Abstract
This review summarizes the current knowledge on the role of alternative σ factors in the highly invasive spirochaete Leptospira interrogans, the causative agent of leptospirosis. This globally distributed zoonosis affects both animals and humans, resulting in substantial public health and economic consequences. Together with the primary σ70, alternative σ factors provide transcriptional flexibility essential for bacterial adaptation to environmental changes and host–pathogen interactions. Comparative genomic analyses have revealed that the L. interrogans genome encodes 14 σ factors, including one housekeeping σ70-like factor and three types of alternative σ factors: σ54, σ28, and 11 predicted extracytoplasmic function (ECF, σᴱ-type) factors. This review discusses the characteristics of these regulators, with particular emphasis on the poorly understood ECF σ factors and their potential roles in gene regulation, adaptive responses, and pathogenicity.
1. Introduction
The Leptospira genus (Spirochaetes phylum) is comprised of pathogenic, intermediate, and saprophytic species. Saprophytic species are free-living and primarily inhabit soil or surface water [1], whereas pathogenic species can infect a wide range of animals and humans. Intermediate species display moderate pathogenicity in both animals and humans [2]. One of the pathogenic species is the highly invasive Leptospira interrogans, which causes leptospirosis, a zoonotic disease that has significant public health and economic impacts worldwide [3,4,5]. This disease is responsible for an estimated one million human cases and about 60,000 deaths annually [6,7]. In livestock, the economic losses are largely driven by reproductive disorders, abortion, reduced fertility, and decreased milk production, while in companion and wild animals, the disease can cause a severe systemic illness [8,9,10,11]. In humans, transmission mainly occurs through direct contact with the urine of infected animals or indirectly via contaminated water or moist soil, reflecting a strong environmental link to this disease [12]. Following host entry, Leptospira disseminates rapidly through the bloodstream and can colonize multiple organs. Clinical presentation ranges from mild flu-like symptoms to life-threatening forms, such as Weil’s syndrome, which is characterized by jaundice, renal and hepatic failure, pulmonary hemorrhage, and in some cases death [4,13]. Despite the global burden of leptospirosis, the molecular mechanisms underlying L. interrogans virulence and disease pathogenesis remain incompletely understood, primarily due to limitations in the genetic manipulation of pathogenic strains. However, recent advances in genetic and genomic tools have begun to provide new insights into these processes [14,15].
The complex life cycle of L. interrogans includes environmental persistence, shedding from chronically infected animals, host acquisition, and dissemination to the kidneys [4]. To survive, these bacteria must cope with diverse environmental and host-induced stressors, including iron limitation, temperature and pH changes, osmotic stress, and host immune defenses. These adaptive responses are largely mediated by transcriptional regulators, among which alternative σ factors play a central role by directing RNA polymerase to specific promoters and orchestrating global transcriptional programs [16,17].
In L. interrogans, comparative genomics and in silico genome-wide analyses have revealed a repertoire of 14 σ factors (Table 1), including one σ70-like housekeeping factor responsible for maintaining basal transcription necessary for growth, as well as a set of alternative σ factors: σ54, σ28, and 11 ECF σᴱ-type, which collectively regulate adaptive responses and contribute to this bacterium’s pathogenic potential [14,15]. Interestingly, unlike many other bacteria, L. interrogans lacks the RpoH (σH/σ32) factor, which is essential for heat shock responses, as well as the RpoS (σS/σ38) factor, a key regulator of general stress responses.

Table 1.
Summary of σ factors present in pathogenic L. interrogans and their corresponding σ factors in E. coli based on [18].
This review summarizes current knowledge on alternative σ factors in L. interrogans, with particular emphasis on the ECF σ factors (ECF σs), which represent the largest and most poorly understood group of transcriptional regulators. It also discusses their potential roles in adaptive responses and host–pathogen interactions, despite the limited number of studies in this research area. Nevertheless, this review organizes existing knowledge and provides valuable insights into this underexplored aspect of Leptospira biology.
2. Diversity of Alternative σ Factors in Bacteria
Alternative σ factors belong to two evolutionarily distinct families: the σ70 family, which includes the primary housekeeping σ70-like factors, and the σ54 family, also known as RpoN. Although both families are essential for transcription initiation, they differ in domain architecture (Figure 1), promoter recognition, regulatory mechanisms, and biological roles, reflecting their specialized functions in bacterial physiology and pathogenicity [19,20]. Members of the σ70 family typically contain several well-conserved regions (σ1.1–σ4), which mediate core RNA polymerase binding, promoter recognition, and DNA melting (Figure 1A). In contrast, σ54 factors have a distinct domain architecture and require interaction with bacterial enhancer-binding proteins (bEBPs) to promote DNA melting and initiate transcription (Figure 1B) [18,20]. Most bacteria possess only one representative of the σ54 family, whereas the σ70 family includes multiple diverse members, reflecting their specialized roles in adaptation to environmental changes.
Promoters recognized by σ70-family proteins bound with core RNAP, i.e., the RNAP-σ70 holoenzyme, typically contain −35 and −10 elements, and transcription initiation proceeds directly upon RNAP binding, without additional activators. In contrast, the σ54 family, recognizes distinct −24/−12 promoter elements and strictly requires bEBPs, which use ATP hydrolysis to drive DNA melting and initiate transcription (Figure 1) [18,20,21]. This energy-dependent activation enables σ54 to regulate highly specialized and often energy-intensive pathways, such as nitrogen assimilation, motility, or host adaptation in bacterial pathogens. Functionally, σ70 family members provide broad transcriptional flexibility, from basal gene expression to stress adaptation, supporting rapid responses to environmental changes.
Within the σ70 family, four major groups have been defined based on structural and functional characteristics [16,22] (Figure 1A): group 1 includes the housekeeping σ70-like factors responsible for basal transcription; group 2 (e.g., σS/RpoS) encompasses related but truncated forms governing stress adaptation and stationary phase adaptation; group 3 includes specialized regulators such as σ28 (FliA), controlling flagellar synthesis and motility, and σ32 (RpoH), which mediates the heat shock response; and group 4, which is the most diverse and numerous, is comprised of extracytoplasmic function ECF σs. Group 4 factors consist primarily of σ2 and σ4 domains, making them the smallest and most specialized σ factors. They regulate extracytoplasmic stress responses and are often controlled by cognate anti-σ factors, typically membrane-bound proteins that transmit extracellular or periplasmic signals to the cytoplasm.
Overall, the σ70 and σ54 families illustrate how bacteria balance broad transcriptional flexibility with pathway-specific regulation, which is crucial for adaptation and virulence in bacterial pathogens, including L. interrogans.
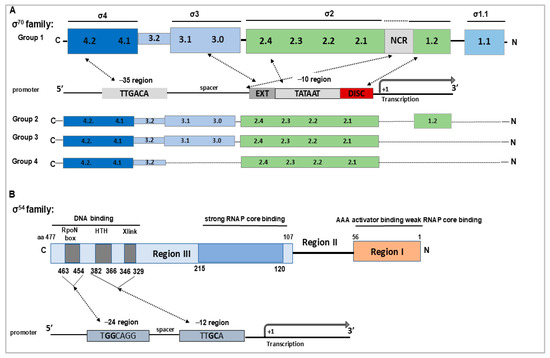
Figure 1.
Schematic representation of the functional and structural domains of the σ70 and σ54 family of σ factors. (A) Domain organization of the four groups of σ factors within the σ70 family based on [18,23]. Key elements of the RNAP-σ70 recognized promoter are also indicated: −35 element, extended −10 element (T−15G−14N), −10 element (T−12ATAAT−7), and the discriminator sequence with the optimal trinucleotide motif 5′-GGG-3′, which is involved in regulation by the stringent response factor ppGpp during nutrient stress [20,24]. Recognition of these promoter elements is mediated by the corresponding σ70 domains, as indicated by arrows based on [18,20]. The σ2 and σ4 domains are critical for binding the RNAP core enzyme and initiating transcription. Group 1 σ factors additionally contain a non-conserved region (NCR) between σ1.2 and σ2.1 implicated in an interaction with promoter DNA just upstream of the −10 element, which facilitates DNA opening at the transcription start site (+1) [25]. (B) Functional domains of σ54 and their roles in transcription initiation based on [26]. Key motifs involved in promoter recognition are also indicated: DNA crosslinking motif (Xlink), a predicted helix-turn-helix (HTH) motif, and the RpoN box motif, all located within Region III. Region II, located between Regions I and III, is not essential for σ54 function, but its deletion—partial or complete—can significantly impair promoter recognition, DNA binding, and formation of the RNAP-σ54 holoenzyme [27].
3. Alternative σ Factors in L. interrogans
3.1. σ54 (RpoN): Metabolic Adaptation and Virulence Potential
Genome-wide in silico analyses conducted by Zhukova et al. [15] identified canonical σ54-binding motifs in the promoter regions of two predicted lipoprotein genes (LIC_12503 and LIC_11935) and the ammonium transporter gene amtB (LIC_10441), which is essential for growth under low-ammonium conditions. These motifs closely resemble the E. coli conserved σ54 promoter elements at positions −24 and −12, as validated by EMSA experiments [15,28]. Additional analyses using Proscan software revealed putative σ54-type promoters upstream of genes involved in glutamine biosynthesis (glnA), alanine racemization (alr), flagellar assembly (fliO), sodium/bile acid transport (ncP1), and lipid A biosynthesis (lpxC), suggesting that σ54 may have broader roles in Leptospira [28].
It is known that transcription initiating from σ54-dependent promoters requires ATP-hydrolyzing EBPs, which remodel the promoter region to facilitate transcription initiation [20]. The L. interrogans genome encodes two such regulators, EbpA (FhlA-like; LIC_10132) and EbpB (NtrC-like; LIC_11549), whereas saprophytic Leptospira species encode only EbpA [14,28]. It has been proposed that EbpA is mainly associated with transcription of genes whose products support environmental survival in both pathogenic and saprophytic species, while EbpB may provide functions relevant to host adaptation in pathogens [14]. Both activators exhibit modular organization, comprising an N-terminal sensory domain, a central AAA+ ATPase domain required for σ54 activation, and a C-terminal helix–turn–helix (HTH) DNA-binding domain that targets enhancer elements (Figure 2) [28,29]. This dual regulatory system enables L. interrogans to respond to diverse environmental signals and coordinate σ54-dependent transcription.
Recent experimental evidence indicates that the EbpA–σ54 pathway in L. interrogans regulates expression of at least three genes, including amtB (an ammonium transport protein gene) and two putative lipoprotein-encoding genes [28]. It was demonstrated that recombinant RpoN and EbpA specifically bind to the promoter and upstream regions of these genes, respectively. Furthermore, genetic disruption of ebpA and subsequent complementation experiments confirmed that this pathway is required for transcription of the three mentioned genes, resulting in impaired growth under nitrogen-limited conditions and markedly reduced survival in water, whereas virulence in animal infection models remained unaffected. Thus, although this regulatory system is not essential for infection of mammalian hosts, it plays a crucial role in bacterial survival in aquatic environments, emphasizing its importance for environmental adaptation during the leptospiral enzootic cycle [28]. Notably, in the saprophytic species L. biflexa, σ54 directly controls a newly identified nitrogen-responsive gene, LEPBI_I1011, and is necessary for long-term environmental persistence [29].
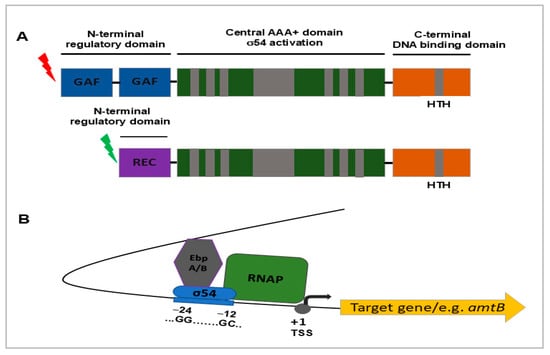
Figure 2.
Schematic representation of domain organization of the EbpA/B transcriptional activators and the predicted EbpA/B-σ54 regulatory mechanism in pathogenic L. interrogans based on [28,29]. (A) Both EbpA/B activators (AAA+-type ATPases) of σ54-dependent transcription are composed of three functional domains: an N-terminal domain that is a “signal receiver”, a central AAA+ domain responsible for σ54 activation, and a C-terminal DNA-binding domain with an HTH (helix-turn-helix) motif that directs EbpA/B to a specific activation region in DNA. EbpA and EbpB have different regulatory/sensory domains and respond to different signals. EbpA contains two putative GAF (cGMP-specific phosphodiesterases, Adenylyl cyclases and FhlA) domains at its N-terminus, while EbpB contains a REC (receiver) domain at its N-terminus that is activated by phosphorylation by a cognate histidine kinase. The central domains of EbpA/B contain seven conserved regions essential for σ54-dependent transcription. (B) Different signals received and transmitted by the N-terminal sensory domain of EbpA/EbpB to the central AAA+ domain trigger their ATPase activity, oligomerization and transcription activation of a specific set of σ54-dependent genes. RNAP—RNA polymerase core enzyme. +1 TSS—transcriptional start site.
Taken together, in L. interrogans, the EbpA–σ54 pathway primarily supports environmental survival and nitrogen metabolism, complementing observations in other spirochaetes (particularly in Borrelia burgdorferi) where similar systems contribute to host adaptation, virulence, and mammalian host infection [30,31,32]. It is worth noting that in B. burgdorferi, the response regulator Rrp2 is the sole EBP that activates RpoN (σ54), thereby initiating the RpoN–RpoS regulatory cascade essential for mammalian infection. This pathway governs expression of key virulence factors, such as OspC, DbpA, and Mlp8, and is crucial for B. burgdorferi survival and adaptation during its enzootic cycle [30,33]. Moreover, acetyl phosphate is suggested to activate the Rrp2-RpoN-RpoS pathway, supporting the concept that this molecule can serve as a global signal in bacterial pathogenesis [34].
3.2. σ28 (FliA): Motility Control and Contribution to Virulence
Genomic analyses have revealed that L. interrogans encodes a σ28 factor (FliA) [35]. It is likely involved in the regulation of late flagellar and chemotaxis genes, consistent with its role in other spirochaetes, with the exception of Borrelia, which lacks σ28. Supporting this hypothesis, genome-wide mapping of transcription start sites in L. interrogans identified σ28-binding motifs upstream of genes encoding structural components of the leptospiral endoflagellum, including sheath protein FlaA1, core proteins FlaB1 and FlaB4, and a flagellin-specific chaperone FliS [15]. The endoflagellum is indispensable for motility, enabling Leptospira to penetrate blood and tissues, thereby facilitating tissue penetration and dissemination within the host [36,37,38]. Notably, FlaA1 is required not only for motility but also plays a key role as a virulence factor during leptospirosis pathogenesis. Its loss disrupts flagellar assembly, leading to non-motile mutants with markedly reduced infectivity in animal models [37]. In addition, up-regulation of fliS expression (~1.98-fold) in L. interrogans cells upon exposure to normal guinea pig serum suggests a protective role of its product (FliS) against stress-induced aggregation and degradation of flagellar proteins [39]. Collectively, these findings indicate that σ28 may contribute to gene regulation during the early phase of spirochetemia infection.
More recently, it has been shown that in another pathogenic spirochaete, Treponema denticola, activity of σ28 is regulated by FlgM, an anti-σ factor that inhibits transcription of the late flagellar genes [40]. This discovery has expanded our understanding of σ28 regulation in spirochaetes. Remarkably, the T. denticola FlgM regulator displays distinct functional properties compared to its homologs in other bacteria: (i) it stabilizes FliA by preventing its degradation; (ii) flgM deletion results in reduced, rather than elevated, flagellin gene expression; (iii) it interacts with other regulators such as CsrA; and (iv) its lack alters flagellar number and polarity, leading to impaired motility. Sequence-based searches using the T. denticola FliA and FlgM as queries revealed FlgM-like genes in Leptospira spp., indicating the presence of a potential anti-σ factor homolog [40]. However, no direct evidence currently supports the presence of a FlgM homolog in L. interrogans. Thus, the existence of an anti-σ factor controlling σ28 activity in L. interrogans remains to be experimentally investigated.
Taken together, these observations suggest that σ28 in L. interrogans may play a similar role in controlling genes essential for motility and virulence, reflecting a conserved regulatory strategy among spirochaetes that links flagellar assembly to environmental sensing and pathogenesis.
3.3. ECF σ Factors: Diversity, Regulatory Mechanisms and Predicted Functions
Genome-wide dRNA-seq analyses have predicted the presence of 469 putative σ24-binding sites in the promoter regions of L. interrogans genes, which may be recognized by ECF σE-type factors [15]. It should be noted that σ24-dependent promoters often exhibit poorly conserved −35 elements in phylogenetically distant bacteria, making accurate prediction of σ24 targets in L. interrogans challenging. Interestingly, the number of ECF σs varies among Leptospira species: pathogenic L. interrogans encodes 11 ECF σs, whereas saprophytic species contain only five [14]. Most of these ECF σ-coding genes are located on the large chromosome (CI), with one on the small chromosome (CII) (Table 2). Based on ECF σ classification performed using the ECF hub web page (an interactive data platform for ECF σ factors) and following Casas-Pastor et al. [41], L. interrogans ECF σ factors were clustered into five groups: ECF208 (2 factors), ECF242 (2 factors), ECF16 (2 factors), ECF229, and ECF246 (one factor each), with three additional ECF σs remaining unclassified (Table 2) [18]. Two ECF σs, i.e., LIC_12490 and LIC_10144, remained ungrouped in the new ECF classification, while another ECF σ factor, LIC_10225, was not included in this re-classification and remained unclassified under the new criteria [18].

Table 2.
ECF σ factors (σE-type) from L. interrogans based on [18].
Table 2.
ECF σ factors (σE-type) from L. interrogans based on [18].
| a Gene ID | ECF Group | ECF Subgroup | NCBI Reference Sequence (Protein ID) | Number of Amino Acid Residues | Calculated Molecular Weight |
|---|---|---|---|---|---|
| Chromosome I (large) | |||||
| LIC_10144 | ungrouped | unsubgrouped | WP_000777857.1 | 174 | 20,096 |
| LIC_10225 | unclassified | unclassified | WP_001054050.1 | 301 (21-194 σ70-ECF family region) | 35,314 |
| LIC_10386 | ECF242 | ECF242s1 | WP_000951509.1 | 182 | 20,970 |
| LIC_10559 | ECF208 | ECF208s1 | WP_000988152.1 | 181 | 21,087 |
| LIC_10644 | ECF229 | ECF229s2 | WP_000777857.1 | 174 | 20,321 |
| LIC_11817 | ECF16 (SigF-like) | ECF16s14 | WP_001274737.1 | 184 | 21,620 |
| b LIC_12490 | ungrouped | ECFs21 | WP_000378482.1 | 206 | 23,881 |
| LIC_12757 | ECF229 | ECF229s1 | WP_000081452.1 | 180 | 21,105 |
| LIC_13266 | ECF246 | ECF246s2 | WP_001209037.1 | 192 | 22,906 |
| LIC_13285 | ECF208 | ECF208s2 | WP_000435437.1 | 169 | 19,550 |
| Chromosome II (small) | |||||
| LIC_20115 | ECF242 | ECF242s2 | WP_001971886.1 | 166 | 19,377 |
Note: a Gene IDs are based on ORFs from the genome sequence of L. interrogans serovar Copenhageni deposited in GenBank under accession numbers AE016823 (chromosome I; 4,277,185 bp) and AE016824 (chromosome II; 350,181 bp) [35]. Genes in bold were found to be up-regulated at elevated temperatures [42]. b ECF present in both the pathogenic L. interrogans and the saprophytic L. biflexa.
Specific functions of individual ECF σs in both pathogenic and saprophytic species remain largely unknown. It has been proposed that ECF σs unique to pathogenic Leptospira contribute to survival and adaptation within mammalian hosts [14]. The above-mentioned re-classification has provided new insights into the potential regulatory mechanisms and functional roles of L. interrogans. ECF σs. Potential functions of these factors include proton-motive force-dependent ion transport (ECF229), detoxification of harmful compounds (ECF242), antibiotic resistance (ECF246), and responses to heavy metal or oxidative stress (ECF16) (Table 2). Notably, two ECF of these σ factors, i.e., LIC_12757 (ECF229s1) and LIC_10559 (ECF208s1), were found to be 1.5-and 2-fold up-regulated at higher temperatures, respectively [42]. LIC_10559 is particularly noteworthy due to its exclusive presence in highly pathogenic Leptospira spp. [14]. We have recently demonstrated that LIC_10559 is able to activate the host’s immune system and elicit a specific antibody response in infected animals [43], providing direct evidence of its functional role during infection. These findings strongly support the involvement of LIC_10559 in leptospiral pathogenesis and highlight its potential as a promising candidate for future diagnostic applications. Furthermore, we have shown that LIC_12757 may regulate, especially under thermal stress, transcription of L. interrogans clpB gene encoding the AAA+ chaperone ClpB [44]. Of note, this chaperone, acting as a disaggregase in bacterial cells [45], is a leptospiral virulence factor [46] and belongs to L. interrogans hub proteins interacting with human proteins [47]. It is important to note that interactions between bacterial proteins and the host are critical for successful evasion of the host’s immune defenses. Consequently, it is plausible that LIC_12757 regulates transcription of genes that enhance bacterial survival under stress and facilitate escape from the host immune responses [18,43]. On the other hand, overexpression of LIC_10144 (ungrouped ECF σ) has similarly elevated clpB promoter activity, indicating that multiple ECF σs may coordinate stress responses in L. interrogans [44].
Our recent studies have also provided insights into regulatory mechanisms of ECF σs in L. interrogans [48,49]. First, we have demonstrated that LIC_12757 is autoregulated at the transcriptional level [48]. Further, our investigations revealed that LIC_12757 interacts with a putative FecR-like regulator, LIC_12756, which functions as both an anti-σ factor and a positive regulator of LIC_12757 under specific environmental conditions [49]. Interestingly, our results suggest that nutrient-limiting conditions, including iron deficiency, may act as specific signals for the LIC_12757activation. As these conditions are also encountered by Leptospira during host infection, it is likely that the LIC_12757–LIC_12756 regulatory system is active under these conditions, controlling expression of a subset of genes essential for bacterial survival within the mammalian host. Precise identification of these target genes will require further studies. According to our recently proposed regulatory model [49], under inducing conditions, LIC_12756 promotes LIC_12757 binding to the RNA polymerase core enzyme to initiate transcription and may undergo membrane-associated processing, releasing its N-terminal fragment together with the associated σ factor LIC_12757 (Figure 3) [50]. We have also shown that LIC_12756 may not only assist LIC_12757 in binding to the RNA polymerase core enzyme, but also stabilize LIC_12757 by preventing its proteolytic degradation. It is also worth mentioning here that its similarity to the E. coli FecI–FecR system suggests that the leptospiral LIC_12757–LIC_12756 regulatory system could involve a TonB-dependent receptor, such as a FecA-like protein, potentially transmitting extracellular signals to LIC_12756 and subsequently to LIC_12757. Nevertheless, previous comparative and functional genomic studies have indicated that none of the predicted TonB-dependent receptors in Leptospira contain the N-terminal extension typical for interaction with an anti-sigma factor, as observed in the E. coli FecI–FecR system [51]. Despite this, exploring such a possibility seems worthwhile.
Overall, a regulatory system in L. interrogans, consisting of the LIC_12757 ECF σ factor and the LIC_12756 FecR-like regulator, represents one of this bacterium’s strategies to appropriately adjust gene expression in response to host-induced stress.
It is worth noting that the L. interrogans genome encodes over 30 predicted regulatory proteins, including anti-σ and anti-anti-σ factors, and their modulators, which may influence ECF σ factor activity in response to environmental stimuli [14] as listed in the review by Kędzierska [18]. Additional regulatory inputs may come from two-component systems (2CS), of which L. interrogans possesses 74 genes, reflecting a complex network of environmental sensing [14].
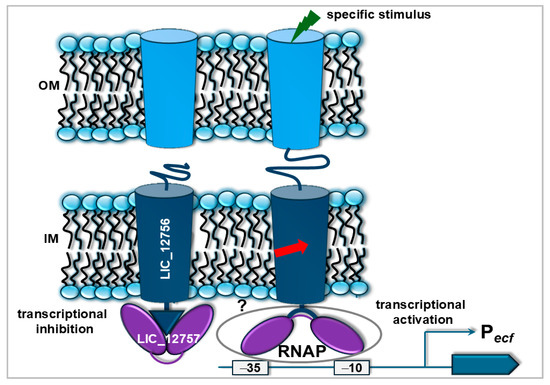
Figure 3.
The proposed model of LIC_12757 regulation mediated by interactions with LIC_12756 is based on [49]. This schematic illustrates the dual regulatory effect of LIC_12756 on the transcriptional activity of LIC_12757 under different environmental conditions. In the absence of a specific external signal, LIC_12756 inhibits the activity of LIC_12757, preventing it from carrying out its σ factor function (transcriptional inhibition). Conversely, when an appropriate environmental stimulus is present (e.g., iron limitation), LIC_12756 activates LIC_12757, possibly by promoting its interaction with the RNA polymerase core enzyme (RNAP). This interaction facilitates RNAP recruitment to target promoters, initiating gene transcription (transcriptional activation). The arrow indicates a potential processing event of the leptospiral FecR-like regulator at the membrane, involving intramembrane proteolysis and release of its N-terminal cytoplasmic fragment together with the associated σ factor [50]. The light blue cylinders denote unidentified receptors involved in both transport and signal transduction (e.g., iron uptake receptors). OM—outer membrane; IM—inner membrane.
Further studies are needed to elucidate the specific roles of each predicted ECF σ factor. In particular, comprehensive functional analyses will be essential to clarify how these factors contribute to leptospiral adaptation under environmental stresses and during mammalian host infection. Elucidating their diverse activities will not only enhance our understanding of leptospiral physiology but may also help identify potential targets for diagnostic or therapeutic applications.
4. Concluding Remarks and Future Perspectives
The presence of σ54, σ28, and multiple ECF σs highlights the layered complexity of transcriptional control in the pathogenic L. interrogans. These alternative σ factors likely act at different stages of the leptospiral life cycle: σ28 is engaged in motility and host entry, σ54 in metabolic adaptation, and ECF σs in environmental stress survival and possibly virulence. The regulatory pair LIC_12757–LIC_12756 demonstrates how a specific ECF σ factor and its cognate regulator may mediate responses to iron and nutrient limitation, as well as to host-induced stress, allowing the pathogen to adjust gene expression accordingly. Unraveling the interplay between these regulatory networks remains a major challenge, but its understanding could reveal novel strategies used by Leptospira to thrive in diverse environments and cause disease.
Despite advances in comparative genomics and bioinformatic predictions, functional understanding and characterization of alternative σ factors and regulatory elements in L. interrogans remains incomplete. The development of more efficient genetic tools, including CRISPR-based mutagenesis and transcriptional reporters, as well as RNA-seq analyses, will be essential to study genes regulated by σ54, σ28, and ECF σs. Characterization of specific ECF group regulons, such as ECF242 (FecI-like), ECF229, and ECF208, may help to uncover novel pathways involved in stress adaptation, environmental persistence, and host colonization. The use of the guinea pig model and the murine macrophage-like cell line J774.1 as an in vitro model could facilitate the investigation of the role of ECF σs in Leptospira virulence and host–pathogen interactions. It has been demonstrated that this cell line is a permissive host cell for virulent leptospires [52]. Moreover, structural and biochemical studies on interactions between ECF σs and their anti-σ partners could provide new insights into the unique regulatory mechanisms of spirochaetes. A deeper understanding of these regulatory elements in L. interrogans will not only advance our knowledge of bacterial transcriptional control but also lay the groundwork for novel strategies to combat leptospirosis.
Author Contributions
Conceptualization, S.K.-M.; Writing—original draft preparation, S.K.-M. and Z.A.; Writing—review and editing, S.K.-M.; Supervision, S.K.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
I am very grateful to Katarzyna Potrykus from the Department of Bacterial Molecular Genetics (University of Gdańsk, Faculty of Biology) for her careful and critical reading of the manuscript and for many helpful suggestions and comments. I would like to thank Tomasz Wenta for his invaluable help in preparing Figure 3.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| EBP | enhancer-binding protein |
| ECF σs | extracytoplasmic function σ factors |
| L. interrogans | Leptospira interrogans |
| RNAP | RNA polymerase core enzyme |
References
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Adler, B. History of Leptospira and leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 1–9. [Google Scholar] [CrossRef]
- Haake, D.A.; Levett, P.N. Human leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [CrossRef]
- Pal, M.; Bulcha, M.R.; Bune, W.M. Leptospirosis and one health perspective. Am. J. Public Health Res. 2021, 9, 180–183. [Google Scholar] [CrossRef]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, S.; Fernando, N.; Dreyfus, A.; Smith, C.; Rodrigo, C. Leptospirosis. Nat. Rev. Dis. Primers 2025, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.G.; Nola, L.; O’Grady, L.; More, S.J.; Doherty, L.M. Seroprevalence of Leptospira Hardjo in the irish suckler cattle population. Ir. Vet. J. 2012, 65, 8. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Animal leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99–137. [Google Scholar] [CrossRef]
- Arent, Z.; Kędzierska-Mieszkowska, S. Seroprevalence study of leptospirosis in horses in northern Poland. Vet. Rec. 2013, 172, 269. [Google Scholar] [CrossRef]
- Orjuela, A.G.; Parra-Arango, J.L.; Sarmiento-Rubiano, L.A. Bovine leptospirosis: Effects on reproduction and an approach to research in Colombia. Trop. Anim. Health Prod. 2022, 54, 251. [Google Scholar] [CrossRef]
- Casanovas-Massana, A.; Pedra, G.G.; Wunder, E.A.; Diggle, P.J.; Begon, M.; Ko, A.I. Quantification of Leptospira interrogans survival in soil and water microcosms. Appl. Environ. Microbiol. 2018, 84, e00507-18. [Google Scholar] [CrossRef]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef]
- Fouts, D.E.; Matthias, M.A.; Adhikarla, H.; Adler, B.; Amorim-Santos, L.; Berg, D.E.; Bulach, D.; Buschiazzo, A.; Chang, Y.F.; Galloway, R.L.; et al. What makes a bacterial species pathogenic?: Comparative genomic analysis of the genus Leptospira. PLoS Negl. Trop. Dis. 2016, 10, e0004403. [Google Scholar] [CrossRef] [PubMed]
- Zhukova, A.; Fernandes, L.G.; Hugon, P.; Pappas, C.J.; Sismeiro, O.; Coppée, J.-Y.; Becavin, C.; Malabat, C.; Eshghi, A.; Zhang, J.J.; et al. Genome-wide transcriptional start site mapping and sRNA identification in the pathogen Leptospira interrogans. Front. Cell. Infect. Microbiol. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gruber, T.M.; Gross, C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003, 57, 441–466. [Google Scholar] [CrossRef]
- Feklístov, A.; Sharon, B.D.; Darst, S.A.; Gross, C.A. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014, 68, 357–376. [Google Scholar] [CrossRef]
- Kędzierska-Mieszkowska, S. Sigma factors of RNA polymerase in the pathogenic spirochaete Leptospira interrogans, the causative agent of leptospirosis. FASEB J. 2023, 37, e23163. [Google Scholar] [CrossRef]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005, 69, 527–543. [Google Scholar] [CrossRef]
- Paget, M.S. Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 2015, 5, 1245–1265. [Google Scholar] [CrossRef]
- Shingler, V. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol. Rev. 2011, 35, 425–440. [Google Scholar] [CrossRef]
- Paget, M.S.; Helmann, J.D. The σ70family of sigma factors. Genome Biol. 2003, 4, 203. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Li, D.; Liu, Y.J.; Bayer, E.A.; Cui, Q.; Feng, Y.; Zhu, P. Structure of the transcription open complex of distinct σI factors. Nat. Commun. 2023, 14, 6455. [Google Scholar] [CrossRef]
- Sanchez- Vazquez, P.; Dewey, C.N.; Kitten, N.; Ross, W.; Gourse, R.L. Genome- wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 8310–8319. [Google Scholar] [CrossRef]
- Narayanan, A.; Vago, F.S.; Li, K.; Qayyum, M.Z.; Yernool, D.; Jiang, W.; Murakami, K.S. Cryo-EM structure of Escherichia coli σ70 RNA polymerase and promoter DNA complex revealed a role of σ non-conserved region during the open complex formation. J. Biol. Chem. 2018, 293, 7367–7375. [Google Scholar] [CrossRef]
- Zhang, N.; Buck, M.A. A perspective on the enhancer dependent bacterial RNA polymerase. Biomolecules 2015, 5, 1012–1019. [Google Scholar] [CrossRef]
- Southern, S.; Merrick, M. The role of region II in the RNA polymerase σ factor σN (σ54). Nucleic Acids Res. 2000, 28, 2563–2570. [Google Scholar] [CrossRef][Green Version]
- Hu, W.L.; Pappas, C.J.; Zhang, J.J.; Yang, Y.Y.; Yan, J.; Picardeau, M.; Yang, X.F. The EbpA- RpoN regulatory pathway of the pathogen Leptospira interrogans is essential for survival in the environment. Appl. Environ. Microbiol. 2017, 83, e02377-16. [Google Scholar] [CrossRef]
- Zhang, J.-J.; Hu, W.L.; Yang, Y.; Li, H.; Picardeau, M.; Yan, J.; Yang, X.F. The sigma factor σ54 is required for the long- term survival of Leptospira biflexa in water. Mol. Microbiol. 2018, 109, 63–77. [Google Scholar] [CrossRef]
- Boardman, B.K.; He, M.; Ouyang, Z.; Xu, H.; Pang, X.; Yang, X.F. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 2008, 76, 3844–3853. [Google Scholar] [CrossRef]
- Dunham-Ems, S.M.; Caimano, M.J.; Eggers, C.H.; Radolf, J.D. Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog. 2012, 8, e1002532. [Google Scholar] [CrossRef]
- Alanazi, F.; Raghunandanan, S.; Priya, R.; Yang, X.F. The Rrp2-RpoN-RpoS pathway plays an important role in the blood-brain barrier transmigration of the Lyme disease pathogen. Infect. Immun. 2023, 91, e0022723. [Google Scholar] [CrossRef]
- Yang, X.F.; Hübner, A.; Popova, T.G.; Hagman, K.E.; Norgard, M.V. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect. Immun. 2003, 71, 5012–5020. [Google Scholar] [CrossRef]
- Xu, H.; Caimano, M.J.; Lin, T.; He, M.; Radolf, J.D.; Norris, S.J.; Gheradini, F.; Wolfe, A.J.; Yang, X.F. Role of acetyl-phosphate in activation of the Rpr2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 2010, 6, e100104. [Google Scholar] [CrossRef]
- Nascimento, A.L.T.O.; Ko, A.I.; Martins, E.A.L.; Monteiro-Vitorello, C.B.; Ho, L.P.; Haake, D.A.; Verjovski-Almeida, S.; Hartskeerl, R.A.; Marques, M.D.; Oliveira, M.C.; et al. Comparative genomics of two Leptospira interrogans reveals novel insight into physiology and pathogenesis. J. Bcateriol. 2004, 186, 2164–2172. [Google Scholar] [CrossRef]
- Nakamura, S. Motility of the Zoonotic Spirochete Leptospira: Insight into Association with Pathogenicity. Int. J. Mol. Sci. 2022, 23, 1859. [Google Scholar] [CrossRef]
- Lambert, A.; Picardeau, M.; Haake, D.A.; Sermswan, R.W.; Srikram, A.; Adler, B.; Murray, G.A. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect. Immun. 2012, 80, 2019–2025. [Google Scholar] [CrossRef]
- Schons-Fonseca, L.; da Silva, J.B.; Milanez, J.S.; Domingos, R.H.; Smith, J.L.; Nakaya, H.I.; Grossman, A.D.; Ho, P.L.; da Costa, R.M. Analysis of LexA binding sites and transcriptomics in response to genotoxic stress in Leptospira interrogans. Nucleic Acids Res. 2016, 44, 1179–1191. [Google Scholar] [CrossRef]
- Patarakul, K.; Lo, M.; Adler, B. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 2010, 10, 31. [Google Scholar] [CrossRef]
- Kurniyati, K.; Chang, Y.; Guo, W.; Liu, J.; Malkowski, M.G.; Li, C. Anti-σ28 factor FlgM regulates flagellin gene expression and flagellar polarity of Treponema denticola. J. Bacteriol. 2023, 205, e00463-22. [Google Scholar] [CrossRef]
- Casas-Pastor, D.; Müller, R.R.; Jaenicke, S.; Brinkrolf, K.; Becker, A.; Buttner, M.J.; Gross, C.A.; Mascher, T.; Goesmann, A.; Fritz, G. Expansion and re-classification of the extracytoplasmic function (ECF) σ factor family. Nucleic Acids Res. 2021, 49, 986–1005. [Google Scholar] [CrossRef]
- Lo, M.; Bulach, D.M.; Powell, D.R.; Haake, D.A.; Matsunaga, J.; Paustian, M.L.; Zuerner, R.L.; Adler, B. Effects of temperature on gene expression patterns in Leptospira interrogans serowar Lai as assessed by whole-genome microarrays. Infect. Immun. 2006, 74, 5848–5859. [Google Scholar] [CrossRef]
- Kędzierska-Mieszkowska, S.; Arent, Z. Immunoreactivity of a putative ECF σ factor, LIC_10559, from Leptospira interrogans with sera from Leptospira-infected animals. Pathogens 2023, 12, 512. [Google Scholar] [CrossRef]
- Kędzierska-Mieszkowska, S.; Potrykus, K.; Arent, Z.; Krajewska, J. Identification of σE-dependent promoter upstream of clpB from the pathogenic spirochaete Leptospira interrogans by applying an E. coli two-plasmid system. Int. J. Mol. Sci. 2019, 20, 6325. [Google Scholar] [CrossRef]
- Krajewska, J.; Modrak-Wójcik, A.; Arent, Z.; Więckowski, D.; Zolkiewski, M.; Bzowska, A. Characterization of the molecular chaperone ClpB from the pathogenic spirochaete Leptospira interrogans. PLoS ONE 2017, 12, e0181118. [Google Scholar] [CrossRef]
- Lourdault, K.; Cerqueira, G.M.; Wunder Jr, E.A.; Picardeau, M. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 2011, 79, 3711–3717. [Google Scholar] [CrossRef]
- Kumar, S.; Lata, K.S.; Sharma, P.; Bhairappanavar, S.B.; Soni, S.; Das, J. Inferring pathogen-host interactions between Leptospira interrogans and Homo sapiens using network theory. Sci. Rep. 2019, 9, 1434. [Google Scholar] [CrossRef]
- Kędzierska-Mieszkowska, S.; Kędzierska, B.; Potrykus, K. LIC_12757 from the pathogenic spirochaetes Leptospira interrogans encodes an autoregulated ECF σE-type factor. Vet. Microbiol. 2024, 293, 110092. [Google Scholar] [CrossRef]
- Kędzierska-Mieszkowska, S.; Kędzierska, B.; Pardyak, L.; Arent, Z. Evidence for a putative regulatory system consisting of an ECF σE-type factor, LIC_12757, and a FecR-like σ factor regulator, LIC_12756, in the pathogenic spirochaetes Leptospira interrogans. Int. J. Mol. Sci. 2025, 26, 4994. [Google Scholar] [CrossRef]
- Yokoyama, T.; Niinae, T.; Tsumagari, K.; Imami, K.; Ishihama, Y.; Hizukuri, Y.; Akiyama, Y. The Escherichia coli S2P intramembrane protease RseP regulates ferric citrate uptake by cleaving the sigma factor regulator FecR. J. Biol. Chem. 2021, 296, 100673. [Google Scholar] [CrossRef]
- Louvel, H.; Bommezzadri, S.; Zidane, N.; Boursaux-Eude, C.; Creno, S.; Magnier, A.; Rouy, Z.; Médigue, C.; Girons, I.S.; Bouchier, C.; et al. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J. Bacteriol. 2006, 188, 7893–7904. [Google Scholar] [CrossRef]
- Merien, F.; Baranton, G.; Perolat, P. Invasion of vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect. Immun. 1997, 65, 729–738. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).