 2024, 15, 92. [Google Scholar] [CrossRef]
- Li, R.; Li, J.; Zhou, X. Lung microbiome: New insights into the pathogenesis of respiratory diseases. Signal Transduct. Target. Ther. 2024, 9, 19. [Google Scholar] [CrossRef]
- Wu, L.; Shen, C.; Chen, Y.; Yang, X.; Luo, X.; Hang, C.; Yan, L.; Xu, X. Follow-up study of airway microbiota in children with persistent wheezing. Respir. Res. 2021, 22, 213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ai, T.; Xie, C.; Xia, W.; Zhang, Y.; Liao, H.; Jia, L.; Fan, Y.; Xu, J. Lower airway microbiome of children with recurrent wheezing: A clinical cohort study. Transl. Pediatr. 2022, 11, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, H.; Hermansen, M.N.; Buchvald, F.; Loland, L.; Halkjaer, L.B.; Bønnelykke, K.; Brasholt, M.; Heltberg, A.; Vissing, N.H.; Thorsen, S.V.; et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007, 357, 1487–1495. [Google Scholar] [CrossRef]
- Kloepfer, K.M.; Lee, W.M.; Pappas, T.E.; Kang, T.J.; Vrtis, R.F.; Evans, M.D.; Gangnon, R.E.; Bochkov, Y.A.; Jackson, D.J.; Lemanske, R.F., Jr.; et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J. Allergy Clin. Immunol. 2014, 133, 1301–1307.e3. [Google Scholar] [CrossRef]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef]
- Coburn, B.; Wang, P.W.; Diaz Caballero, J.; Clark, S.T.; Brahma, V.; Donaldson, S.; Zhang, Y.; Surendra, A.; Gong, Y.; Elizabeth Tullis, D.; et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015, 5, 10241. [Google Scholar] [CrossRef]
- Linnane, B.; Walsh, A.M.; Walsh, C.J.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; McDermott, M.; Renwick, J.; McNally, P. The Lung Microbiome in Young Children with Cystic Fibrosis: A Prospective Cohort Study. Microorganisms 2021, 9, 492. [Google Scholar] [CrossRef]
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Nina, P.B.; Jp, D.; Kumar, S.; Singh, B.; Tiwari, R.R. Futuristic non-antibiotic therapies to combat antibiotic resistance: A review. Front. Microbiol. 2021, 12, 609459. [Google Scholar]
- Niculescu, A.-G.; Mük, G.R.; Avram, S.; Vlad, I.M.; Limban, C.; Nuta, D.; Grumezescu, A.M.; Chifiriuc, M.-C. Novel strategies based on natural products and synthetic derivatives to overcome resistance in Mycobacterium tuberculosis. Eur. J. Med. Chem. 2024, 269, 116268. [Google Scholar] [CrossRef]
- Canaparo, R.; Foglietta, F.; Giuntini, F.; Della Pepa, C.; Dosio, F.; Serpe, L. Recent Developments in Antibacterial Therapy: Focus on Stimuli-Responsive Drug-Delivery Systems and Therapeutic Nanoparticles. Molecules 2019, 24, 1991. [Google Scholar] [CrossRef] [PubMed]
- Merker, M.; Tueffers, L.; Vallier, M.; Groth, E.E.; Sonnenkalb, L.; Unterweger, D.; Baines, J.F.; Niemann, S.; Schulenburg, H. Evolutionary approaches to combat antibiotic resistance: Opportunities and challenges for precision medicine. Front. Immunol. 2020, 11, 568485. [Google Scholar]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Mullany, P. Functional metagenomics for the investigation of antibiotic resistance. Virulence 2014, 5, 443–447. [Google Scholar] [CrossRef]
- Sherrard, L.J.; Tunney, M.M.; Elborn, J.S. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 2014, 384, 703–713. [Google Scholar] [CrossRef]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Endale, H.; Mathewos, M.; Abdeta, D. Potential causes of spread of antimicrobial resistance and preventive measures in one health perspective-a review. Infect. Drug Resist. 2023, 16, 7515–7545. [Google Scholar]
- Cookson, W.O.C.M.; Cox, M.J.; Moffatt, M.F. New opportunities for managing acute and chronic lung infections. Nat. Rev. Microbiol. 2018, 16, 111–120. [Google Scholar] [CrossRef]
- Relman, D.A.; Lipsitch, M. Microbiome as a tool and a target in the effort to address antimicrobial resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12902–12910. [Google Scholar] [CrossRef]
- Zumla, A.; Memish, Z.A.; Maeurer, M.; Bates, M.; Mwaba, P.; Al-Tawfiq, J.A.; Denning, D.W.; Hayden, F.G.; Hui, D.S. Emerging novel and antimicrobial-resistant respiratory tract infections: New drug development and therapeutic options. Lancet. Infect. Dis. 2014, 14, 1136–1149. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Despotovic, M.; de Nies, L.; Busi, S.B.; Wilmes, P. Reservoirs of antimicrobial resistance in the context of One Health. Curr. Opin. Microbiol. 2023, 73, 102291. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L.; Heyderman, R. The challenges of defining the human nasopharyngeal resistome. Trends Microbiol. 2023, 31, 816–831. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.A.; Miravitlles, M. Bronchiectasis in COPD patients: More than a comorbidity? Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Kitsios, G.D.; Sayed, K.; Fitch, A.; Yang, H.; Britton, N.; Shah, F.; Bain, W.; Evankovich, J.W.; Qin, S.; Wang, X.; et al. Prognostic Insights from Longitudinal Multicompartment Study of Host-Microbiota Interactions in Critically Ill Patients. Res. Sq. 2023, rs-3. [Google Scholar] [CrossRef]
- Rosenwasser, Y.; Berger, I.; Loewy, Z.G. Therapeutic Approaches for Chronic Obstructive Pulmonary Disease (COPD) Exacerbations. Pathogens. 2022, 11, 1513. [Google Scholar] [CrossRef]
- Good, A.; Olans, R. CE: Pediatric Antibiotic Stewardship. AJN Am. J. Nurs. 2021, 121, 38–43. [Google Scholar] [CrossRef]
- Cox, J.A.; Vlieghe, E.; Mendelson, M.; Wertheim, H.; Ndegwa, L.; Villegas, M.V.; Gould, I.; Levy Hara, G. Antibiotic stewardship in low- and middle-income countries: The same but different? Clin. Microbiol. Infect. 2017, 23, 812–818. [Google Scholar] [CrossRef]
- Feldstein, D.; Sloane, P.D.; Feltner, C. Antibiotic Stewardship Programs in Nursing Homes: A Systematic Review. J. Am. Med. Dir. Assoc. 2018, 19, 110–116. [Google Scholar] [CrossRef]
- Doernberg, S.B.; Abbo, L.M.; Burdette, S.D.; Fishman, N.O.; Goodman, E.L.; Kravitz, G.R.; Leggett, J.E.; Moehring, R.W.; Newland, J.G.; Robinson, P.A. Essential resources and strategies for antibiotic stewardship programs in the acute care setting. Clin. Infect. Dis. 2018, 67, 1168–1174. [Google Scholar] [CrossRef]
- Stenehjem, E.; Hersh, A.L.; Buckel, W.R.; Jones, P.; Sheng, X.; Evans, R.S.; Burke, J.P.; Lopansri, B.K.; Srivastava, R.; Greene, T.; et al. Impact of Implementing Antibiotic Stewardship Programs in 15 Small Hospitals: A Cluster-Randomized Intervention. Clin. Infect. Dis. 2018, 67, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dunbar, S.; Tang, Y.W. Laboratory Diagnosis of Respiratory Tract Infections in Children—The State of the Art. Front Microbiol. 2018, 9, 2478. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Buttrini, M.; Farina, B.; Montecchini, S.; De Conto, F.; Chezzi, C. Respiratory Tract Infections and Laboratory Diagnostic Methods: A Review with A Focus on Syndromic Panel-Based Assays. Microorganisms 2022, 10, 1856. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wu, L.; Xu, W.; Wei, J.; Niu, X.; Liu, G.; Yu, L.; Wu, Y.; Zhou, Q.; Liu, L. Diagnostic techniques for critical respiratory infections: Update on current methods. Heliyon 2023, 9, e18957. [Google Scholar] [CrossRef]
- Skevaki, C.L.; Papadopoulos, N.G.; Tsakris, A.; Johnston, S.L. Microbiologic Diagnosis of Respiratory Illness: Practical Applications. In Kendig & Chernick’s Disorders of the Respiratory Tract in Children; Elsevier: Amsterdam, The Netherlands, 2012; pp. 399–423. [Google Scholar] [CrossRef]
- Zilberberg, M.D.; Nathanson, B.H.; Sulham, K.; Fan, W.; Shorr, A.F. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit. Care. 2016, 20, 221. [Google Scholar] [CrossRef]
- Lin, W.H.; Chiu, H.C.; Chen, K.F.; Tsao, K.C.; Chen, Y.Y.; Li, T.H.; Huang, Y.C.; Hsieh, Y.C. Molecular detection of respiratory pathogens in community-acquired pneumonia involving adults. J. Microbiol. Immunol. Infect. 2022, 55, 829–837. [Google Scholar] [CrossRef]
- Stojanovic, Z.; Gonçalves-Carvalho, F.; Marín, A.; Abad Capa, J.; Domínguez, J.; Latorre, I.; Lacoma, A.; Prat-Aymerich, C. Advances in diagnostic tools for respiratory tract infections: From tuberculosis to COVID-19—Changing paradigms? ERJ Open Res. 2022, 8, 00113–02022. [Google Scholar] [CrossRef]
- Noviello, S.; Huang, D.B. The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections. Diagnostics 2019, 9, 37. [Google Scholar] [CrossRef]
- Dimeas, I.E.; Kotsiou, O.S.; Salgkami, P.; Poulakida, I.; Boutlas, S.; Daniil, Z.; Papadamou, G.; Gourgoulianis, K.I. Real-Life Insights into Pertussis Diagnosis: High Yield of PCR Testing and Clinical Outcomes-An Emerging Old Enemy or Just a Sign of PCR Times? J. Pers. Med. 2024, 14, 1116. [Google Scholar] [CrossRef]
- Tellapragada, C.; Giske, C.G. The Unyvero Hospital-Acquired pneumonia panel for diagnosis of secondary bacterial pneumonia in COVID-19 patients. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2479–2485. [Google Scholar] [CrossRef]
- Gastli, N.; Loubinoux, J.; Daragon, M.; Lavigne, J.P.; Saint-Sardos, P.; Pailhoriès, H.; Lemarié, C.; Benmansour, H.; d’Humières, C.; Broutin, L.; et al. Multicentric evaluation of BioFire FilmArray Pneumonia Panel for rapid bacteriological documentation of pneumonia. Clin. Microbiol. Infect. 2021, 27, 1308–1314. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.G.; Ralph, E.; Norville, I.H.; Prior, J.L.; Atkins, T.P. Comparison of PCR and Viable Count as a Method for Enumeration of Bacteria in an A/J Mouse Aerosol Model of Q Fever. Front. Microbiol. 2019, 10, 1552. [Google Scholar] [CrossRef]
- Chapman, R.; Jones, L.; D’Angelo, A.; Suliman, A.; Anwar, M.; Bagby, S. Nanopore-Based Metagenomic Sequencing in Respiratory Tract Infection: A Developing Diagnostic Platform. Lung 2023, 201, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Zhao, Q.; Liu, J.; Wang, S.; Wu, W.; Miao, L.; Zhan, P.; Chen, X.; Huang, M.; Ye, M.; et al. Utilizing metagenomic next-generation sequencing for pathogen detection and diagnosis in lower respiratory tract infections in real-world clinical practice. Infection 2024, 52, 625–636. [Google Scholar] [CrossRef]
- Pardo-Freire, M.; Domingo-Calap, P. Phages and nanotechnology: New insights against multidrug-resistant bacteria. BioDesign Res. 2023, 5, 0004. [Google Scholar] [CrossRef]
- Chee, E.; García, A.J. Biomaterial therapeutic strategies for treatment of bacterial lung infections. Biofilm 2023, 5, 100111. [Google Scholar] [CrossRef]
- Li, J.; Zheng, H.; Leung, S.S.Y. Pulmonary delivery of emerging antibacterials for bacterial lung infections treatment. Pharm. Res. 2023, 40, 1057–1072. [Google Scholar]
- Caselli, L.; Rodrigues, G.R.; Franco, O.L.; Malmsten, M. Pulmonary delivery systems for antimicrobial peptides. Crit. Rev. Biotechnol. 2024, 44, 963–980. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef]
- Loh, B.; Gondil, V.S.; Manohar, P.; Khan, F.M.; Yang, H.; Leptihn, S. Encapsulation and delivery of therapeutic phages. Appl. Environ. Microbiol. 2021, 87, e01979-20. [Google Scholar]
- Paczesny, J.; Bielec, K. Application of Bacteriophages in Nanotechnology. Nanomaterials 2020, 10, 1944. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xie, S.; Wei, J.; Song, X.; Ding, Z.; Li, X. Antibacterial Micelles with Vancomycin-Mediated Targeting and pH/Lipase-Triggered Release of Antibiotics. ACS Appl. Mater. Interfaces 2018, 10, 36814–36823. [Google Scholar] [CrossRef] [PubMed]
- Mercan, D.-A.; Niculescu, A.-G.; Grumezescu, A.M. Nanoparticles for Antimicrobial Agents Delivery—An Up-to-Date Review. Int. J. Mol. Sci. 2022, 23, 13862. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Delshadi, R.; Jafari, S.M. Active delivery of antimicrobial nanoparticles into microbial cells through surface functionalization strategies. Trends Food Sci. Technol. 2020, 99, 217–228. [Google Scholar] [CrossRef]
- Spirescu, V.A.; Chircov, C.; Grumezescu, A.M.; Andronescu, E. Polymeric Nanoparticles for Antimicrobial Therapies: An up-to-date Overview. Polymers 2021, 13, 724. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Lee, M.-Y.; Yang, C.-H.; Huang, K.-S. Active targeted drug delivery for microbes using nano-carriers. Curr. Top. Med. Chem. 2015, 15, 1525–1531. [Google Scholar] [CrossRef]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-strategies to fight multi-drug resistant bacteria-“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef]
- Masri, A.; Anwar, A.; Khan, N.A.; Siddiqui, R. The Use of Nanomedicine for Targeted Therapy against Bacterial Infections. Antibiotics 2019, 8, 260. [Google Scholar] [CrossRef]
- Yetisgin, A.A.; Cetinel, S.; Zuvin, M.; Kosar, A.; Kutlu, O. Therapeutic Nanoparticles and Their Targeted Delivery Applications. Molecules 2020, 25, 2193. [Google Scholar] [CrossRef]
- Smerkova, K.; Dolezelikova, K.; Bozdechova, L.; Heger, Z.; Zurek, L.; Adam, V. Nanomaterials with active targeting as advanced antimicrobials. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1636. [Google Scholar] [CrossRef]
- Zhao, Z.; Ukidve, A.; Kim, J.; Mitragotri, S. Targeting Strategies for Tissue-Specific Drug Delivery. Cell 2020, 181, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, Y.; Zhang, Y.; Zhang, Q.; Zhang, L. Nanoparticle-based local antimicrobial drug delivery. Adv. Drug Deliv. Rev. 2018, 127, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Samrot, A.V.; Sahithya, C.S.; Sruthi, P.D.; Selvarani, A.J.; Raji, P.; Prakash, P.; Ponnaiah, P.; Petchi, I.; Pattammadath, S.; Purayil, S.K.; et al. Itraconazole Coated Super Paramagnetic Iron Oxide Nanoparticles for Antimicrobial Studies. Biointerface Res. Appl. Chem. 2020, 10, 6262–6269. [Google Scholar] [CrossRef]
- Malviya, R. Exploration of neem gum-chitosan and kheri gum-chitosan polyelectrolyte complex based film for transdermal delivery of protein/peptide. Biointerface Res. Appl. Chem. 2020, 10, 5860–5868. [Google Scholar] [CrossRef]
- Román, L.E.; Gomez, E.D.; Solís, J.L.; Gómez, M.M. Antibacterial Cotton Fabric Functionalized with Copper Oxide Nanoparticles. Molecules 2020, 25, 5802. [Google Scholar] [CrossRef]
- Niculescu, A.-G.; Grumezescu, A.M. Polymer-Based Nanosystems—A Versatile Delivery Approach. Materials 2021, 14, 6812. [Google Scholar] [CrossRef]
- Walvekar, P.; Gannimani, R.; Salih, M.; Makhathini, S.; Mocktar, C.; Govender, T. Self-assembled oleylamine grafted hyaluronic acid polymersomes for delivery of vancomycin against methicillin resistant Staphylococcus aureus (MRSA). Colloids Surf. B Biointerfaces 2019, 182, 110388. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, D.; Sui, G.; Wang, D.; Wu, M.; Han, L.; Mu, H.; Duan, J. Gentamicin decorated phosphatidylcholine-chitosan nanoparticles against biofilms and intracellular bacteria. Int. J. Biol. Macromol. 2020, 156, 640–647. [Google Scholar] [CrossRef]
- Ciro, Y.; Rojas, J.; Oñate-Garzon, J.; Salamanca, C.H. Synthesis, Characterisation and Biological Evaluation of Ampicillin–Chitosan–Polyanion Nanoparticles Produced by Ionic Gelation and Polyelectrolyte Complexation Assisted by High-Intensity Sonication. Polymers 2019, 11, 1758. [Google Scholar] [CrossRef]
- Kaur, J.; Kour, A.; Panda, J.J.; Harjai, K.; Chhibber, S. Exploring Endolysin-Loaded Alginate-Chitosan Nanoparticles as Future Remedy for Staphylococcal Infections. AAPS PharmSciTech 2020, 21, 233. [Google Scholar] [CrossRef]
- Scolari, I.R.; Páez, P.L.; Musri, M.M.; Petiti, J.P.; Torres, A.; Granero, G.E. Rifampicin loaded in alginate/chitosan nanoparticles as a promising pulmonary carrier against Staphylococcus aureus. Drug Deliv. Transl. Res. 2020, 10, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Falciani, C.; Zevolini, F.; Brunetti, J.; Riolo, G.; Gracia, R.; Marradi, M.; Loinaz, I.; Ziemann, C.; Cossío, U.; Llop, J.; et al. Antimicrobial Peptide-Loaded Nanoparticles as Inhalation Therapy for Pseudomonas aeruginosa Infections. Int. J. Nanomed. 2020, 15, 1117–1128. [Google Scholar] [CrossRef] [PubMed]
- Macêdo, D.C.d.S.; Cavalcanti, I.D.L.; Medeiros, S.M.d.F.R.d.S.; Souza, J.B.d.; Lira Nogueira, M.C.d.B.; Cavalcanti, I.M.F. Nanotechnology and tuberculosis: An old disease with new treatment strategies. Tuberculosis 2022, 135, 102208. [Google Scholar] [CrossRef]
- Hatae, A.C.; Roque-Borda, C.A.; Pavan, F.R. Strategies for lipid-based nanocomposites with potential activity against Mycobacterium tuberculosis: Microbial resistance challenge and drug delivery trends. OpenNano 2023, 13, 100171. [Google Scholar] [CrossRef]
- Chakravarty, M.; Vora, A. Nanotechnology-based antiviral therapeutics. Drug Deliv. Transl. Res. 2021, 11, 748–787. [Google Scholar]
- Patil, T.S.; Deshpande, A.S. Nanostructured lipid carrier-mediated lung targeted drug delivery system to enhance the safety and bioavailability of clofazimine. Drug Dev. Ind. Pharm. 2021, 47, 385–393. [Google Scholar] [CrossRef]
- Mukai, H.; Ogawa, K.; Kato, N.; Kawakami, S. Recent advances in lipid nanoparticles for delivery of nucleic acid, mRNA, and gene editing-based therapeutics. Drug Metab. Pharmacokinet. 2022, 44, 100450. [Google Scholar] [CrossRef]
- Aldosari, B.N.; Alfagih, I.M.; Almurshedi, A.S. Lipid nanoparticles as delivery systems for RNA-based vaccines. Pharmaceutics 2021, 13, 206. [Google Scholar] [CrossRef]
- Bailey-Hytholt, C.M.; Ghosh, P.; Dugas, J.; Zarraga, I.E.; Bandekar, A. Formulating and Characterizing Lipid Nanoparticles for Gene Delivery using a Microfluidic Mixing Platform. J. Vis. Exp. Jove 2021, 168, e62226. [Google Scholar]
- Wang, H.-L.; Wang, Z.-G.; Liu, S.-L. Lipid Nanoparticles for mRNA Delivery to Enhance Cancer Immunotherapy. Molecules 2022, 27, 5607. [Google Scholar] [CrossRef]
- Curley, S.M.; Putnam, D. Biological nanoparticles in vaccine development. Front. Bioeng. Biotechnol. 2022, 10, 867119. [Google Scholar] [CrossRef] [PubMed]
- Lozano, D.; Larraga, V.; Vallet-Regí, M.; Manzano, M. An Overview of the Use of Nanoparticles in Vaccine Development. Nanomaterials 2023, 13, 1828. [Google Scholar] [CrossRef] [PubMed]
- Croitoru, G.-A.; Niculescu, A.-G.; Epistatu, D.; Mihaiescu, D.E.; Antohi, A.M.; Grumezescu, A.M.; Nicolae, C.-L. Nanostructured Drug Delivery Systems in Immunotherapy: An Updated Overview of Nanotechnology-Based Therapeutic Innovations. Appl. Sci. 2024, 14, 8948. [Google Scholar] [CrossRef]
- Waiezi, S.; Malek, N.; Asraf, M.H.; Sani, N.S. Preparation, characterization, and antibacterial activity of green-biosynthesised silver nanoparticles using Clinacanthus nutans extract. Biointerface Res. Appl. Chem. 2023, 13, 171. [Google Scholar]
- Bharathi, D.; Thiruvengadam Nandagopal, J.G.; Lee, J.; Ranjithkumar, R. Facile Synthesis and Characterization of Chitosan Functionalized Silver Nanoparticles for Antibacterial and Anti-Lung Cancer Applications. Polymers 2023, 15, 2700. [Google Scholar] [CrossRef]
- Bîrcă, A.C.; Gherasim, O.; Niculescu, A.-G.; Grumezescu, A.M.; Neacșu, I.A.; Chircov, C.; Vasile, B.Ș.; Oprea, O.C.; Andronescu, E.; Stan, M.S.; et al. A Microfluidic Approach for Synthesis of Silver Nanoparticles as a Potential Antimicrobial Agent in Alginate–Hyaluronic Acid-Based Wound Dressings. Int. J. Mol. Sci. 2023, 24, 11466. [Google Scholar] [CrossRef]
- Pokhrel, L.R.; Williams, F.; Cook, P.P.; O’Rourke, D.; Murray, G.; Akula, S.M. Preclinical efficacy and safety of novel SNAT against SARS-CoV-2 using a hamster model. Drug Deliv. Transl. Res. 2022, 12, 3007–3016. [Google Scholar] [CrossRef]
- Souza, F.R.; Fornasier, F.; Carvalho, A.S.; Silva, B.M.; Lima, M.C.; Pimentel, A.S. Polymer-coated gold nanoparticles and polymeric nanoparticles as nanocarrier of the BP100 antimicrobial peptide through a lung surfactant model. J. Mol. Liq. 2020, 314, 113661. [Google Scholar] [CrossRef]
- Samsulkahar, N.F.; Hadi, A.A.; Shamsuddin, M.; Nik, N.A.N. Biosynthesis of Gold Nanoparticles Using Strobilanthes crispa Aqueous Leaves Extract and Evaluation of Its Antibacterial Activity. Biointerface Res. Appl. Chem. 2023, 13, 63. [Google Scholar]
- Changsan, N.; Atipairin, A.; Muenraya, P.; Sritharadol, R.; Srichana, T.; Balekar, N.; Sawatdee, S. In Vitro Evaluation of Colistin Conjugated with Chitosan-Capped Gold Nanoparticles as a Possible Formulation Applied in a Metered-Dose Inhaler. Antibiotics 2024, 13, 630. [Google Scholar] [CrossRef]
- Caciandone, M.; Niculescu, A.-G.; Grumezescu, V.; Bîrcă, A.C.; Ghica, I.C.; Vasile, B.Ș.; Oprea, O.; Nica, I.C.; Stan, M.S.; Holban, A.M.; et al. Magnetite Nanoparticles Functionalized with Therapeutic Agents for Enhanced ENT Antimicrobial Properties. Antibiotics 2022, 11, 623. [Google Scholar] [CrossRef] [PubMed]
- Abdussalam-Mohammed, W.; Abraheem, M.S.; Mezoughi, A.B.; Mohamed, L.; Alwahsh, M.A.A. Comparative Analysis of Novel Iron Oxide Nanoparticles Synthesized by Different Approaches with Evaluation of Their Antibacterial Activities. Biointerface Res. Appl. Chem. 2023, 13, 317. [Google Scholar] [CrossRef]
- Ezealigo, U.S.; Ezealigo, B.N.; Aisida, S.O.; Ezema, F.I. Iron oxide nanoparticles in biological systems: Antibacterial and toxicology perspective. JCIS Open 2021, 4, 100027. [Google Scholar] [CrossRef]
- Celebi, D.; Celebi, O.; Taghizadehghalehjoughi, A.; Baser, S.; Aydın, E.; Calina, D.; Charvalos, E.; Docea, A.O.; Tsatsakis, A.; Mezhuev, Y. Activity of zinc oxide and zinc borate nanoparticles against resistant bacteria in an experimental lung cancer model. DARU J. Pharm. Sci. 2024, 32, 197–206. [Google Scholar]
- Grămadă, A.M.; Stoica, A.-E.; Niculescu, A.-G.; Bîrcă, A.C.; Vasile, B.Ș.; Holban, A.M.; Mihaiescu, T.; Șerban, A.I.; Ciceu, A.; Balta, C.; et al. Zinc Oxide-Loaded Recycled PET Nanofibers for Applications in Healthcare and Biomedical Devices. Polymers 2025, 17, 45. [Google Scholar] [CrossRef]
- Rayyif, S.M.I.; Mohammed, H.B.; Curuțiu, C.; Bîrcă, A.C.; Grumezescu, A.M.; Vasile, B.Ș.; Dițu, L.M.; Lazăr, V.; Chifiriuc, M.C.; Mihăescu, G.; et al. ZnO Nanoparticles-Modified Dressings to Inhibit Wound Pathogens. Materials 2021, 14, 3084. [Google Scholar] [CrossRef]
- Serov, D.A.; Gritsaeva, A.V.; Yanbaev, F.M.; Simakin, A.V.; Gudkov, S.V. Review of Antimicrobial Properties of Titanium Dioxide Nanoparticles. Int. J. Mol. Sci. 2024, 25, 10519. [Google Scholar] [CrossRef]
- Younis, A.B.; Haddad, Y.; Kosaristanova, L.; Smerkova, K. Titanium dioxide nanoparticles: Recent progress in antimicrobial applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnology 2023, 15, e1860. [Google Scholar] [CrossRef]
- Mansoor, A.; Mehmood, M.; Ul Hassan, S.M.; Ali, M.I.; Badshah, M.; Jamal, A. Anti-Bacterial Effect of Titanium-Oxide Nanoparticles and their Application as Alternative to Antibiotics. Pak. Vet. J. 2023, 43, 269–275. [Google Scholar] [CrossRef]
- León-Gutiérrez, G.; Elste, J.E.; Cabello-Gutiérrez, C.; Millán-Pacheco, C.; Martínez-Gómez, M.H.; Mejía-Alvarez, R.; Tiwari, V.; Mejía, A. A potent virucidal activity of functionalized TiO2 nanoparticles adsorbed with flavonoids against SARS-CoV-2. Appl. Microbiol. Biotechnol. 2022, 106, 5987–6002. [Google Scholar] [CrossRef]
- Ritsema, J.A.S.; der Weide, H.v.; Te Welscher, Y.M.; Goessens, W.H.F.; Van Nostrum, C.F.; Storm, G.; Bakker-Woudenberg, I.A.J.M.; Hays, J.P. Antibiotic-nanomedicines: Facing the challenge of effective treatment of antibiotic-resistant respiratory tract infections. Future Microbiol. 2018, 13, 1683–1692. [Google Scholar] [PubMed]
- Trivedi, R.; Barve, K. Gut microbiome a promising target for management of respiratory diseases. Biochem. J. 2020, 477, 2679–2696. [Google Scholar] [CrossRef] [PubMed]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Bucurica, S.; Lupanciuc, M.; Ionita-Radu, F.; Stefan, I.; Munteanu, A.E.; Anghel, D.; Jinga, M.; Gaman, E.L. Estrobolome and Hepatocellular Adenomas—Connecting the Dots of the Gut Microbial β-Glucuronidase Pathway as a Metabolic Link. Int. J. Mol. Sci. 2023, 24, 16034. [Google Scholar] [CrossRef]
- van Beveren, G.J.; Said, H.; van Houten, M.A.; Bogaert, D. The respiratory microbiome in childhood asthma. J. Allergy Clin. Immunol. 2023, 152, 1352–1367. [Google Scholar] [CrossRef]
- Depner, M.; Ege, M.J.; Cox, M.J.; Dwyer, S.; Walker, A.W.; Birzele, L.T.; Genuneit, J.; Horak, E.; Braun-Fahrländer, C.; Danielewicz, H.; et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J. Allergy Clin. Immunol. 2017, 139, 826–834.e813. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef]
- Hoen, A.G.; Li, J.; Moulton, L.A.; O’Toole, G.A.; Housman, M.L.; Koestler, D.C.; Guill, M.F.; Moore, J.H.; Hibberd, P.L.; Morrison, H.G.; et al. Associations between Gut Microbial Colonization in Early Life and Respiratory Outcomes in Cystic Fibrosis. J. Pediatr. 2015, 167, 138–147.e133. [Google Scholar] [CrossRef]
- Vinayamohan, P.; Joseph, D.; Viju, L.S.; Baskaran, S.A.; Venkitanarayanan, K. Efficacy of Probiotics in Reducing Pathogenic Potential of Infectious Agents. Fermentation 2024, 10, 599. [Google Scholar] [CrossRef]
- Chunxi, L.; Haiyue, L.; Yanxia, L.; Jianbing, P.; Jin, S. The Gut Microbiota and Respiratory Diseases: New Evidence. J. Immunol. Res. 2020, 2020, 2340670. [Google Scholar] [CrossRef]
- Xie, Y.; Xue, Q.; Jiao, W.; Wu, J.; Yu, Y.; Zhao, L.; Xu, Y.; Deng, X.; Fang, G.; Zheng, Y.; et al. Associations Between Sputum Torque Teno Virus Load and Lung Function and Disease Severity in Patients With Chronic Obstructive Pulmonary Disease. Front. Med. 2021, 8, 618757. [Google Scholar]
- Wang, W.-W.; Mao, B.; Liu, Y.; Gu, S.-Y.; Lu, H.-W.; Bai, J.-W.; Liang, S.; Yang, J.-W.; Li, J.-X.; Su, X.; et al. Altered fecal microbiome and metabolome in adult patients with non-cystic fibrosis bronchiectasis. Respir. Res. 2022, 23, 317. [Google Scholar] [CrossRef]
- Xia, Q.; Chen, G.; Ren, Y.; Zheng, T.; Shen, C.; Li, M.; Chen, X.; Zhai, H.; Li, Z.; Xu, J.; et al. Investigating efficacy of “microbiota modulation of the gut-lung Axis” combined with chemotherapy in patients with advanced NSCLC: Study protocol for a multicenter, prospective, double blind, placebo controlled, randomized trial. BMC Cancer 2021, 21, 721. [Google Scholar] [CrossRef]
- Hegazy, M.A.; Fouad, M.M.; Abd elshafy, S.A.; Abdelfatah, D.; Lithy, R.M.; Abdelghani, A.; Ashoush, O.A. Beneficial role of healthy eating Index-2015 score & physical activity on COVID-19 outcomes. BMC Nutr. 2023, 9, 113. [Google Scholar] [CrossRef]
- Pacheco, A.R.; Segrè, D. A multidimensional perspective on microbial interactions. FEMS Microbiol. Lett. 2019, 366, fnz125. [Google Scholar] [CrossRef]
- Liu, W.; Pi, Z.; Wang, X.; Shang, C.; Song, C.; Wang, R.; He, Z.; Zhang, X.; Wan, Y.; Mao, W. Microbiome and lung cancer: Carcinogenic mechanisms, early cancer diagnosis, and promising microbial therapies. Crit. Rev. Oncol./Hematol. 2024, 196, 104322. [Google Scholar] [CrossRef]
- Busnatu, Ș.S.; Niculescu, A.-G.; Bolocan, A.; Andronic, O.; Pantea Stoian, A.M.; Scafa-Udriște, A.; Stănescu, A.M.; Păduraru, D.N.; Nicolescu, M.I.; Grumezescu, A.M.; et al. A Review of Digital Health and Biotelemetry: Modern Approaches towards Personalized Medicine and Remote Health Assessment. J. Pers. Med. 2022, 12, 1656. [Google Scholar] [CrossRef]
- Acharjee, A.; Singh, U.; Choudhury, S.P.; Gkoutos, G.V. The diagnostic potential and barriers of microbiome based therapeutics. Diagnosis 2022, 9, 411–420. [Google Scholar] [CrossRef]

| Term | Definition |
|---|---|
| Microbiota | Living microorganisms coexisting in a defined habitat; a dynamic microbial community confined within a specified environment (e.g., human body and oral cavity) [12] |
| Microbiome | Collection of genomes from all the microorganisms in the environment, comprising the genetic information and inferred physicochemical properties of the gene products of a microbiota [13] |
| Metagenomics | A process of randomly sequencing the entire DNA present in a sample (including DNA from the host organism and microorganisms), which is further analyzed, organized, and identified by means of sequence databases and computational tools to highlight the genetic potential of the population [14] |
| Dysbiosis | Modification in microbiota composition linked to perturbation of local ecological conditions, usually linked with impaired host–microbe interactions [15] |
| Microdysbiosis | Ecosystem disturbance produced by the destruction of the microecological balance [16] |
| Antibiotic resistance | Ability of bacteria to develop defense mechanisms and evade the activity of antibiotic drugs [17] |
| Resistome | Assembly of quantity, identity, and functions of ARGs [18] |
| Prebiotics | Gut microbiota-accessible dietary fibers are able to nourish and promote beneficial bacteria growth [19] |
| Probiotics | Beneficial live microorganisms present in fermented foods and dietary supplements [20] |
| Antibiotic | Disease | Analyzed Sample | Impact of Antibiotics on the Diversity of the Microbiome | Impact on the Relative Abundance of Bacterial Taxa |
|---|---|---|---|---|
| Oral macrolides | Bronchiectasis | Oropharyngeal swab | No impact on α-diversity measures | Difference between treated and placebo groups: ↓ Actinomyces and Streptococcus ↑ Haemophilus after 48 weeks of treatment |
| Sputum | Increase in genus richness between baseline and 48 weeks in the treated group, but no difference with the placebo group | No change in the composition of the airway microbiome in the P. aeruginosa-dominated subgroup ↓ H. influenzae and ↑ P. aeruginosa in non-P. aeruginosa-dominated subgroup | ||
| Severe asthma | Oropharyngeal swab | Impact on β-diversity measures | ↓ Fusobacteria ↑ Firmicutes during treatment compared with the untreated group, but return to the pre-treatment state after a 1-month washout period | |
| Sputum | ↓ Faith’s phylogenetic diversity | ↓ Gammaproteobacteria (including H. influenzae) after 48 weeks of azithromycin treatment compared with a placebo group | ||
| Moderate and severe asthma | Bronchoalveolar lavage of the right upper lung lobe | ↓ Shannon’s diversity index | ↓ Prevotella, Staphylococcus, and Haemophilus ↑ Anaerococcus between the pre- and post-treatment states | |
| Chronic obstructive pulmonary disease | Sputum | - | ↓ of multiple taxa, mainly Proteobacteria | |
| β-lactams | Cystic fibrosis | Nasal swabs | ↑ Shannon’s diversity index | ↓ Moraxellaceae ↑ other bacterial families (this increase was verified after more than one antibiotic treatment) |
| Bronchoalveolar lavage, sputum, or deep throat swabs | ↓ α-diversity between exacerbation and treatment ↓, α-diversity at therapeutic doses between baseline and treatment ↑ α-diversity at sub-therapeutic doses at the same time points | ↓ Haemophilus, Clostridiales, and Lachnospiraceae ↑ Fusobacterium and Pseudomonas in the group treated at therapeutic doses between baseline and treatment samples No difference was observed in the sub-therapeutic group at the same time points No difference in the bacterial composition was observed in the two groups between post-recovery and baseline samples or between exacerbation and treatment samples | ||
| Sputum | ↓/↑ Shannon’s diversity index (depending on the type of treatment) ↓ Bray–Curtis β-diversity with ↑ AZLI cycles | ↓ Relative abundance of some low-abundance taxa with antibiotic treatment: Gemella, two Pasteurella operational taxonomic units, two Streptococcus operational taxonomic units, Oribacterium and Neisseria ↓ P. aeruginosa ↑ anaerobes (Prevotella and Veillonella) in the first 72 h of treatment, but return to the baseline state after 8–10 days of treatment | ||
| Aminoglycosides | Cystic fibrosis | Sputum | ↓ average species richness (Shannon and Simpson diversity indices) after 1 week of therapy. Return to baseline state after the end of TIP therapy No difference in Shannon’s diversity index | Most changes noticed between the baseline state and the first week of treatment occurred among low-abundance taxa, mostly facultative and obligate anaerobes (Neisseria, Megasphaera, Granulicatella, Haemophilus, Streptococcus, Gemella, Rothia, Veillonella, Oribacterium) ↓ Parvimonas |
| Association of different antibiotic classes in the treatment of cystic fibrosis exacerbation episodes | Cystic fibrosis | Sputum | ↓ α-diversity (inverse Simpson index) ↑ Species richness through PEx and treatment periods, but returns to the baseline state during the recovery period | ↓ Prevotella melaninogenica and S. sanguinis ↑ Veillonella parvula during the treatment period, but returns to the baseline state in post-recovery samples |
| ClinicalTrials.Gov Identifier | Official Title | Study Type | Intervention/ Treatment | Phase | (Estimated) Completion Date | References |
|---|---|---|---|---|---|---|
| NCT06271213 | The Gut–Lung Axis and Respiratory Illness in Children | Observational | - | - | 1 May 2028 | - |
| NCT04813718 | Post-COVID-19 Syndrome: A Pilot Study to Explore the Gut–Lung Axis | Interventional | Dietary Supplement: Omni-Biotic Pro Vi 5 Dietary Supplement: Placebo | N/A | 31 December 2023 | - |
| NCT03236480 | Dynamic Changes in the Respiratory Microbiota and Its Relationship to Fecal Microbiota in Chronic Obstructive Pulmonary Disease | Observational | - | - | 1 January 2019 | [163] |
| NCT05623007 | Dietary Modulation of Gut Microbiota on Nutritional Status and COVID-19 Infection in Adolescents: Gut–Lung Axis | Interventional | Dietary Supplement: Probiotics Behavioral: Counseling On Healthy Eating, Physical Activity, and Psychosocial Stimulation Dietary Supplement: Placebo Probiotics | 2 | 2 November 2025 | - |
| NCT05937815 | Monitoring of the Intestine–Lung Axis of Cystic Fibrosis Patients Treated with the Combination Elexacaftor/Tezacaftor/Ivacaftor: Study of the Pulmonary and Gut Microbiota and Inflammation | Interventional | Procedure: Sample Collection | N/A | 13 September 2024 | - |
| NCT04490447 | Identification of Microbiome and Metabolome of Bronchiectasis in Chinese Population and Role of the “Gut-lung Axis” in Chronic Respiratory Infection with P. aeruginosa. | Observational | - | - | 1 September 2021 | [164] |
| NCT04979065 | Effect of Probiotic and Vitamin D Supplementation in Modulating Gut Dysbiosis, Nutrition, Inflammation, and Immune Status and Reduce Risk of COVID-19 in Obese People: Gut–Lung Axis Randomized Trial | Interventional | Dietary Supplement: Probiotics, Vitamin D Other: Placebo | N/A | 30 December 2022 | - |
| NCT03642548 | A Prospective Multicenter Double-blind Randomized Clinical Trial of Probiotics Combined with Chemotherapy in the Treatment of Patients with Advanced Non-Small Cell Lung Cancer | Interventional | Drug: Bifico Drug: Placebo | 3 | 1 March 2024 | [165] |
| NCT05164445 | Observational Participants Not Assigned to Intervention(s) Based on a Protocol, typically in the context of routine care | Interventional | Diagnostic Test: Transbronchial Forceps Biopsy Diagnostic Test: Transbronchial Forceps Biopsy + Transbronchial Cryobiopsy | N/A | 31 August 2023 | - |
| NCT04960878 | The Effect of Synbiotics on the Upper Respiratory Tract Infection in Healthy Subjects: A Randomized Double-Blind Trial | Interventional | Dietary Supplement: Synbiotic Dietary Supplement: Placebo | N/A | 5 January 2021 | - |
| NCT04824222 | Two-stage Study: Phase II/III—With a Pilot Safety Assessment in an Open-label Study and Phase III—a Multicenter, Randomized, Double-blind, Placebo-Controlled Evaluation of the Effect of Fecal Microbiota Transplantation as an Immunomodulation, in Addition to Standard Therapy, on the Risk Reduction in COVID-19 Disease Progression with Escalating Cytokine Storm and Inflammation | Interventional | Drug: Human Fecal Microbiota, MBiotix HBI Drug: Placebo Drug: SOC | 3 | December 2022 | - |
| NCT04447144 | Nutritional Habits: Do They Affect Coronavirus Disease 2019 (COVID-19) Infection Outcome? An Egyptian Experience | Observational | - | - | 1 September 2020 | [166] |
| NCT06348212 | Effect of Probiotic Strain Lactobacillus Paracasei PS23 on Brain Fog in People with Long COVID | Interventional | Dietary Supplement: Lactobacillus Paracasei PS23 Dietary Supplement: Microcrystalline Cellulose | N/A | December 2025 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niculescu, A.-G.; Mitache, M.M.; Grumezescu, A.M.; Chifiriuc, M.C.; Mihai, M.M.; Tantu, M.M.; Tantu, A.C.; Popa, L.G.; Grigore, G.A.; Cristian, R.-E.; et al. From Microbial Ecology to Clinical Challenges: The Respiratory Microbiome’s Role in Antibiotic Resistance. Pathogens 2025, 14, 355. https://doi.org/10.3390/pathogens14040355
Niculescu A-G, Mitache MM, Grumezescu AM, Chifiriuc MC, Mihai MM, Tantu MM, Tantu AC, Popa LG, Grigore GA, Cristian R-E, et al. From Microbial Ecology to Clinical Challenges: The Respiratory Microbiome’s Role in Antibiotic Resistance. Pathogens. 2025; 14(4):355. https://doi.org/10.3390/pathogens14040355
Chicago/Turabian StyleNiculescu, Adelina-Gabriela, Mihaela Magdalena Mitache, Alexandru Mihai Grumezescu, Mariana Carmen Chifiriuc, Mara Madalina Mihai, Monica Marilena Tantu, Ana Catalina Tantu, Loredana Gabriela Popa, Georgiana Alexandra Grigore, Roxana-Elena Cristian, and et al. 2025. "From Microbial Ecology to Clinical Challenges: The Respiratory Microbiome’s Role in Antibiotic Resistance" Pathogens 14, no. 4: 355. https://doi.org/10.3390/pathogens14040355
APA StyleNiculescu, A.-G., Mitache, M. M., Grumezescu, A. M., Chifiriuc, M. C., Mihai, M. M., Tantu, M. M., Tantu, A. C., Popa, L. G., Grigore, G. A., Cristian, R.-E., Popa, M. I., & Vrancianu, C. O. (2025). From Microbial Ecology to Clinical Challenges: The Respiratory Microbiome’s Role in Antibiotic Resistance. Pathogens, 14(4), 355. https://doi.org/10.3390/pathogens14040355











