One Health Surveillance for SARS-CoV-2 in Non-Human Primates and Small Mammals in Minas Gerais, Brazil
Abstract
1. Introduction
2. Materials and Methods
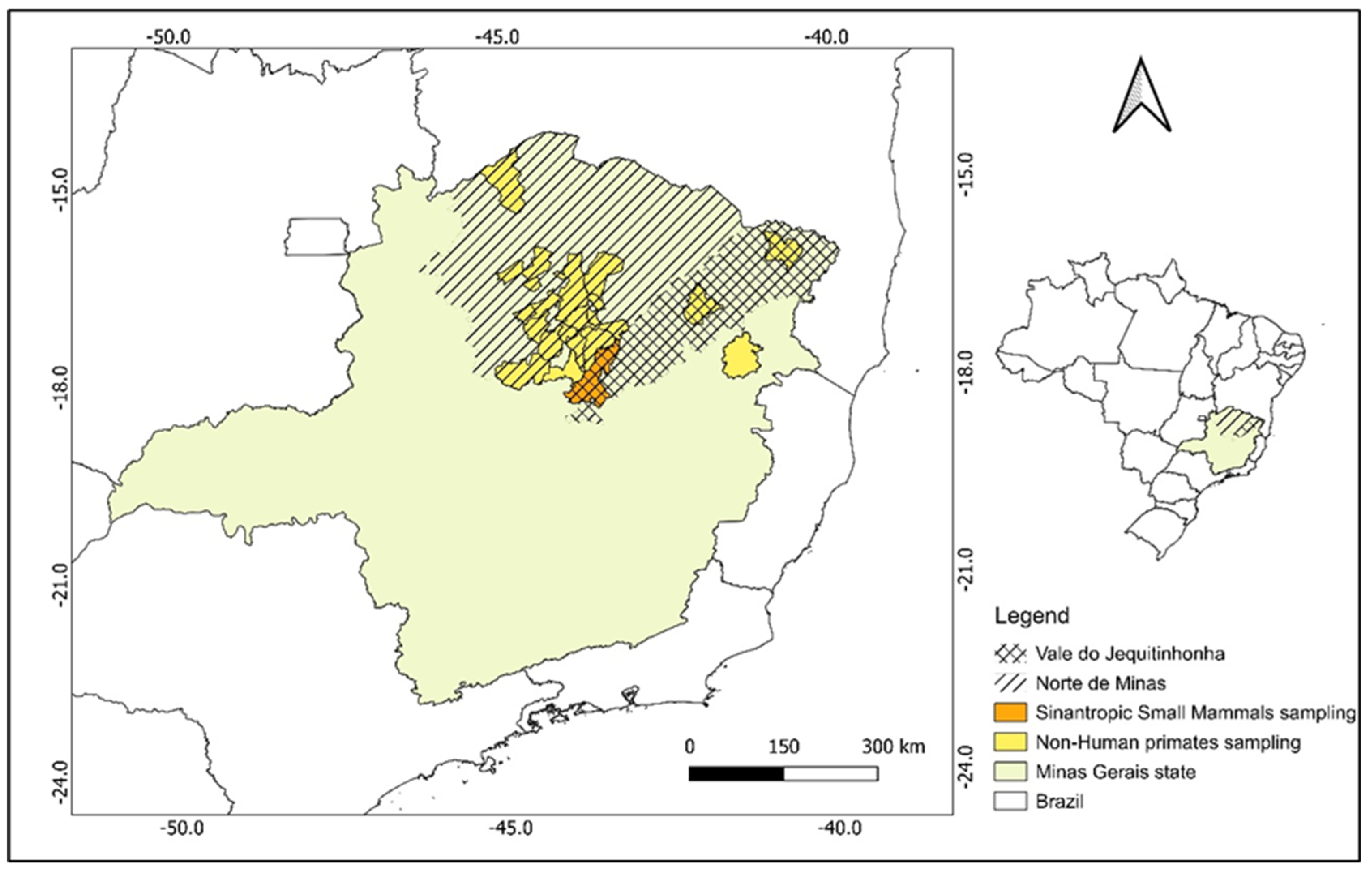
2.1. Capture Efforts and Sampling Methods
2.2. RNA Extraction and Molecular Detection by RT-qPCR
2.3. Serological Assay-Plaque Reduction Neutralization Test (PRNT) Anti-SARS-CoV-2
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICTV. ICTV Genus: Betacoronavirus. Available online: https://ictv.global/report/chapter/coronaviridae/coronaviridae/betacoronavirus (accessed on 23 March 2025).
- Oliveira, W.K.d.; Duarte, E.; França, G.V.A.d.; Garcia, L.P. Como o Brasil Pode Deter a COVID-19. Epidemiol. Serviços Saúde 2020, 29, e2020044. [Google Scholar] [CrossRef]
- WHO Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (accessed on 10 September 2024).
- WHO COVID-19 Cases. WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/cases?n=o (accessed on 27 January 2025).
- Calvet, G.A.; Pereira, S.A.; Ogrzewalska, M.; Pauvolid-Corrêa, A.; Resende, P.C.; Tassinari, W.d.S.; Costa, A.d.P.; Keidel, L.O.; da Rocha, A.S.B.; da Silva, M.F.B.; et al. Investigation of SARS-CoV-2 Infection in Dogs and Cats of Humans Diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS ONE 2021, 16, e0250853. [Google Scholar] [CrossRef] [PubMed]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of Dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Fonager, J.; Rasmussen, M.; Mundbjerg, K.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Alfaro-Núñez, A.; et al. SARS-CoV-2 Transmission between Mink (Neovison vison) and Humans, Denmark. Emerg. Infect. Dis. 2021, 27, 547–551. [Google Scholar] [CrossRef]
- Dusseldorp, F.; Bruins-van-Sonsbeek, L.G.R.; Buskermolen, M.; Niphuis, H.; Dirven, M.; Whelan, J.; Oude Munnink, B.B.; Koopmans, M.; Fanoy, E.B.; Sikkema, R.S.; et al. SARS-CoV-2 in Lions, Gorillas and Zookeepers in the Rotterdam Zoo, the Netherlands, a One Health Investigation, November 2021. Eurosurveillance 2023, 28, 2200741. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.H.B.; Vasconcelos, A.L.; Silva, V.L.B.; Nogueira, B.S.; Silva, A.C.P.; Pacheco, R.C.; Souza, M.A.; Colodel, E.M.; Ubiali, D.G.; Biondo, A.W.; et al. Natural SARS-CoV-2 Infection in a Free-Ranging Black-Tailed Marmoset (Mico Melanurus) from an Urban Area in Mid-West Brazil. J. Comp. Pathol. 2022, 194, 22–27. [Google Scholar] [CrossRef]
- Orlando, S.A.; Mera, M.D.; Mora Jaramillo, N.; Leon-Sosa, A.; Calderon, J.; Rodriguez-Pazmiño, A.S.; Garcia-Bereguiain, M.A. SARS-CoV-2 Infection in Synanthropic Rats from Guayaquil City (Ecuador) during COVID-19 Pandemic: A Proxy to Prevent Wild Reservoirs in the Tropics. Acta Trop. 2024, 259, 107371. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on Mink Farms between Humans and Mink and Back to Humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple Spillovers from Humans and Onward Transmission of SARS-CoV-2 in White-Tailed Deer. Proc. Natl. Acad. Sci. USA 2022, 119, e2121644119. [Google Scholar] [CrossRef]
- McBride, D.S.; Garushyants, S.K.; Franks, J.; Magee, A.F.; Overend, S.H.; Huey, D.; Williams, A.M.; Faith, S.A.; Kandeil, A.; Trifkovic, S.; et al. Accelerated Evolution of SARS-CoV-2 in Free-Ranging White-Tailed Deer. Nat. Commun. 2023, 14, 5105. [Google Scholar] [CrossRef]
- de Abreu, F.V.S.; Macedo, M.V.; da Silva, A.J.J.; de Oliveira, C.H.; de Ottone, V.O.; de Almeida, M.A.B.; dos Santos, E.; da Cardoso, J.C.; Campos, A.S.; da Silva, C.M.D.; et al. No Evidence of SARS-CoV-2 Infection in Neotropical Primates Sampled During COVID-19 Pandemic in Minas Gerais and Rio Grande Do Sul, Brazil. EcoHealth 2021, 18, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Delahay, R.J.; de la Fuente, J.; Smith, G.C.; Sharun, K.; Snary, E.L.; Flores Girón, L.; Nziza, J.; Fooks, A.R.; Brookes, S.M.; Lean, F.Z.X.; et al. Assessing the Risks of SARS-CoV-2 in Wildlife. One Health Outlook 2021, 3, 7. [Google Scholar] [CrossRef]
- Leroy, E.M.; Ar Gouilh, M.; Brugère-Picoux, J. The Risk of SARS-CoV-2 Transmission to Pets and Other Wild and Domestic Animals Strongly Mandates a One-Health Strategy to Control the COVID-19 Pandemic. One Health 2020, 10, 100133. [Google Scholar] [CrossRef]
- Milich, K.M.; Morse, S.S. The Reverse Zoonotic Potential of SARS-CoV-2. Heliyon 2024, 10, e33040. [Google Scholar] [CrossRef] [PubMed]
- Sparrer, M.N.; Hodges, N.F.; Sherman, T.; VandeWoude, S.; Bosco-Lauth, A.M.; Mayo, C.E. Role of Spillover and Spillback in SARS-CoV-2 Transmission and the Importance of One Health in Understanding the Dynamics of the COVID-19 Pandemic. J. Clin. Microbiol. 2023, 61, e01610-22. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Goldberg, A.R.; Langwig, K.E.; Brown, K.L.; Marano, J.M.; Rai, P.; King, K.M.; Sharp, A.K.; Ceci, A.; Kailing, C.D.; Kailing, M.J.; et al. Widespread Exposure to SARS-CoV-2 in Wildlife Communities. Nat. Commun. 2024, 15, 6210. [Google Scholar] [CrossRef]
- Diaz, E.A.; Sáenz, C.; Cabrera, F.; Rodríguez, J.; Carvajal, M.; Barragán, V. COVID-19 in a Common Woolly Monkey (Lagothrix lagothricha): First Evidence of Fatal Outcome in a Nonhuman Primate after Natural SARS-CoV-2 Infection. Am. J. Primatol. 2024, 86, e23654. [Google Scholar] [CrossRef]
- Carvajal, M.; Saenz, C.; Fuentes, N.; Guevara, R.; Muñoz, E.; Prado-Vivar, B.; Diaz, E.; Alfonso-Cortes, F.; Coloma, J.; Grunauer, M.; et al. SARS-CoV-2 Infection in Brown-Headed Spider Monkeys (Ateles fusciceps) at a Wildlife Rescue Center on the Coast of Ecuador—South America. Microbiol. Spectr. 2024, 12, e02741-23. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Pemberton, A.C.; Kelleher, J.; Ahmed, W. The Comparison of Decay Rates of Infectious SARS-CoV-2 and Viral RNA in Environmental Waters and Wastewater. Sci. Total Environ. 2024, 946, 174379. [Google Scholar] [CrossRef] [PubMed]
- Koza, S.R.; Li, Z. Presence, Transmission, and Management of the SARS-CoV-2 in Wastewater: A Brief Review. Int. J. Environ. Sci. Technol. 2024, 21, 9719–9742. [Google Scholar] [CrossRef]
- Saulnier, A.; Wendling, J.-M.; Hermant, B.; Lepelletier, D. SARS-CoV-2 Transmission Modes: Why and How Contamination Occurs around Shared Meals and Drinks? Food Microbiol. 2023, 114, 104297. [Google Scholar] [CrossRef]
- Abreu, F.V.S.; dos Santos, E.; Gomes, M.Q.; Vargas, W.P.; Oliveira Passos, P.H.; Nunes e Silva, C.; Araújo, P.C.; Pires, J.R.; Romano, A.P.M.; Teixeira, D.S.; et al. Capture of Alouatta guariba clamitans for the Surveillance of Sylvatic Yellow Fever and Zoonotic Malaria: Which Is the Best Strategy in the Tropical Atlantic Forest? Am. J. Primatol. 2019, 81, e23000. [Google Scholar] [CrossRef] [PubMed]
- Brasil. Guia de Vigilância de Epizootias Em Primatas Não Humanos e Entomologia Aplicada à Vigilância Da Febre Amarela. Guide to Surveillance of Epizootics in Non-Human Primates and Entomology Applied to Surveillance of Yellow Fever. Available online: http://vigilancia.saude.mg.gov.br/index.php/download/guia-de-epizootias-febre-amarela-2a-edicao-2017/?wpdmdl=3736 (accessed on 23 March 2025).
- Silva, T.G.M. Vigilância de Sars-Cov-2 em Águas Residuais e a Inter-Relação Com Indicadores Epidemiológicos da COVID-19 em Diamantina-Mg; Universidade Federal de Diamantina: Diamantina, Brazil, 2023. [Google Scholar]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Stoffella-Dutra, A.G.; de Campos, B.H.; Bastos e Silva, P.H.; Dias, K.L.; da Silva Domingos, I.J.; Hemetrio, N.S.; Xavier, J.; Iani, F.; Fonseca, V.; Giovanetti, M.; et al. SARS-CoV-2 Spillback to Wild Coatis in Sylvatic–Urban Hotspot, Brazil. Emerg. Infect. Dis. 2023, 29, 664–667. [Google Scholar] [CrossRef]
- Sacchetto, L.; Chaves, B.A.; Costa, E.R.; de Menezes Medeiros, A.S.; Gordo, M.; Araújo, D.B.; Oliveira, D.B.L.; da Silva, A.P.B.; Negri, A.F.; Durigon, E.L.; et al. Lack of Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spillover in Free-Living Neotropical Non-Human Primates, Brazil. Viruses 2021, 13, 1933. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; Ma, C.; et al. Comparison of Nonhuman Primates Identified the Suitable Model for COVID-19. Signal Transduct. Target. Ther. 2020, 5, 157. [Google Scholar] [CrossRef]
- Singh, D.K.; Singh, B.; Ganatra, S.R.; Gazi, M.; Cole, J.; Thippeshappa, R.; Alfson, K.J.; Clemmons, E.; Gonzalez, O.; Escobedo, R.; et al. Responses to Acute Infection with SARS-CoV-2 in the Lungs of Rhesus Macaques, Baboons and Marmosets. Nat. Microbiol. 2020, 6, 73–86. [Google Scholar] [CrossRef]
- Cano-Terriza, D.; Beato-Benítez, A.; Fernández-Bastit, L.; Segalés, J.; Vergara-Alert, J.; Martínez-Nevado, E.; Carretero, A.; Crailsheim, D.; Soriano, P.; Planas, J.; et al. SARS-CoV-2 in Captive Nonhuman Primates, Spain, 2020–2023. Emerg. Infect. Dis. 2024, 30, 1253. [Google Scholar] [CrossRef]
- Tavera Gonzales, A.; Bazalar Gonzales, J.; Silvestre Espejo, T.; Leiva Galarza, M.; Rodríguez Cueva, C.; Carhuaricra Huamán, D.; Luna Espinoza, L.; Maturrano Hernández, A. Possible Spreading of SARS-CoV-2 from Humans to Captive Non-Human Primates in the Peruvian Amazon. Animals 2024, 14, 732. [Google Scholar] [CrossRef] [PubMed]
- ICMBio/MMA. Livro Vermelho Fauna Brasileira Ameaçada de Extinção Ii—Mamíferos; Instituto Chico Mendes de Conservação da Biodiversidade: Brasília, Brazil, 2018. [Google Scholar]
- Yen, H.-L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Law, P.Y.T.; Leung, G.M.; Peiris, M.; Poon, L.L.M.; et al. Transmission of SARS-CoV-2 Delta Variant (AY.127) from Pet Hamsters to Humans, Leading to Onward Human-to-Human Transmission: A Case Study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hernández, F.; Gonzalez-Arenas, N.R.; Cervantes, J.A.O.; Villalobos, G.; Olivo-Diaz, A.; Rendon-Franco, E.; Maravilla, P.; Valdovinos, M.R.; Muñoz-Garcia, C.I. Identification of SARS-CoV-2 in Urban Rodents from Southern Mexico City at the Beginning of the COVID-19 Pandemic. Rev. Inst. Med. Trop. Sao Paulo 2024, 66, e8. [Google Scholar] [CrossRef]
- Colombo, V.C.; Sluydts, V.; Mariën, J.; Vanden Broecke, B.; Van Houtte, N.; Leirs, W.; Jacobs, L.; Iserbyt, A.; Hubert, M.; Heyndrickx, L.; et al. SARS-CoV-2 Surveillance in Norway Rats (Rattus norvegicus) from Antwerp Sewer System, Belgium. Transbound. Emerg. Dis. 2022, 69, 3016–3021. [Google Scholar] [CrossRef] [PubMed]
- IBGE. IBGE|Cidades@|Minas Gerais|Diamantina|Panorama. Available online: https://cidades.ibge.gov.br/brasil/mg/diamantina/panorama (accessed on 27 January 2025).
| Mammal | Cod. | Species | Sample | Municipality | Sampling Point | Lat | Long | Environment | Sampling Date |
|---|---|---|---|---|---|---|---|---|---|
| NHP | 097 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 17 March 2022 |
| 098 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 17 March 2022 | |
| 099 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 17 March 2022 | |
| 100 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 22 March 2022 | |
| 101 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 22 March 2022 | |
| 102 | C. penicillata | L | Francisco Dumont | random | −17.27788 | −44.23403 | Rural | 30 March 2022 | |
| 103 | C. penicillata | L | Bocaiuva | random | −17.07593 | −43.93418 | Rural | 30 March 2022 | |
| 104 | C. penicillata | L | Jequitaí | random | −16.98432 | −44.35691 | Rural | 30 March 2022 | |
| 105 | C. penicillata | L | São João da Lagoa | random | −16.82729 | −44.32653 | Sylvatic | 30 March 2022 | |
| 106 | C. penicillata | L | São João da Lagoa | random | −16.82729 | −44.32653 | Sylvatic | 30 March 2022 | |
| 107 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 4 April 2022 | |
| 108 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 4 April 2022 | |
| 109 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 7 April 2022 | |
| 110 | C. penicillata | O | Salinas | random | −16.15637 | −42.30730 | Rural | 7 April 2022 | |
| 111 | C. penicillata | O | Buenópolis | random | −17.87375 | −44.18200 | Urban | 26 May 2022 | |
| 112 | C. penicillata | O | Buenópolis | random | −17.87375 | −44.18200 | Urban | 26 May 2022 | |
| 113 | C. penicillata | O | Buenópolis | random | −17.87375 | −44.18200 | Urban | 26 May 2022 | |
| 114 | C. penicillata | L | Salinas | random | −16.15053 | −42.30761 | Rural | 20 June 2022 | |
| 115 | C. penicillata | O; L | Salinas | random | −16.15053 | −42.30761 | Rural | 20 June 2022 | |
| 116 | C. penicillata | L | Ubaí | random | −16.28660 | −44.77993 | Urban | 29 June 2022 | |
| 117 | C. penicillata | L | Ubaí | random | −16.28660 | −44.77993 | Urban | 8 June 2022 | |
| 118 | C. penicillata | L | Ubaí | random | −16.29680 | −44.78357 | Urban | 17 June 2022 | |
| 119 | C. penicillata | L | Ubaí | random | −16.29680 | −44.78357 | Urban | 27 June 2022 | |
| 120 | C. penicillata | L | Montes Claros | random | −16.65341 | −43.89827 | Sylvatic | 6 March 2022 | |
| 121 | C. penicillata | L | Guaraciama | random | −17.01723 | −43.68094 | Rural | 27 April 2022 | |
| 122 | C. penicillata | L | Olhos D’água | random | −17.39881 | −43.56943 | Urban | 7 June 2022 | |
| 123 | C. penicillata | L | Lagoa dos Patos | random | −17.01663 | −44.78229 | Rural | 15 June 2022 | |
| 124 | C. penicillata | L | Engenheiro Navarro | random | −17.28509 | −43.95465 | Urban | 7 July 2022 | |
| 125 | C. penicillata | L | Lassance | random | −17.88429 | −44.57745 | Urban | 8 June 2022 | |
| 126 | C. penicillata | L | Montes Claros | random | −16.74956 | −43.89984 | Rural | 4 May 2022 | |
| 127 | C. penicillata | L | Francisco Dumont | random | −17.50288 | −44.12984 | Rural | 7 July 2022 | |
| 128 | C. penicillata | L | Francisco Dumont | random | −17.31509 | −44.23189 | Urban | 7 July 2022 | |
| 129 | C. penicillata | L | Francisco Dumont | random | −17.50288 | −44.12984 | Rural | 10 July 2022 | |
| 130 | C. penicillata | L | Montes Claros | random | −16.73397 | −43.87953 | Urban | 12 July 2022 | |
| 131 | C. penicillata | L | Brasília de Minas | random | −16.21072 | −44.43640 | Urban | 7 July 2022 | |
| 132 | C. penicillata | L | Salinas | random | −16.15637 | −42.30730 | Rural | 13 July 2022 | |
| 137 | C. penicillata | L | Ubaí | random | −16.28485 | −42.26081 | Rural | 11 September 2022 | |
| 138 | C. penicillata | L | Ubaí | random | −16.28534 | −44.78366 | Rural | 12 September 2022 | |
| 139 | C. penicillata | L | Ubaí | random | −16.28504 | −44.78340 | Urban | 13 September 2022 | |
| 140 | C. penicillata | L | Ubaí | random | −16.28469 | −44.78361 | Urban | 14 September 2022 | |
| 141 | C. penicillata | L | Montes Claros | random | −16.72416 | −43.83795 | Urban | 20 October 2022 | |
| 142 | C. penicillata | L | Guaraciama | random | −17.01429 | −43.66866 | Urban | 20 October 2022 | |
| 143 | C. penicillata | L | Juramento | random | −16.84982 | −43.58690 | Urban | 20 October 2022 | |
| 144 | C. penicillata | L | São João da Lagoa | random | −16.85326 | −44.34971 | Urban | 20 October 2022 | |
| 145 | C. penicillata | L | São João da Lagoa | random | −16.85275 | −44.35031 | Urban | 20 October 2022 | |
| 146 | C. penicillata | L | São João da Lagoa | random | −16.85341 | −44.34989 | Urban | 20 October 2022 | |
| 147 | C. penicillata | L | São João da Lagoa | random | −16.85311 | −44.35059 | Urban | 20 October 2022 | |
| 148 | C. penicillata | L | Francisco Sá | random | −16.26377 | −43.58839 | Rural | 20 October 2022 | |
| 149 | C. penicillata | L | Francisco Sá | random | −16.26385 | −43.58842 | Rural | 20 October 2022 | |
| 150 | C. penicillata | L | São João da Lagoa | random | −16.85277 | −44.35047 | Urban | 23 August 2022 | |
| 151 | C. penicillata | L | São João da Lagoa | random | −16.85101 | −44.34995 | Urban | 23 August 2022 | |
| 152 | C. penicillata | L | São João do Pacuí | random | −16.53487 | −44.53158 | Urban | 22 September 2022 | |
| 153 | C. penicillata | L | São João do Pacuí | random | −16.53506 | −44.53145 | Urban | 29 August 2022 | |
| 154 | C. penicillata | L | São João do Pacuí | random | −16.53537 | −44.53156 | Urban | 2 September 2022 | |
| 155 | C. penicillata | L | São João da Lagoa | random | −16.85102 | −44.34995 | Urban | 15 September 2022 | |
| 156 | C. penicillata | L | Jequitaí | random | −17.23142 | −44.44344 | Urban | 20 October 2022 | |
| 157 | C. penicillata | L | Jequitaí | random | −17.23142 | −44.44344 | Urban | 20 October 2022 | |
| 158 | C. penicillata | L | São João da Lagoa | random | −16.85309 | −44.35072 | Urban | 22 October 2022 | |
| 159 | C. penicillata | L | Jequitaí | random | −17.23079 | −44.44417 | Urban | 24 October 2022 | |
| 160 | C. penicillata | L | Jequitaí | random | −17.23079 | −44.44417 | Urban | 25 October 2022 | |
| 161 | A. caraya | L | Salinas | random | −16.16133 | −42.31082 | Rural | 26 October 2022 | |
| 162 | C. penicillata | L | Salinas | random | −16.16287 | −42.29934 | Urban | 14 October 2022 | |
| 166 | C. penicillata | O | Bonito de Minas | random | −15.34867 | −44.90012 | Sylatic | 10 February 2023 | |
| 167 | C. penicillata | O | Bonito de Minas | random | −15.34867 | −44.90012 | Sylatic | 10 February 2023 | |
| 168 | C. penicillata | O | Bonito de Minas | random | −15.34867 | −44.90012 | Sylatic | 10 February 2023 | |
| 169 | C. geoffroyi | O | Teófilo Otoni | random | −17.89125 | −41.52602 | Rural | 8 March 2023 | |
| 170 | C. geoffroyi | O | Teófilo Otoni | random | −17.89125 | −41.52602 | Rural | 8 March 2023 | |
| 171 | C. geoffroyi | O | Teófilo Otoni | random | −17.89125 | −41.52602 | Rural | 8 March 2023 | |
| 172 | C. geoffroyi | O | Teófilo Otoni | random | −17.89125 | −41.52602 | Rural | 8 March 2023 | |
| 173 | A. caraya | O | Almenara | random | −16.18280 | −40.69075 | Urban | 12 March 2023 | |
| 174 | A. caraya | O | Almenara | random | −16.18280 | −40.69075 | Urban | 12 March 2023 | |
| 175 | C. kuhlii | O | Almenara | random | −16.15744 | −40.69302 | Urban | 13 March 2023 | |
| 176 | C. kuhlii | O | Almenara | random | −16.15744 | −40.69302 | Urban | 13 March 2023 | |
| 177 | A. caraya | O | Almenara | random | −16.15744 | −40.69302 | Urban | 13 March 2023 | |
| 178 | C. kuhlii | O | Almenara | random | −16.18280 | −40.69075 | Urban | 14 March 2023 | |
| 179 | C. penicillata | O | Salinas | random | −16.15475 | −42.30760 | Rural | 23 February 2023 | |
| 180 | C. penicillata | O | Salinas | random | −16.06398 | −42.24160 | Rural | 18 May 2023 | |
| 182 | C. penicillata | O | Salinas | random | −16.15475 | −42.30760 | Rural | 20 June 2023 | |
| 183 | C. penicillata | O | Salinas | random | −16.15475 | −42.30760 | Rural | 13 July 2023 | |
| 185 | C. penicillata | O | Salinas | random | −16.15475 | −42.30760 | Rural | 25 July 2023 | |
| 186 | C. penicillata | O | Araçuaí | random | −16.73491 | −42.06367 | Rural | 20 September 2023 | |
| 187 | C. penicillata | L | Salinas | random | −16.15475 | −42.30760 | Rural | 5 October 2023 | |
| SSMs | D1 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P1 | −18.24400 | −43.62300 | Urban | 25 June 2021 |
| D2 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 25 June 2021 | |
| D3 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P6 | −18.25185 | −43.58418 | Urban | 25 June 2021 | |
| D4 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 25 June 2021 | |
| D5 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 2 July 2021 | |
| D6 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 2 July 2021 | |
| D7 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 2 July 2021 | |
| D8 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P2 | −18.24907 | −43.61668 | Urban | 9 July 2021 | |
| D9 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 9 July 2021 | |
| D10 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 9 July 2021 | |
| D11 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P6 | −18.25185 | −43.58418 | Urban | 9 July 2021 | |
| D12 | R. novergicus | O; BS; B; L; S; Lv; F | Diamantina | P9 | −18.22757 | −43.61225 | Urban | 16 July 2021 | |
| D13 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P2 | −18.24907 | −43.61668 | Urban | 16 July 2021 | |
| D14 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 16 July 2021 | |
| D15 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 16 July 2021 | |
| D16 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 16 July 2021 | |
| D17 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P6 | −18.25185 | −43.58418 | Urban | 16 July 2021 | |
| D18 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P7 | −18.24375 | −43.59173 | Urban | 16 July 2021 | |
| D19 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 23 July 2021 | |
| D20 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 29 July 2021 | |
| D21 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 29 July 2021 | |
| D22 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P6 | −18.25185 | −43.58418 | Urban | 29 July 2021 | |
| D23 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 30 July 2021 | |
| D24 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P7 | −18.24375 | −43.59173 | Urban | 6 August 2021 | |
| D25 | R. rattus | O; AS; B; L; S; Lv; F | Diamantina | P8 | −18.23735 | −43.59505 | Urban | 6 August 2021 | |
| D26 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P8 | −18.23735 | −43.59505 | Urban | 27 August 2021 | |
| D27 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P2 | −18.24907 | −43.61668 | Urban | 2 September 2021 | |
| D28 | M. musculus | O; BS; B; L; S; Lv; F | Diamantina | P2 | −18.24907 | −43.61668 | Urban | 2 September 2021 | |
| D29 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P3 | −18.25642 | −43.60078 | Urban | 2 September 2021 | |
| D30 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P4 | −18.25900 | −43.58435 | Urban | 2 September 2021 | |
| D31 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 2 September 2021 | |
| D32 | D. albiventris | O; BS; B; L; S; Lv; F | Diamantina | P5 | −18.25650 | −43.58195 | Urban | 2 September 2021 | |
| D33 | R. rattus | O; BS; B; L; S; Lv; F | Diamantina | P8 | −18.23735 | −43.59505 | Urban | 3 September 2021 | |
| D34 | R. novergicus | O; BS; B; L; S; Lv; F | Diamantina | P9 | −18.22757 | −43.61225 | Urban | 15 September 2021 | |
| D35 | R. novergicus | O; BS; B; L; S; Lv; F | Diamantina | P9 | −18.22757 | −43.61225 | Urban | 15 September 2021 | |
| D36 | R. novergicus | O; BS; B; L; S; Lv; F | Diamantina | P10 | −18.22757 | −43.61225 | Urban | 16 September 2021 | |
| D37 | R. novergicus | O; BS; B; L; S; Lv; F | Diamantina | P10 | −18.24907 | −43.61668 | Urban | 24 September 2021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida-Souza, P.A.; Silva, T.G.M.; Penha, G.B.; de Jesus Teixeira, T.; Oliveira-Silva, R.; Celestino, I.A.; Gonçalves-dos-Santos, M.E.; de Oliveira, C.H.; dos Santos Nunes Ferreira, A.; Gusmão, E.M.; et al. One Health Surveillance for SARS-CoV-2 in Non-Human Primates and Small Mammals in Minas Gerais, Brazil. Pathogens 2025, 14, 356. https://doi.org/10.3390/pathogens14040356
Almeida-Souza PA, Silva TGM, Penha GB, de Jesus Teixeira T, Oliveira-Silva R, Celestino IA, Gonçalves-dos-Santos ME, de Oliveira CH, dos Santos Nunes Ferreira A, Gusmão EM, et al. One Health Surveillance for SARS-CoV-2 in Non-Human Primates and Small Mammals in Minas Gerais, Brazil. Pathogens. 2025; 14(4):356. https://doi.org/10.3390/pathogens14040356
Chicago/Turabian StyleAlmeida-Souza, Pedro Augusto, Thamires Gabriele Macedo Silva, Gabriele Barbosa Penha, Thaynara de Jesus Teixeira, Ramon Oliveira-Silva, Iago Alves Celestino, Maria Eduarda Gonçalves-dos-Santos, Cirilo Henrique de Oliveira, Alice dos Santos Nunes Ferreira, Emerson Márcio Gusmão, and et al. 2025. "One Health Surveillance for SARS-CoV-2 in Non-Human Primates and Small Mammals in Minas Gerais, Brazil" Pathogens 14, no. 4: 356. https://doi.org/10.3390/pathogens14040356
APA StyleAlmeida-Souza, P. A., Silva, T. G. M., Penha, G. B., de Jesus Teixeira, T., Oliveira-Silva, R., Celestino, I. A., Gonçalves-dos-Santos, M. E., de Oliveira, C. H., dos Santos Nunes Ferreira, A., Gusmão, E. M., Ottone, V. d. O., Simonini-Teixeira, D., Campos, F. S., Roehe, P. M., de Oliveira, L. C., Teixeira, M. M., Abreu, F. V. S. d., & Oliveira, D. B. d. (2025). One Health Surveillance for SARS-CoV-2 in Non-Human Primates and Small Mammals in Minas Gerais, Brazil. Pathogens, 14(4), 356. https://doi.org/10.3390/pathogens14040356








