Resistance Landscape and Clonal Dynamics of ESKAPE Pathogens in Bloodstream Infections: A Multicenter Study from Mexico
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Setting
2.2. Bacterial Isolation
2.3. Antimicrobial Susceptibility Testing
2.4. Genetic Identification of Resistance Genes in ESKAPE Bacteria
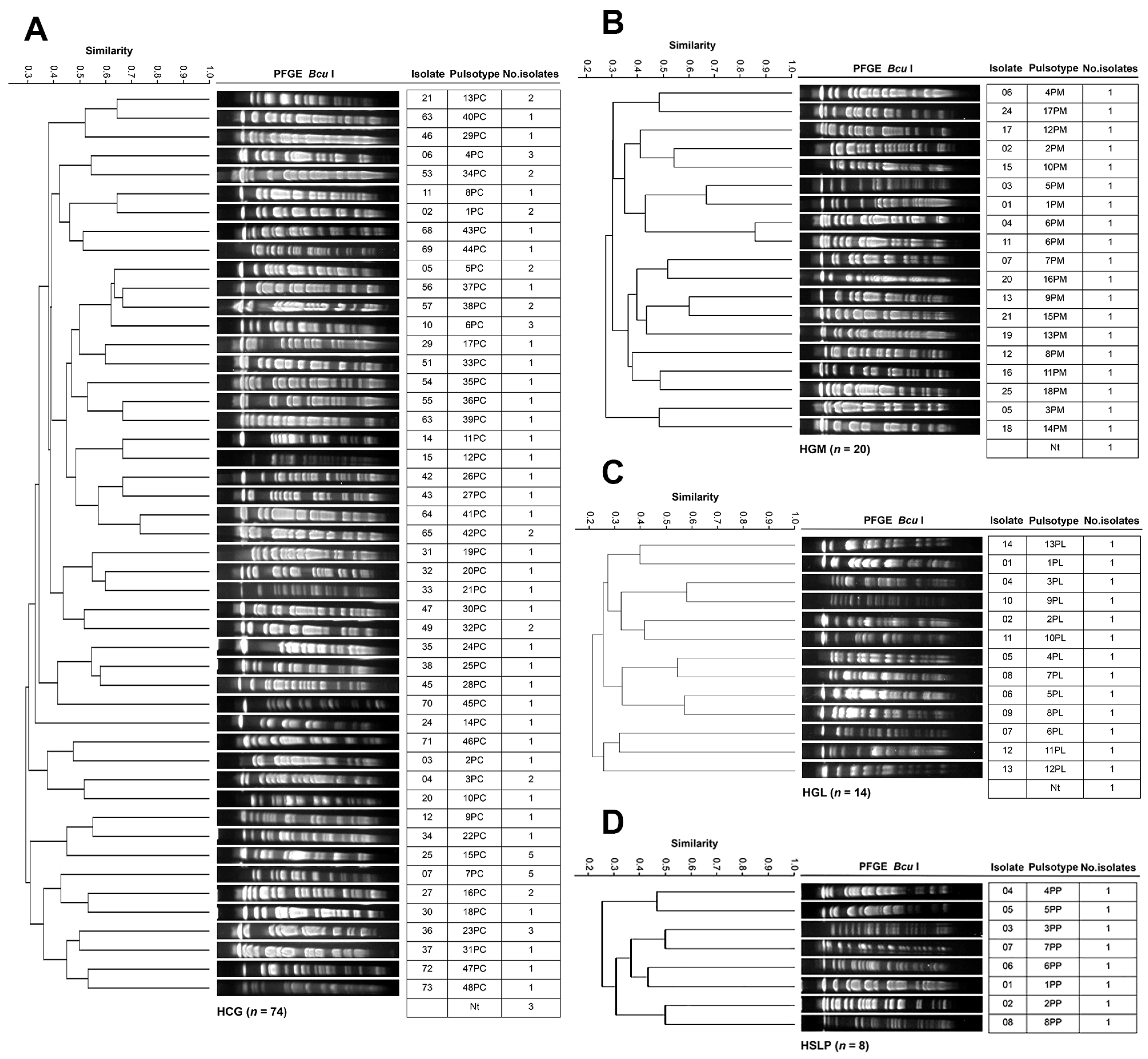
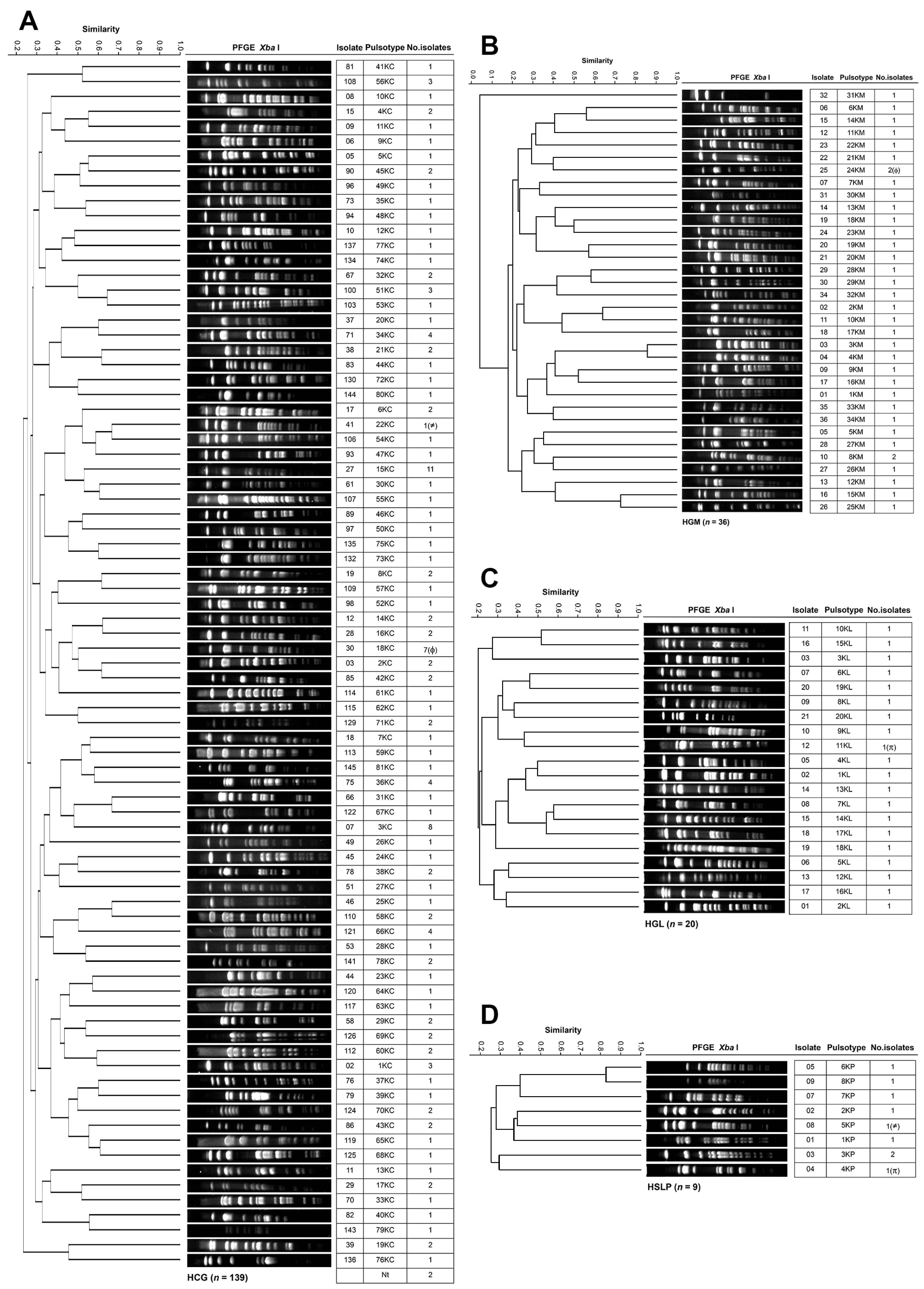
2.5. Pulsed-Field Gel Electrophoresis (PFGE)
2.6. Statistical Analysis
3. Results
3.1. Clinical Isolates of ESKAPE Pathogens
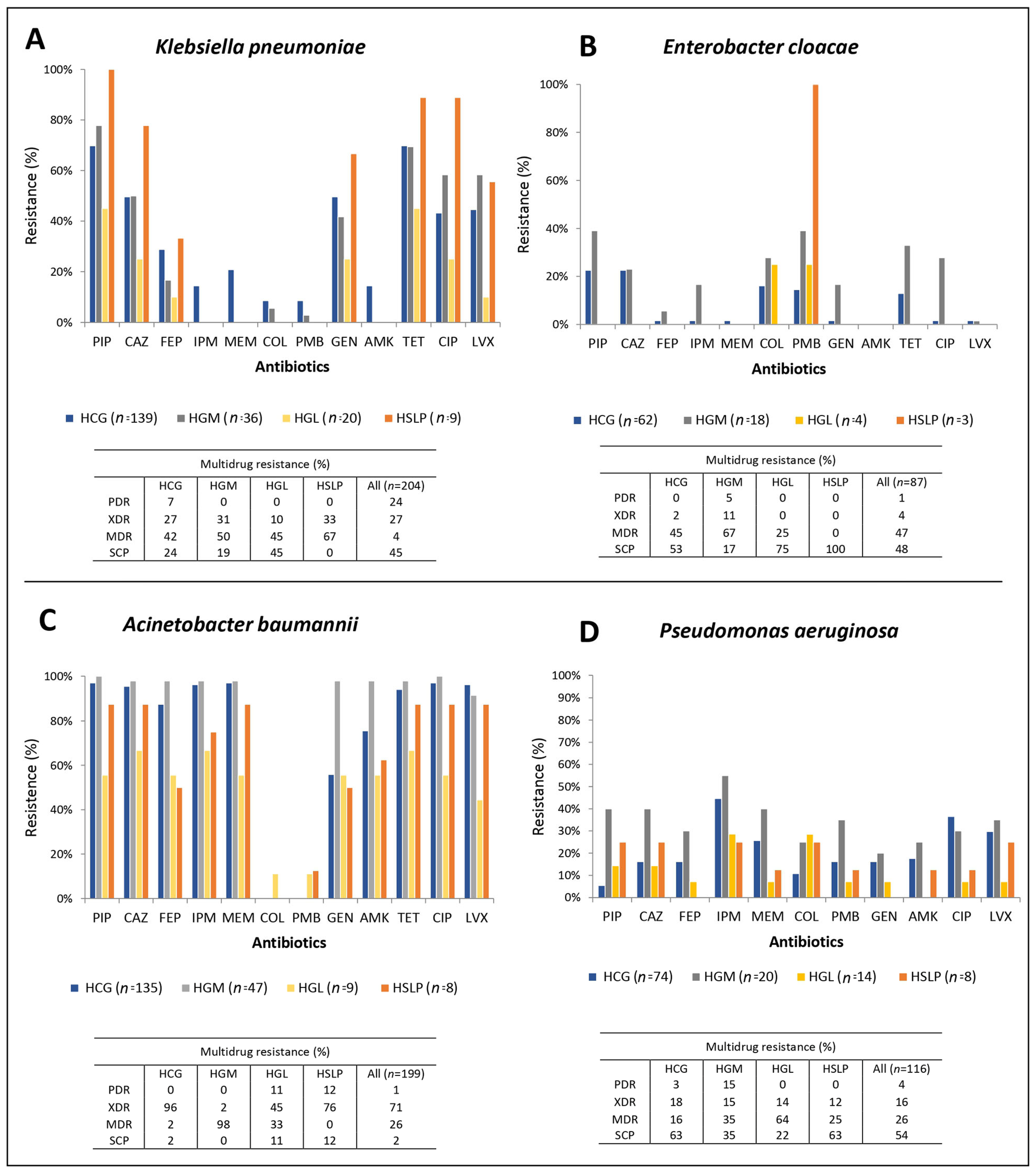
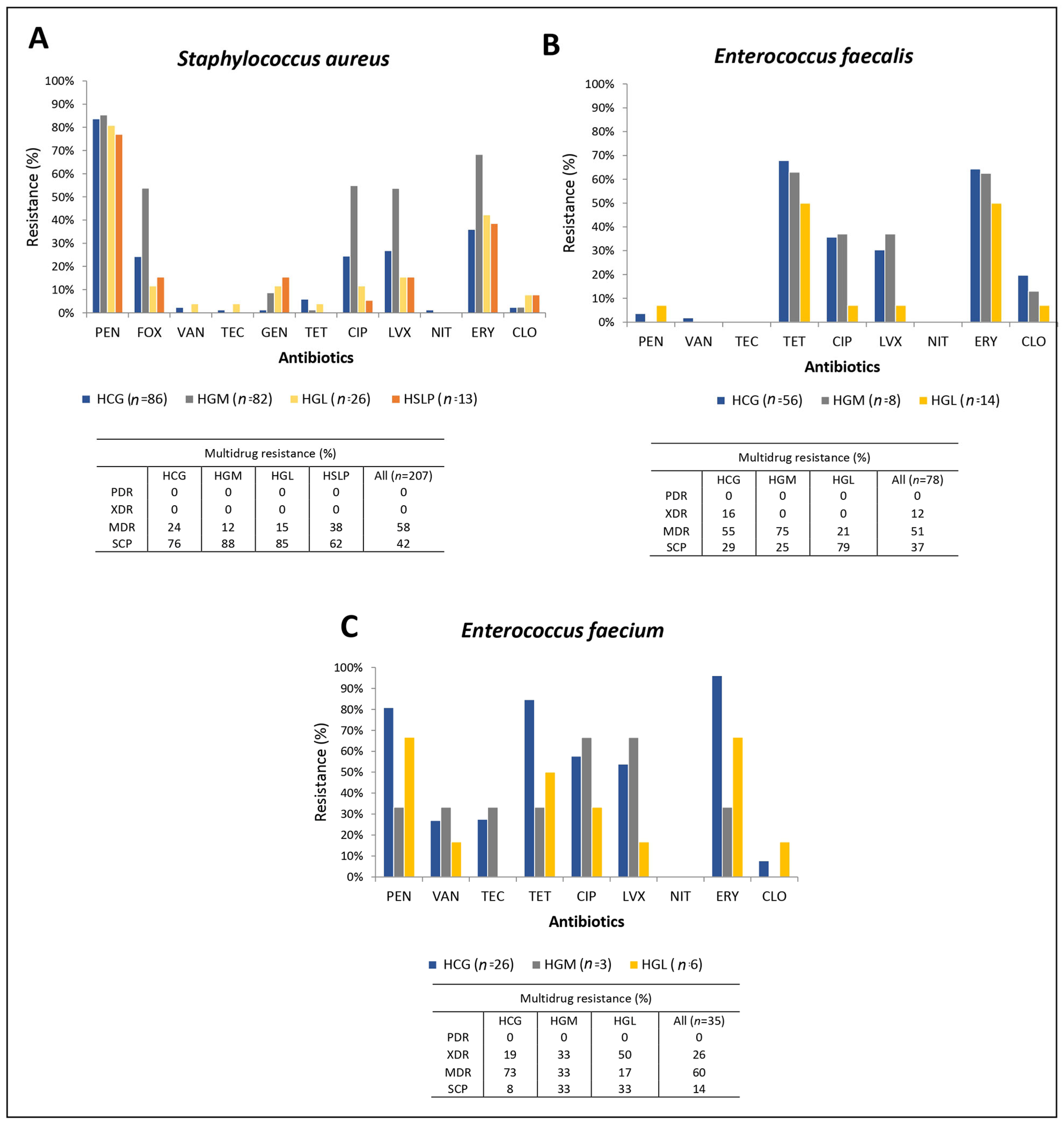
3.2. Antimicrobial Susceptibility in the ESKAPE Pathogens
3.3. Characterization of Resistance Genes in the ESKAPE Pathogens
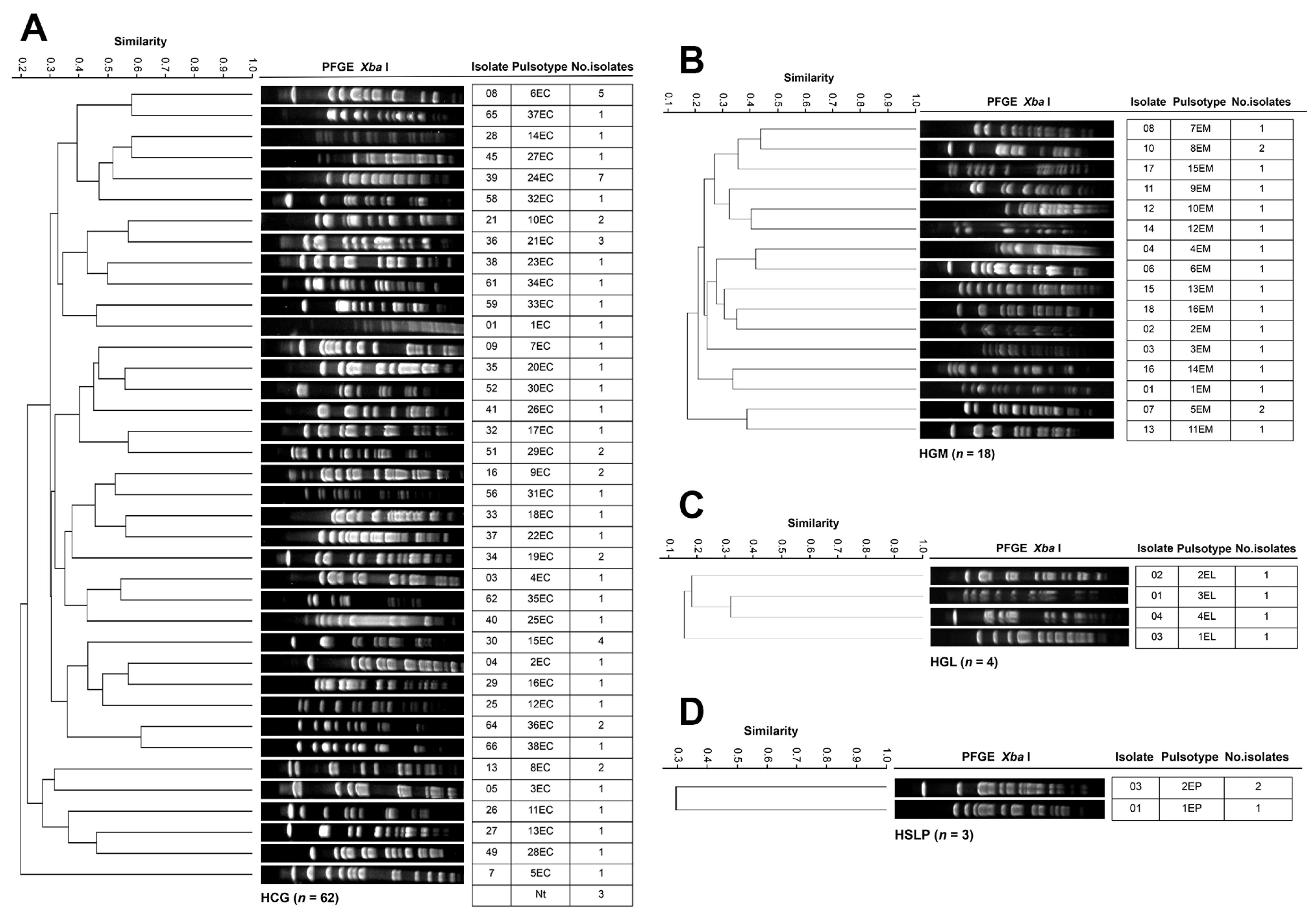
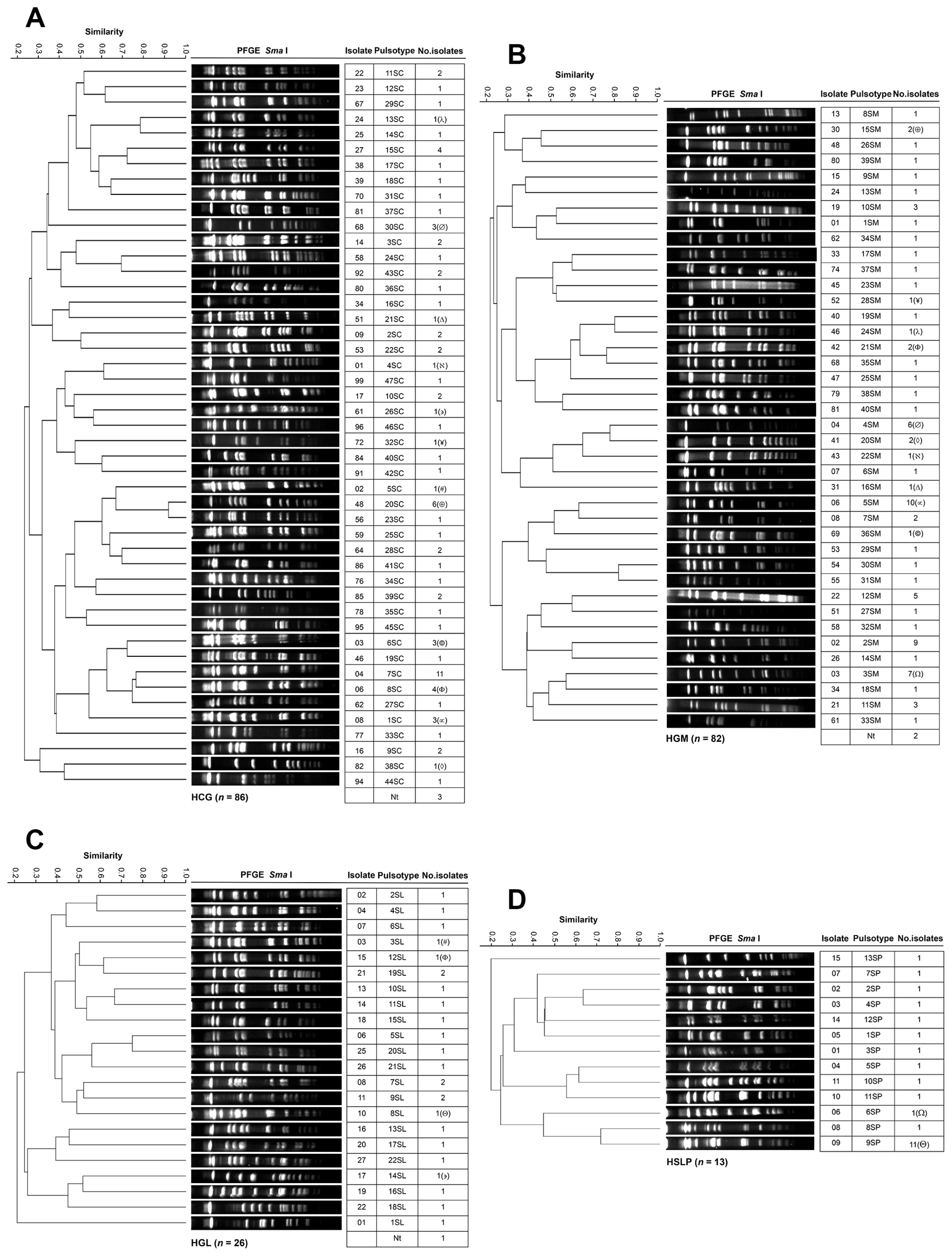
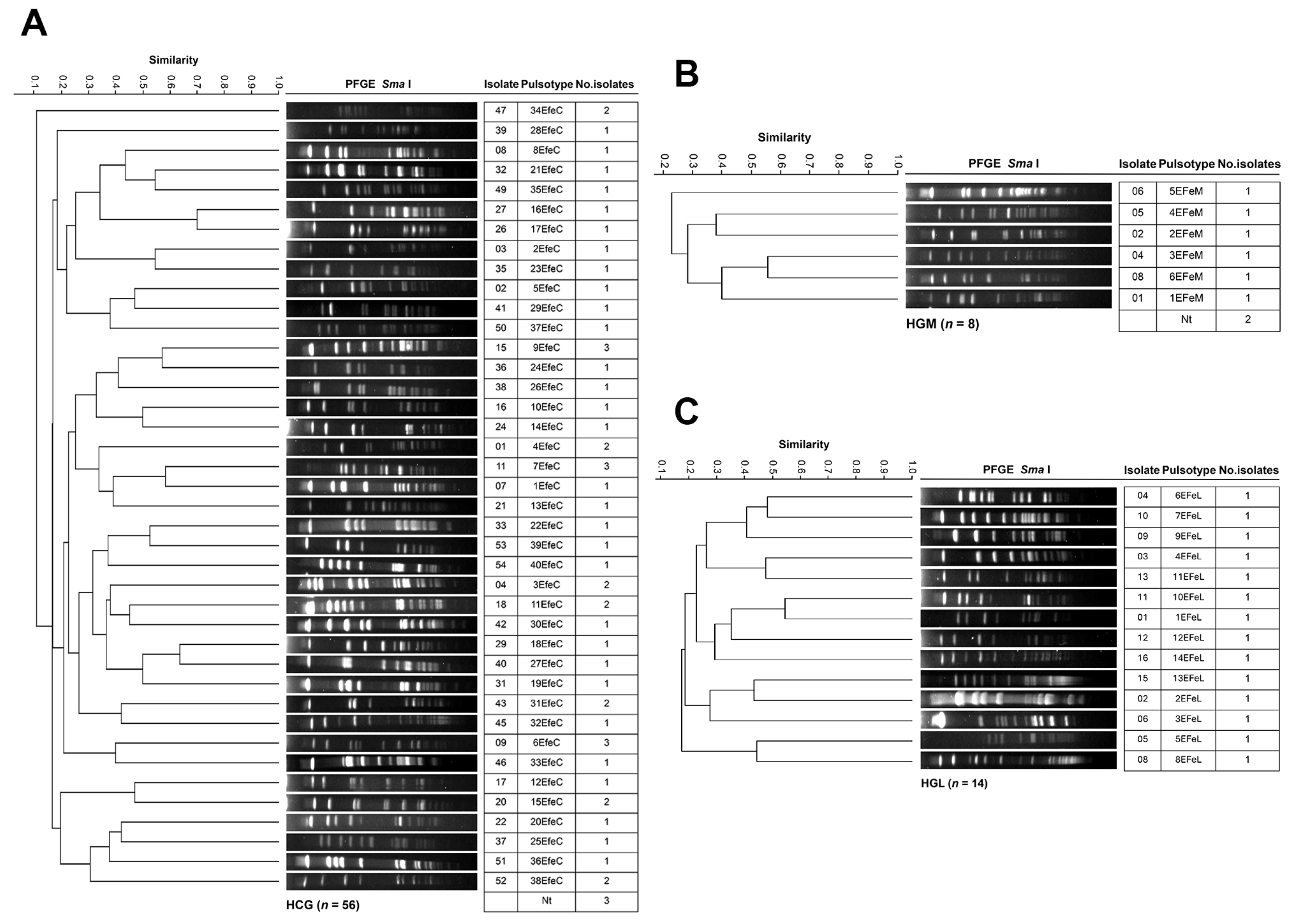
3.4. Genotyping and Clonal Diversity of ESKAPE Bacteria Across Four Mexican Hospitals
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohd Asri, N.A.; Ahmad, S.; Mohamud, R.; Mohd Hanafi, N.; Mohd Zaidi, N.F.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Mohd Noor, N.; Mustafa, F.H.; et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef]
- Aguilar, G.R.; Swetschinski, L.R.; Weaver, N.D.; Ikuta, K.S.; Mestrovic, T.; Gray, A.P.; Chung, E.; Wool, E.E.; Han, C.; Hayoon, A.G.; et al. The Burden of Antimicrobial Resistance in the Americas in 2019: A Cross-Country Systematic Analysis. Lancet Reg. Health-Am. 2023, 25, 100561. [Google Scholar] [CrossRef] [PubMed]
- De Paula, L.; Lopes, G.F.M.; Santos, F.R.D.S.; Oliveira, A.L.B.; De Pilla Varotti, F.; Da Cruz Nizer, W.S.; De Paiva, M.C.; Figueiredo, R.C.; Ferreira, J.M.S. Prevalence of ESKAPEE Pathogens as Agents of Healthcare-Related Infection in Brazil. Microbe 2025, 7, 100326. [Google Scholar] [CrossRef]
- De Prisco, M.; Manente, R.; Santella, B.; Serretiello, E.; Dell’Annunziata, F.; Santoro, E.; Bernardi, F.F.; D’Amore, C.; Perrella, A.; Pagliano, P.; et al. Impact of ESKAPE Pathogens on Bacteremia: A Three-Year Surveillance Study at a Major Hospital in Southern Italy. Antibiotics 2024, 13, 901. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: Early Implementation 2020; World Health Organization: Geneva, Switzerland, 2020; CC BY-NC-SA 3.0 IGO. [Google Scholar]
- De La Cruz-Hernández, I.; Cornejo-Juárez, P.; Tellez-Miranda, O.; Barrera-Pérez, L.; Sandoval-Hernández, S.; Vilar-Compte, D.; Velázquez-Acosta, C.; Volkow, P. Microbiology and Prevalence of E2SKAPE-Resistant Strains in Catheter-Related Bloodstream Infections in Patients with Cancer. Am. J. Infect. Control 2020, 48, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Ainza, M.L.; Fong-Coronado, P.A.; Ruiz-Bustos, E.; Castillón-Campaña, L.G.; Quintero-Reyes, I.E.; Duarte-Zambrano, L.A.; Bolado-Martínez, E. Antibiotic Resistance of ESKAPE Group-Microorganisms in Health Institutions from Hermosillo and Ciudad Obregón, Sonora, México. Front. Cell. Infect. Microbiol. 2024, 14, 1348093. [Google Scholar] [CrossRef]
- Cureño-Díaz, M.A.; Plascencia-Nieto, E.S.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Nolasco-Rojas, A.E.; Durán-Manuel, E.M.; Ibáñez-Cervantes, G.; Gómez-Zamora, E.; Tamayo-Ordóñez, M.C.; Tamayo-Ordóñez, Y.D.J.; et al. Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital. Pathogens 2024, 13, 50. [Google Scholar] [CrossRef]
- Javidnia, S.; Talebi, M.; Saifi, M.; Katouli, M.; Rastegar Lari, A.; Pourshafie, M.R. Clonal Dissemination of Methicillin-Resistant Staphylococcus aureus in Patients and the Hospital Environment. Int. J. Infect. Dis. 2013, 17, e691–e695. [Google Scholar] [CrossRef]
- Ibik, Y.E.; Ejder, N.; Sevim, E.; Rakici, E.; Tanriverdi, E.S.; Copur Cicek, A. Evaluating Molecular Epidemiology of Carbapenem Non-Susceptible Klebsiella pneumoniae Isolates with MLST, MALDI-TOF MS, PFGE. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 93. [Google Scholar] [CrossRef]
- Alcántar-Curiel, M.D.; Rosales-Reyes, R.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Toledano-Tableros, J.E.; Giono-Cerezo, S.; Garza-Villafuerte, P.; López-Huerta, A.; Vences-Vences, D.; et al. Carbapenem-Resistant Acinetobacter baumannii in Three Tertiary Care Hospitals in Mexico: Virulence Profiles, Innate Immune Response and Clonal Dissemination. Front. Microbiol. 2019, 10, 2116. [Google Scholar] [CrossRef]
- Ansari, S.; Hays, J.P.; Kemp, A.; Okechukwu, R.; Murugaiyan, J.; Ekwanzala, M.D.; Ruiz Alvarez, M.J.; Paul-Satyaseela, M.; Iwu, C.D.; Balleste-Delpierre, C.; et al. The Potential Impact of the COVID-19 Pandemic on Global Antimicrobial and Biocide Resistance: An AMR Insights Global Perspective. JAC-Antimicrob. Resist. 2021, 3, dlab038. [Google Scholar] [CrossRef] [PubMed]
- López-Jácome, L.E.; Fernández-Rodríguez, D.; Franco-Cendejas, R.; Camacho-Ortiz, A. Increment Antimicrobial Resistance During the COVID-19 Pandemic: Results from the Invifar Network. Microb. Drug Resist. 2022, 28, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Horan, T.C.; Andrus, M.; Dudeck, M.A. CDC/NHSN Surveillance Definition of Health Care–Associated Infection and Criteria for Specific Types of Infections in the Acute Care Setting. Am. J. Infect. Control 2008, 36, 309–332. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing Suppl. M100-Ed32; CLSI: Malvern, PA, USA, 2022. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: https://www.eucast.org (accessed on 7 September 2022).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Celenza, G.; Pellegrini, C.; Caccamo, M.; Segatore, B.; Amicosante, G.; Perilli, M. Spread of blaCTX-M-Type and blaPER-2 β-Lactamase Genes in Clinical Isolates from Bolivian Hospitals. J. Antimicrob. Chemother. 2006, 57, 975–978. [Google Scholar] [CrossRef]
- Bonnet, R.; Sampaio, J.L.M.; Labia, R.; Chanal, C.; Sirot, J. A Novel CTX-M b-Lactamase (CTX-M-8) in Cefotaxime-Resistant Enterobacteriaceae Isolated in Brazil. Antimicrob. Agents Chemother. 2000, 44, 1936–1942. [Google Scholar] [CrossRef]
- Alcántar-Curiel, M.D.; García-Torres, L.F.; González-Chávez, M.I.; Morfín-Otero, R.; Gayosso-Vázquez, C.; Jarillo-Quijada, M.D.; Fernández-Vázquez, J.L.; Giono-Cerezo, S.; Rodríguez-Noriega, E.; Santos-Preciado, J.I. Molecular Mechanisms Associated with Nosocomial Carbapenem-Resistant Acinetobacter baumannii in Mexico. Arch. Med. Res. 2014, 45, 553–560. [Google Scholar] [CrossRef]
- Vannuffel, P.; Gigi, J.; Ezzedine, H.; Vandercam, B.; Delmee, M.; Wauters, G.; Gala, J.-L. Specific Detection of Methicillin-Resistant Staphylococcus Species by Multiplex PCR. J. Clin. Microbiol. 1995, 33, 2864–2867. [Google Scholar] [CrossRef]
- Li, J.; Yang, L.; Huang, X.; Wen, Y.; Zhao, Q.; Huang, X.; Xia, J.; Huang, Y.; Cao, S.; Du, S.; et al. Molecular Characterization of Antimicrobial Resistance and Virulence Factors of Enterococcus faecalis from Ducks at Slaughterhouses. Poult. Sci. 2022, 101, 101646. [Google Scholar] [CrossRef]
- Sawant, A.; Gillespie, B.; Oliver, S. Antimicrobial Susceptibility of Coagulase-Negative Staphylococcus Species Isolated from Bovine Milk. Vet. Microbiol. 2009, 134, 73–81. [Google Scholar] [CrossRef]
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J.W. Multiplex PCR for Detection of Aminoglycoside Resistance Genes in Enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
- Vliegenthart, J.S.; Gaalen, P.A.G.K.; Van De Klundert, J.A.M. Identification of Three Genes Coding for Aminoglycoside-Modifying Enzymes by Means of the Polymerase Chain Reaction. J. Antimicrob. Chemother. 1990, 25, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Donabedian, S.M.; Thal, L.A.; Hershberger, E.; Perri, M.B.; Chow, J.W.; Bartlett, P.; Jones, R.; Joyce, K.; Rossiter, S.; Gay, K.; et al. Molecular Characterization of Gentamicin-Resistant Enterococci in the United States: Evidence of Spread from Animals to Humans through Food. J. Clin. Microbiol. 2003, 41, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Agerso, Y.; Gerner–Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of Antimicrobial Resistance Phenotypes and Resistance Genes in Enterococcus faecalis and Enterococcus faecium from Humans in the Community, Broilers, and Pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef]
- CDC. Modified Pulse-Net Procedure for Pulsed-field Gel Electrophoresis of Select Gram Negative Bacilli. In Document. No.: CEM.TE.C.0002; Center for Disease Control and Prevention: Atlanta, GA, USA, 2014. [Google Scholar]
- CDC. Unified Pulsed-Field Gel Electrophoresis (PFGE) Protocol for Gram Positive Bacteria. In Division of Healthcare Quality Promotion; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2012. [Google Scholar]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- PUCRA, UNAM Resistencia Antimicrobiana 2017–2022. Boletín de Vigilancia Epidemiológica de La Red PUCRA 2023. Available online: https://www.puis.unam.mx/divulgacion/docs/pucra2023.pdf (accessed on 18 May 2024).
- Garza-González, E.; Morfín-Otero, R.; Mendoza-Olazarán, S.; Bocanegra-Ibarias, P.; Flores-Treviño, S.; Rodríguez-Noriega, E.; Ponce-de-León, A.; Sanchez-Francia, D.; Franco-Cendejas, R.; Arroyo-Escalante, S.; et al. A Snapshot of Antimicrobial Resistance in Mexico. Results from 47 Centers from 20 States during a Six-Month Period. PLoS ONE 2019, 14, e0209865. [Google Scholar] [CrossRef]
- Alcántar-Curiel, M.D.; Huerta-Cedeño, M.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Hernández-Medel, M.L.; Zavala-Pineda, M.; Morales-Gil, M.Á.; Hernández-Guzmán, V.A.; Bolaños-Hernández, M.I.; et al. Gram-Negative ESKAPE Bacteria Bloodstream Infections in Patients during the COVID-19 Pandemic. PeerJ 2023, 11, e15007. [Google Scholar] [CrossRef]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef]
- Golli, A.L.; Popa, S.G.; Ghenea, A.E.; Turcu, F.L. The Impact of the COVID-19 Pandemic on the Antibiotic Resistance of Gram-Negative Pathogens Causing Bloodstream Infections in an Intensive Care Unit. Biomedicines 2025, 13, 379. [Google Scholar] [CrossRef]
- Miranda-Novales, M.G.; Flores-Moreno, K.; López-Vidal, Y.; Rodríguez-Álvarez, M.; Solórzano-Santos, F.; Soto-Hernández, J.L.; Ponce De León-Rosales, S.; Network, U. Antimicrobial Resistance and Antibiotic Consumption in Mexican Hospitals. Salud Publica Mex. 2019, 62, 42. [Google Scholar] [CrossRef] [PubMed]
- Badran, A.A.; Elgayar, F.A.; Gouda, M.K.; Halfawy, N.M.E. Incidence of Extended Spectrum β-Lactamase Genes (ESBLs) among Community and Health Care Infection in Mansoura University Hospital, Egypt. BMC Microbiol. 2025, 25, 316. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Larios, F.; Martínez-Guerra, B.A.; López-Jácome, L.E.; Bolado-Martínez, E.; Vázquez-Larios, M.D.R.; Velázquez-Acosta, M.D.C.; Romero-Romero, D.; Mireles-Dávalos, C.D.; Quintana-Ponce, S.; Feliciano-Guzmán, J.M.; et al. Active Surveillance of Antimicrobial Resistance and Carbapenemase-Encoding Genes According to Sites of Care and Age Groups in Mexico: Results from the INVIFAR Network. Pathogens 2023, 12, 1144. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Santiago, J.; Alvarado-Delgado, A.; Rodríguez-Medina, N.; Garza-González, E.; Tellez-Sosa, J.; Duarte-Zambrano, L.; Nava-Domínguez, N.; Sohlenkamp, C.; Vences-Guzmán, M.A.; López-Jácome, L.E.; et al. Colistin-Resistant Klebsiella pneumoniae Species Complex: The Scenario in Mexico. J. Glob. Antimicrob. Resist. 2025, 43, 203–212. [Google Scholar] [CrossRef]
- De Araujo, L.G.; Cedeño, K.; Bomfim, A.P.; De Oliveira Silva, M.; Mendes, A.V.; Barberino, M.G.; Gouveia, E.L.; Bahia, F.M.M.; Dos Reis, M.G.; Reis, J.N. Carbapenem-Resistant Enterobacterales Bloodstream Infections Related to Death in Two Brazilian Tertiary Hospitals. BMC Infect. Dis. 2025, 25, 725. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Cao, X.; Zheng, J.; Zhang, Y.; Xie, H.; Li, C.; Liu, C.; Shen, H. Genome-Wide Identification and Oxacillinase OXA Distribution Characteristics of Acinetobacter spp. Based on a Global Database. Front. Microbiol. 2023, 14, 1174200. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Narimisa, N.; Mirshekar, M.; Dadgar-Zankbar, L.; Taki, E.; Navidifar, T.; Darban-Sarokhalil, D. Prevalence of Colistin Resistance in Clinical Isolates of Acinetobacter baumannii: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2024, 13, 24. [Google Scholar] [CrossRef]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Hushyar, S.; Doghaheh, H.P.; Arzanlou, M. Evaluation of Aminoglycoside- and Methicillin-Resistant Staphylococcus aureus: Phenotypic and Genotypic Insights from Clinical Specimens in Ardabil, Iran. BMC Infect. Dis. 2025, 25, 285. [Google Scholar] [CrossRef]
- Pardo, L.; Machado, V.; Cuello, D.; Aguerrebere, P.; Seija, V.; Braga, V.; Varela, G. Macrolide-Lincosamide-Streptogramin B Resistance Phenotypes and Their Associated Genotypes in Staphylococcus aureus Isolates from a Tertiary Level Public Hospital of Uruguay. Rev. Argent. Microbiol. 2020, 52, 202–210. [Google Scholar] [CrossRef]
- Rios, R.; Reyes, J.; Carvajal, L.P.; Rincon, S.; Panesso, D.; Echeverri, A.M.; Dinh, A.; Kolokotronis, S.-O.; Narechania, A.; Tran, T.T.; et al. Genomic Epidemiology of Vancomycin-Resistant Enterococcus faecium (VREfm) in Latin America: Revisiting The Global VRE Population Structure. Sci. Rep. 2020, 10, 5636. [Google Scholar] [CrossRef] [PubMed]
- Koleini, M.; Mosadegh, A.; Madadizadeh, F.; Heidari, H. Assessment of Factors Contributing to Infection Severity and High Levels of Drug Resistance in Clinical Enterococcus Isolates. Clin. Lab. Anal. 2025, 39, e70063. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Wei, Q.; Hu, Q.; Lin, X.; Chen, M.; Ye, R.; Lv, H. Characterization of Aminoglycoside Resistance and Virulence Genes among Enterococcus spp. Isolated from a Hospital in China. Int. J. Environ. Res. Public Health 2015, 12, 3014–3025. [Google Scholar] [CrossRef]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, Efflux Pumps and Multidrug Resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef] [PubMed]


| Antimicrobial Resistance | Gene | Sequence 5′–3′ | Amplicon Length (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|
| Primer sequences for Gram-negative bacteria | |||||
| Class A β-lactamase | blaTEM | F: ATGAGTATTCAACATTTTCG R: TTACCAATGCTTAATCAGTGAG | 861 | 55 | [19] |
| blaCTX−M−type | F: CGCTTTGCGATGTGCAG R: ACCGCGATATCGTTGGT | 550 | 52 | [20] | |
| blaSHV | F: ATGCGTTATATTCGCCTGTGTATT R: TTAGCGTTGCCAGTGCTCGATC | 861 | 50 | [12] | |
| Class A Carbapenemase | blaKPC | F: TCACTGTATCGCCGTCTAGTTCTG R: TTACTGCCCGTTGACGCCCAATC | 875 | 58 | [12] |
| Class B β-lactamase | blaVIM | F: GAGTGGTGAGTATCCGACAGTCAACGAAAT R: AGAGTCCTTCTAGAGAATGCGTGGGAATCT | 389 | 58 | [12] |
| blaNDM | F: GTCTGGCAGCACACTTCCTATCTC R: GTAGTGCTCAGTGTCGGCATCACC | 516 | 58 | [12] | |
| blaIMP | F: GCATTGCTACCGCAGCAGAGTCTTTG R: GCTCTAATGTAAGTTTCAAGAGTGATGC | 647 | 58 | [12] | |
| Class D β-lactamase | blaOXA−40−like | F: TCTAGTTTCTCTCAGTGCATGTTCATC R: CATTACGAATAGAACCAGACATTCC | 749 | 58 | [12] |
| blaOXA−51−like | F: ATGAACATTMAARCRCTCTTACTTA R: CTATAAAATACCTAATTMTTCTAA | 825 | 50 | [21] | |
| blaOXA-23-like | F: ATATTTTACTTGCTATGTGGTTGCTTC R: ATAATTCATTACGTATAGATGCCGGCA | 752 | 55 | This study | |
| Primer sequences for Gram-positive bacteria | |||||
| β-lactam resistance | mecA | F: TGGCTATCGTGTCACAATCG R: CTGGAACTTGTTGAGCAGAG | 310 | 52 | [22] |
| blaZ | F: AACACCTGCTGCTTTC R: CTCTTGGCGGTTTCAC | 312 | 49 | [23] | |
| Macrolides | ermA | F: ATCGGATCAGGAAAAGGACA R: CACGATATTCACGGTTTACCC | 486 | 52 | [24] |
| ermB | F: AAGGGCATTTAACGACGAAA R: CTGTGGTATGGCGGGTAAGT | 423 | 50 | [24] | |
| ermC | F: TGAAATCGGCTCAGGAAAAG R: CAAACCCGTATTCCACGATT | 272 | 52 | [24] | |
| msrA | F: TGGTACTGGCAAAACCACAT R: AAACGTCACGCATGTCTTCA | 1000 | 52 | [24] | |
| Aminoglycoside | aac(6′)-Ie-aph(2″)-Ia | F: CAGAGCCTTGGGAAGATGAAG R: CCTCGTGTAATTCATGTTCTGGC | 348 | 55 | [25] |
| aph(3)-IIIa | F: GGCTAAAATGAGAATATCACCGG R: CTTTAAAAAATCATACAGCTCGCG | 523 | 55 | [25] | |
| ant(4)-Ia | F: CAAACTGCTAAATCGGTAGAAGCC R: GGAAAGTTGACCAGACATTACGAACT | 294 | 55 | [25] | |
| acc(6′)-aph(2) | F: CCAAGAGCAATAAGGGCATA R: CACTATCATAACCACTACCG | 220 | 50 | [26] | |
| aph(3′)-III | F: GCCGATGTGGATTGCGAAAA R: GCTTGATCCCCAGTAAGTCA | 292 | 50 | [23] | |
| aph(2″)-Ic | F: GAAGTGATGGAAATCCCTTCGTG R: GCTCTAACCCTTCAGAAATCCAGTC | 627 | 55 | [27] | |
| Vancomycin | vanA | F: GCTATTCAGCTGTACTC R: CAGCGGCCATCATACGG | 783 | 54 | [23] |
| Tetracycline | tetM | F: GTGTGACGAACTTTACCGAA R: GCTTTGTATCTCCAAGAACAC | 501 | 55 | [28] |
| Microorganism | Klebsiella pneumoniae | Enterobacter cloacae | Acinetobacter baumannii | Pseudomonas aeruginosa | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital (n) | HCG (139) | HGM (36) | HGL (20) | HSLP (9) | All (204) | HCG (62) | HGM (18) | All (80) | HCG (135) | HGM (47) | HGL (9) | HSLP (8) | All (199) | HCG (74) | HGM (20) | HGL (14) | HSLP (8) | All (116) |
| β-lactam-intermediate and resistant isolates (n, %) | 97, 69.8 | 29, 80.5 | 10, 50 | 9, 100 | 143, 70 | 16, 25.8 | 7, 38.9 | 23, 28.8 | 134, 99.6 | 47, 100 | 8, 88.9 | 7, 87.5 | 196, 98.5 | 14, 18.9 | 11, 55 | 3, 21.4 | 2, 25 | 30, 25.9 |
| ESBL phenotype (%) | 67.0 | 96.5 | 40 | 100 | 72.7 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Carbapenem-intermediate and resistant isolates (n, %) | 31, 22.3 | 0, 0 | 0,0 | 0,0 | 31, 15.2 | 7, 11.3 | 9, 50 | 16, 20 | 132, 98 | 46, 97 | 6, 66.6 | 7, 87.5 | 196, 98.5 | 36, 48.6 | 17, 85 | 14, 100 | 2, 25 | 69, 59.5 |
| mCIM phenotype (%) | 100 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Microorganism | Klebsiella pneumoniae | Enterobacter cloacae | Acinetobacter baumannii | Pseudomonas aeruginosa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital | HCG | HGM | HGL | HSLP | All | HCG | HGM | All | HCG | HGM | HGL | HSLP | All | HCG | HGM | HGL | HSLP | All | |
| ESBL genes (n, %) | blaTEM | 64, 66 | 17, 58.6 | 3, 30 | 6, 66.7 | 90, 62.9 | 2, 12.5 | 1, 14.3 | 3, 13 | 42, 31 | 11, 23.4 | 4, 50 | 2, 28.6 | 59, 30.1 | 0 | 0 | 0 | 0 | 0 |
| blaSHV | 91, 93.8 | 24, 82.8 | 6, 60 | 7, 77.8 | 128, 89.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| blaCTX-M | 89, 91.8 | 25, 86.2 | 0 | 0 | 114, 79.7 | 2, 12.5 | 4, 57.1 | 6, 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Carbapenemase genes (n, %) | blaOXA-24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 72, 54 | 1, 2 | 1, 17 | 1, 14.3 | 75, 38.3 | 0 | 0 | 0 | 0 | 0 |
| blaOXA-23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15, 11 | 19, 41 | 0 | 0 | 34, 17.3 | 0 | 0 | 0 | 0 | 0 | |
| blaNDM | 21, 67.7 | 0 | 0 | 0 | 21, 67.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| blaVIM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10, 28 | 2, 11.8 | 0 | 0 | 12, 17.4 | |
| blaIMP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| blaKPC | 2, 6.5 | 0 | 0 | 0 | 2, 6.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Microorganism | Staphylococcus aureus | Enterococcus faecium | Enterococcus faecalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital | HCG (n = 86) | HGM (n = 82) | HGL (n = 26) | HSLP (n = 13) | All (207) | HCG (n = 26) | HGM (n = 3) | HGL (6) | All (n = 35) | HCG (n = 56) | HGM (n = 8) | HGL (n = 14) | All (n = 78) | |
| Cefoxitina (%) | 24.4 | 53.7 | 11.6 | 15.4 | 35.5 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Cefinase test (%) | 91.7 | 97.1 | 85.7 | 80 | 87.9 | |||||||||
| D-test (%) | 22.6 | 8.9 | 18.1 | 0 | 13.6 | |||||||||
| HLAR Test (%) | N/A | N/A | N/A | N/A | N/A | 38.5 | 0 | 16.6 | 31.4 | 30.4 | 37.5 | 35.7 | 32 | |
| Microorganism | Staphylococcus aureus | Enterococcus faecium | Enterococcus faecalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital | HCG | HGM | HGL | HSLP | All | HCG | HGM | HGL | All | HCG | HGM | HGL | All | |
| Vancomycin resistance | vanA (%) | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 100 |
| β-lactam resistance | mecA (%) | 61.9 | 100 | 66.6 | 75 | 88.6 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| blaZ (%) | 81.3 | 84.1 | 80.7 | 84.6 | 98.8 | 76.19 | 100 | 100 | 80.8 | 100 | 0 | 100 | 100 | |
| Aminoglycoside resistance | aph(3′)-IIIa (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ant(4´)-Ia (%) | 0 | 11 | 23 | 0 | 14.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| acc(6′)-aph(2) (%) | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 17 | 62.5 | 65 | 67 | 23 | 48.5 | |
| aph(3′)-III (%) | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 67 | 81.3 | 76 | 67 | 46 | 60.6 | |
| acc(6′)-Ie-aph(2’’)-Ia (%) | 0 | 89 | 16 | 0 | 37 | 80 | 0 | 17 | 56.3 | 65 | 67 | 23 | 48.5 | |
| aph(2’’)-Ic (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 6.0 | |
| Macrolide resistance | ermA (%) | 65.4 | 86 | 50 | 75 | 78.9 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D |
| ermB (%) | 3.9 | 0 | 0 | 0 | 1.1 | 80 | 100 | 80 | 81.2 | 91.7 | 43 | 55 | 77.8 | |
| ermC (%) | 11.5 | 2 | 100 | 50 | 11.1 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D | |
| msrA (%) | 0 | 4 | 25 | 0 | 3.3 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D | |
| Tetracycline resistance | tetM (%) | N/A | N/A | N/A | N/A | N/A | 100 | 100 | 100 | 100 | 97 | 57 | 72 | 87.7 |
| Genes | Hospital | ||||||
|---|---|---|---|---|---|---|---|
| HCG | HGM | HGL | HSLP | HCG | HGM | HCG | |
| K. pneumoniae | E. cloacae | A. baumannii | |||||
| blaCTX-M+ blaTEM | 0 | 2 | 0 | 0 | 1 | 1 | 0 |
| blaTEM+ blaSHV | 0 | 0 | 3 | 6 | 0 | 0 | 0 |
| blaCTX-M+ blaSHV | 19 | 7 | 0 | 0 | 0 | 0 | 0 |
| blaCTX-M+ blaSHV+ blaTEM | 48 | 14 | 0 | 0 | 0 | 0 | 0 |
| blaNDM+ blaCTX-M+ blaTEM | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| blaNDM+ blaCTX-M+ blaSHV | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| blaNDM+ blaCTX-M+ blaSHV+ blaTEM | 14 | 0 | 0 | 0 | 0 | 0 | 0 |
| blaKPC + blaCTX-M + blaSHV + blaTEM | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| blaOXA-23 + blaTEM | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| blaOXA-24 + blaTEM | 0 | 0 | 0 | 0 | 0 | 0 | 18 |
| blaOXA-23 + blaOXA-24 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| blaOXA-23 + blaOXA-24 + blaTEM | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Genes | Hospital | |||
|---|---|---|---|---|
| HCG | HGM | HGL | HSLP | |
| blaZ + mecA | 0 | 1 | 0 | 0 |
| blaZ + mecA + ermA | 11 | 42 | 0 | 2 |
| blaZ + ermA | 5 | 0 | 0 | 0 |
| blaZ + ermB | 1 | 0 | 0 | 0 |
| blaZ + ermC | 2 | 1 | 1 | 1 |
| blaZ + mecA + ermA + ermC | 0 | 0 | 2 | 1 |
| mecA + msrA | 0 | 1 | 0 | 0 |
| blaZ + ermC + msrA | 0 | 0 | 1 | 0 |
| blaZ + aac(6′)-Ie-aph(2″)-Ia + ant (4′)-Ia | 0 | 1 | 2 | 0 |
| blaZ + aac(6′)-Ie-aph(2″)-Ia | 0 | 5 | 0 | 0 |
| blaZ + ant(4′)-Ia | 0 | 0 | 1 | 0 |
| Genes | Hospital | |||||
|---|---|---|---|---|---|---|
| HCG | HGM | HGL | HCG | HGM | HGL | |
| E. faecium | E. faecalis | |||||
| vanA + tetM | 2 | 0 | 0 | 0 | 0 | 0 |
| vanA + tetM + ermB | 2 | 0 | 0 | 0 | 0 | 0 |
| vanA + ermB | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + tetM | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + tetM + ermB | 5 | 0 | 0 | 1 | 1 | 0 |
| ermB + tetM | 3 | 1 | 0 | 17 | 1 | 1 |
| blaZ + acc(6′)-aph(2) + aph(3′)-III + tetM + ermB | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + acc(6′)-aph(2) + acc(6′)-Ie-aph(2″)-Ia + ermB | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + aph(3′)-III + tetM + ermB | 1 | 0 | 3 | 0 | 0 | 0 |
| blaZ + vanA + acc(6′)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + tetM | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + vanA + acc(6´)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + ermB | 0 | 0 | 1 | 1 | 0 | 0 |
| blaZ + vanA + acc(6´)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + ermB + tetM | 1 | 0 | 0 | 0 | 0 | 0 |
| blaZ + acc(6′)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + ermB + tetM | 5 | 0 | 0 | 0 | 0 | 1 |
| aph(3′)-III + ermB + tetM | 0 | 0 | 0 | 1 | 0 | 0 |
| aph(3′)-III + aph(2”)-lc + ermB + tetM | 0 | 0 | 0 | 0 | 0 | 2 |
| acc(6′)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + ermB | 0 | 0 | 0 | 0 | 1 | 0 |
| acc(6′)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + tetM | 0 | 0 | 0 | 2 | 0 | 0 |
| acc(6′)-aph(2) + aph(3′)-III + acc(6′)-Ie-aph(2″)-Ia + ermB + tetM | 0 | 0 | 0 | 8 | 1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcántar-Curiel, M.D.; Morfín-Otero, R.; Jarillo-Quijada, M.D.; Fernández-Vázquez, J.L.; Gayosso-Vázquez, C.; Hernández-Medel, M.L.; Zavala-Pineda, M.; Morales-Gil, M.Á.; Osorio-Guzmán, M.; Quevedo-Ramos, M.A.; et al. Resistance Landscape and Clonal Dynamics of ESKAPE Pathogens in Bloodstream Infections: A Multicenter Study from Mexico. Pathogens 2025, 14, 1187. https://doi.org/10.3390/pathogens14111187
Alcántar-Curiel MD, Morfín-Otero R, Jarillo-Quijada MD, Fernández-Vázquez JL, Gayosso-Vázquez C, Hernández-Medel ML, Zavala-Pineda M, Morales-Gil MÁ, Osorio-Guzmán M, Quevedo-Ramos MA, et al. Resistance Landscape and Clonal Dynamics of ESKAPE Pathogens in Bloodstream Infections: A Multicenter Study from Mexico. Pathogens. 2025; 14(11):1187. https://doi.org/10.3390/pathogens14111187
Chicago/Turabian StyleAlcántar-Curiel, María Dolores, Rayo Morfín-Otero, Ma Dolores Jarillo-Quijada, José Luis Fernández-Vázquez, Catalina Gayosso-Vázquez, María Luisa Hernández-Medel, Manuelita Zavala-Pineda, Miguel Ángel Morales-Gil, Mónica Osorio-Guzmán, María Angelina Quevedo-Ramos, and et al. 2025. "Resistance Landscape and Clonal Dynamics of ESKAPE Pathogens in Bloodstream Infections: A Multicenter Study from Mexico" Pathogens 14, no. 11: 1187. https://doi.org/10.3390/pathogens14111187
APA StyleAlcántar-Curiel, M. D., Morfín-Otero, R., Jarillo-Quijada, M. D., Fernández-Vázquez, J. L., Gayosso-Vázquez, C., Hernández-Medel, M. L., Zavala-Pineda, M., Morales-Gil, M. Á., Osorio-Guzmán, M., Quevedo-Ramos, M. A., Pérez-González, L. F., Flores-Santos, A., Esparza-Ahumada, S., Escobedo-Sánchez, R., Rosales-Reyes, R., Toledano-Tableros, J. E., Giono-Cerezo, S., Santos-Preciado, J. I., & Rodríguez-Noriega, E. (2025). Resistance Landscape and Clonal Dynamics of ESKAPE Pathogens in Bloodstream Infections: A Multicenter Study from Mexico. Pathogens, 14(11), 1187. https://doi.org/10.3390/pathogens14111187








