Genome-Based In Silico Analysis of the Structural and Functional Characteristics of the Type Three Secretion System (T3SS) and Core Effector Proteins in Enteropathogenic Escherichia coli (EPEC) Strains Isolated from Food-Producing Animals and Products of Animal Origin
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Determination of LEE Effector Proteins
2.3. Functional Annotation of Proteins
2.4. Validation of SMART
2.5. Analysis of Physiochemical Properties
2.6. Protein–Protein Interactions
2.7. Determination of Transmembrane Domains
2.8. Protein Structure Prediction
2.9. Multiple Sequence Alignment
3. Results
3.1. Analysis of Virulence Genes
3.2. Determination of Protein Domains
3.3. Analysis of SMART Validation
3.4. Physiochemical Properties of Encoded T3SS Proteins
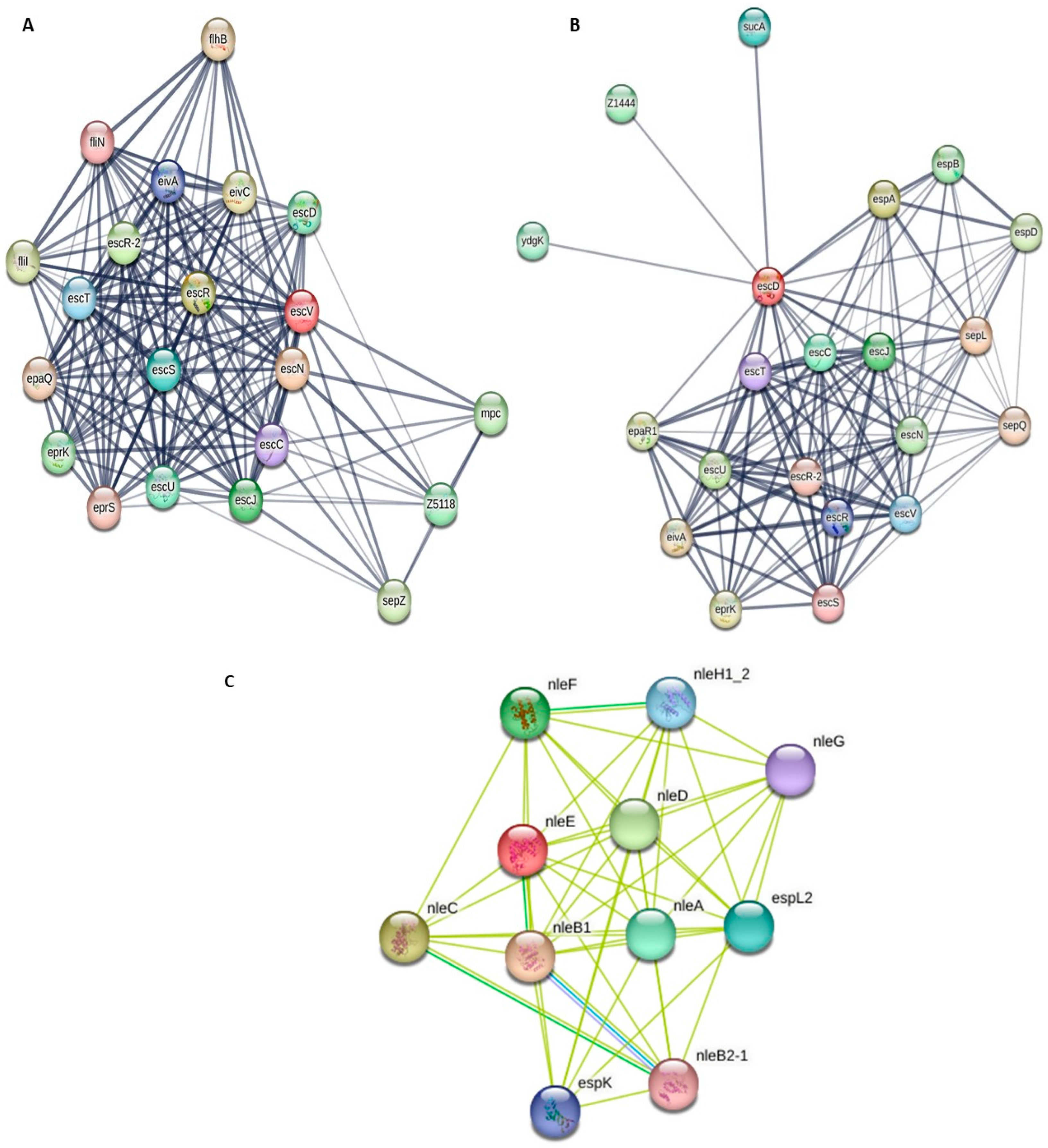
3.5. Analysis of Protein–Protein Interactions
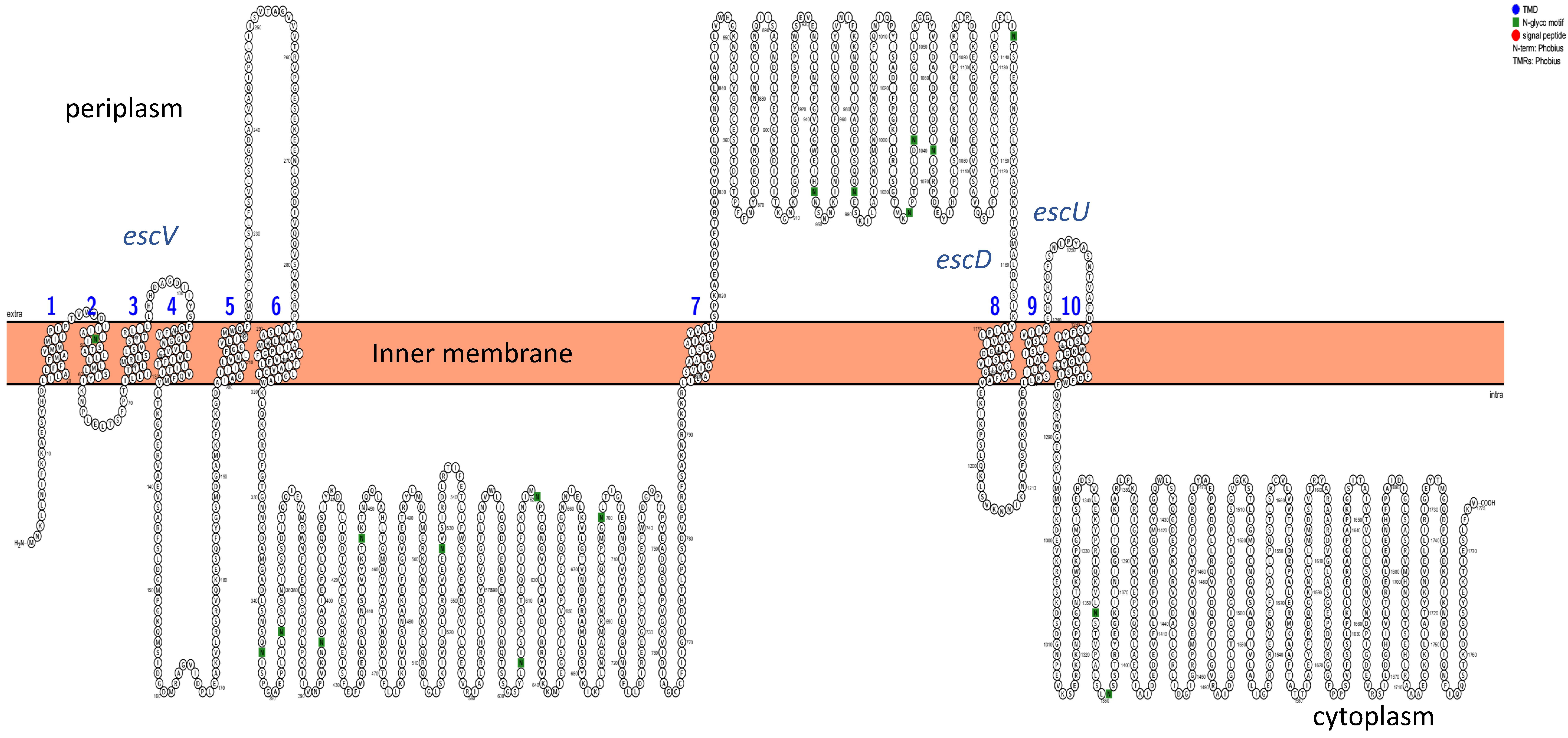
3.6. Analysis of Transmembrane Domains (TMD)
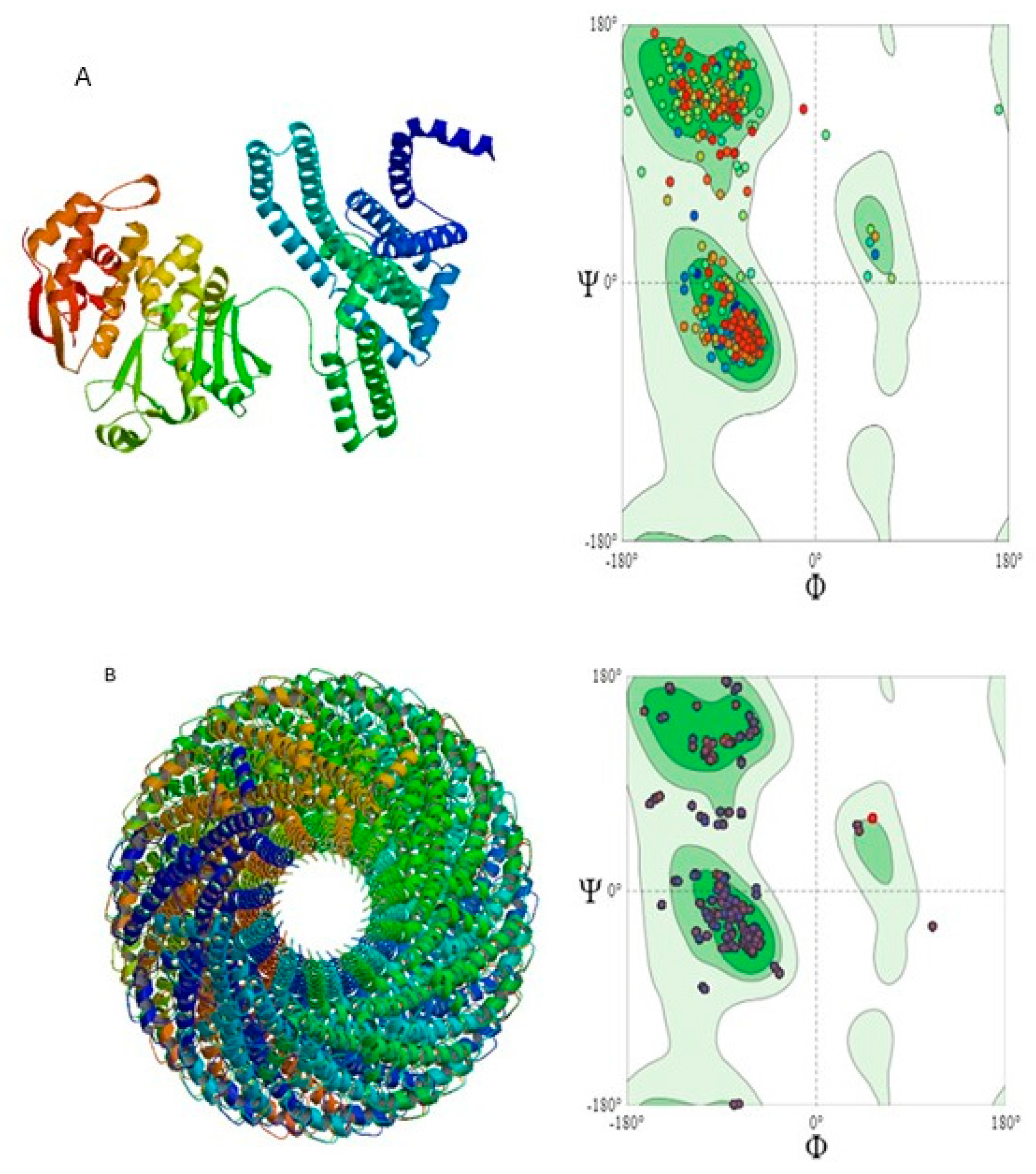
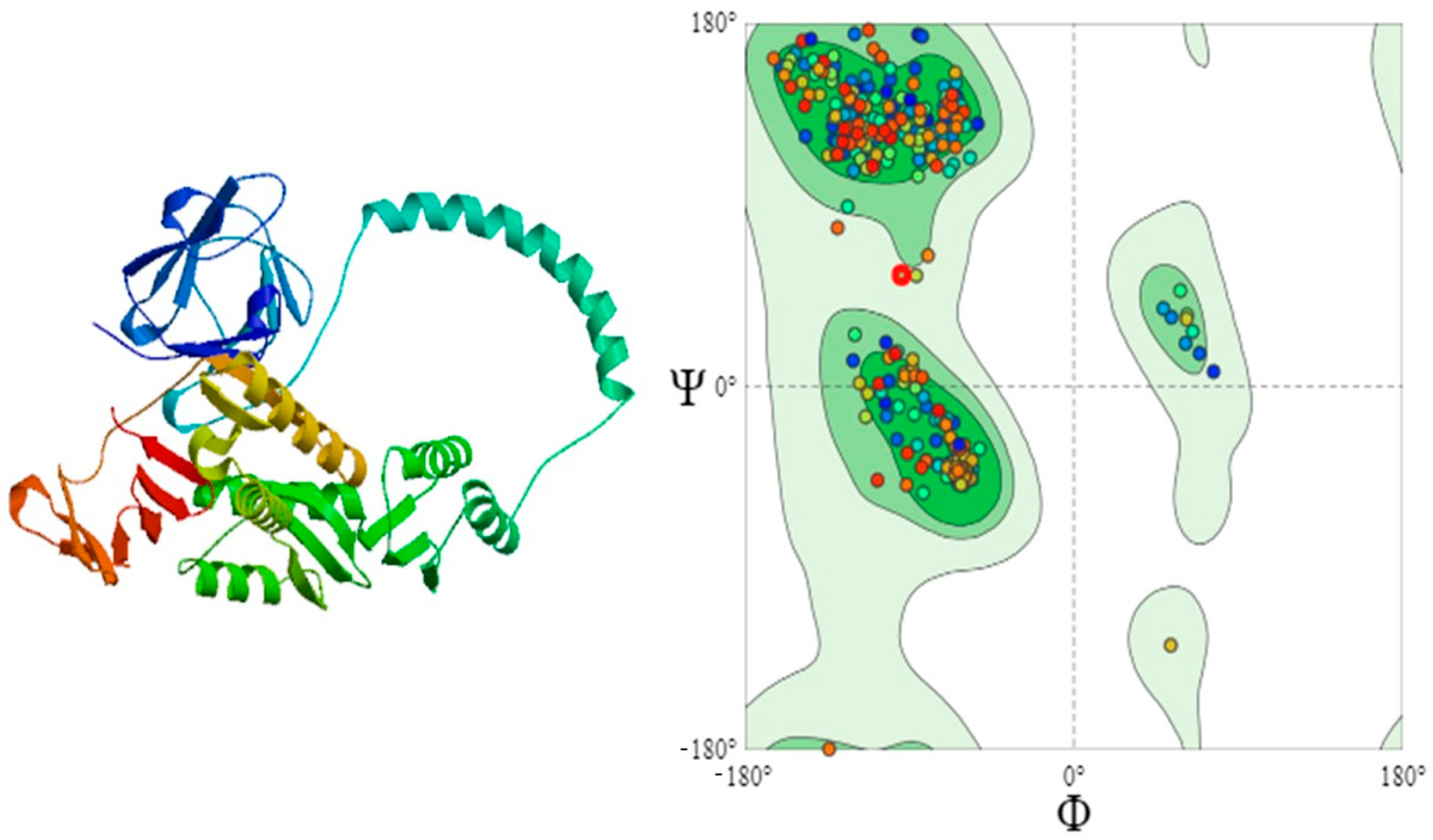
3.7. Predicted Protein Structures
3.8. Quality Assessment of 3D-Structures
3.9. Secondary Structure of the Proteins
3.10. Conservation of Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shen, J.; Zhi, S.; Guo, D.; Jiang, Y.; Xu, X.; Zhao, L.; Lv, J. Prevalence, antimicrobial resistance, and whole genome sequencing analysis of shiga toxin-producing Escherichia coli (STEC) and enteropathogenic Escherichia coli (EPEC) from imported foods in China during 2015–2021. Toxins 2022, 14, 68. [Google Scholar] [CrossRef]
- Lee, W.; Sung, S.; Ha, J.; Kim, E.; An, E.S.; Kim, S.H.; Kim, S.H.; Kim, H.Y. Molecular and genomic analysis of the virulence factors and potential transmission of hybrid enteropathogenic and enterotoxigenic Escherichia coli (EPEC/ETEC) strains isolated in South Korea. Int. J. Mol. Sci. 2023, 24, 12729. [Google Scholar] [CrossRef]
- Luna-Guevara, J.; Arenas-Hernandez, M.M.; Martínez de la Peña, C.; Silva, J.L.; Luna-Guevara, M.L. The role of pathogenic E. coli in fresh vegetables: Behavior, contamination factors, and preventive measures. Int. J. Microbiol. 2019, 2019, 2894328. [Google Scholar] [CrossRef]
- Lindberg, A.M.; Ljungh, Å.; Ahrne, S.; Löfdahl, S.; Molin, G. Enterobacteriaceae found in high numbers in fish, minced meat and pasteurised milk or cream and the presence of toxin encoding genes. Int. J. Food Microbiol. 1998, 39, 11–17. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [PubMed]
- Norazah, A.; Rahizan, I.; Zainuldin, T.; Rohani, M.Y.; Kamel, A.G.M. Enteropathogenic Escherichia coli in raw and cooked food. Southeast Asian J. Trop. Med. Public Health 1998, 29, 91–93. [Google Scholar] [PubMed]
- da Silva, Z.N.; da Cunha, A.S.; Lins, M.C.; Carneiro, L.D.A.; Almeida, A.C.D.F.; Queiroz, M.L. Isolation and serological identification of enteropathogenic Escherichia coli in pasteurized milk in Brazil. Rev. Saude Publica 2001, 35, 375–379. [Google Scholar] [CrossRef]
- Araújo, V.S.; Pagliares, V.A.; Queiroz, M.L.P.; Freitas-Almeida, A.C. Occurrence of Staphylococcus and enteropathogens in soft cheese commercialized in the city of Rio de Janeiro, Brazil. J. Appl. Microbiol. 2002, 92, 1172–1177. [Google Scholar] [CrossRef]
- Bloomfield, S.F.; Aiello, A.E.; Cookson, B.; O’Boyle, C.; Larson, E.L. The effectiveness of hand hygiene procedures in reducing the risks of infections in home and community settings including handwashing and alcohol-based hand sanitizers. Am. J. Infect. Control. 2007, 35, S27–S64. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.D.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β-lactamase (ESBL) production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, S.E.; Abia, A.L.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. Food animals as reservoirs and potential sources of multidrug-resistant diarrheagenic E. coli pathotypes: Focus on intensive pig farming in South Africa. Onderstepoort J. Vet. Res. 2022, 89, 1963. [Google Scholar] [CrossRef] [PubMed]
- Cepeda-Molero, M.; Schüller, S.; Frankel, G.; Fernández, L.Á.; Cepeda-Molero, M.; Schüller, S.; Fernández, L.Á. Systematic deletion of type III secretion system effectors in Enteropathogenic E. coli unveils the role of non-LEE effectors in a/E lesion formation. In E. coli Infections-Importance of Early Diagnosis and Efficient Treatment; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Serapio-Palacios, A.; Finlay, B.B. Dynamics of expression, secretion and translocation of type III effectors during enteropathogenic Escherichia coli infection. Curr. Opin. Microbiol. 2020, 54, 67–76. [Google Scholar] [CrossRef]
- Wagner, S.; Grin, I.; Malmsheimer, S.; Singh, N.; Torres-Vargas, C.E.; Westerhausen, S. Bacterial type III secretion systems: A complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol. Lett. 2018, 365, fny201. [Google Scholar] [CrossRef]
- Elliott, S.J.; Sperandio, V.; Girón, J.A.; Shin, S.; Mellies, J.L.; Wainwright, L.; Hutcheson, S.W.; McDaniel, T.K.; Kaper, J.B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE-and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 2000, 68, 6115–6126. [Google Scholar] [CrossRef]
- Franzin, F.M.; Sircili, M.P. Locus of enterocyte effacement: A pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. BioMed Res. Int. 2015, 2015, 534738. [Google Scholar] [CrossRef]
- Tejeda-Dominguez, F.; Huerta-Cantillo, J.; Chavez-Dueñas, L.; Navarro-Garcia, F. A novel mechanism for protein delivery by the type 3 secretion system for extracellularly secreted proteins. mBio 2017, 8, e00184-17. [Google Scholar] [CrossRef]
- Santos, A.S.; Finlay, B.B. Bringing down the host: Enteropathogenic and enterohaemorrhagic Escherichia coli effector-mediated subversion of host innate immune pathways. Cell. Microbiol. 2015, 17, 318–332. [Google Scholar] [CrossRef]
- Díaz-Guerrero, M.; Gaytán, M.O.; Soto, E.; Espinosa, N.; García-Gómez, E.; Marcos-Vilchis, A.; Andrade, A.; González-Pedrajo, B. CesL regulates type III secretion substrate specificity of the enteropathogenic E. coli injectisome. Microorganisms 2021, 9, 1047. [Google Scholar] [CrossRef]
- Mitrović, B.; Lezerovich, S.; Sal-Man, N. The role of the membrane-associated domain of the export apparatus protein, EscV (SctV), in the activity of the type III secretion system. Front. Microbiol. 2021, 12, 719469. [Google Scholar] [CrossRef] [PubMed]
- Portaliou, A.G.; Tsolis, K.C.; Loos, M.S.; Balabanidou, V.; Rayo, J.; Tsirigotaki, A.; Crepin, V.F.; Frankel, G.; Kalodimos, C.G.; Karamanou, S.; et al. Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 2017, 36, 3517–3531. [Google Scholar] [CrossRef] [PubMed]
- Kuhlen, L.; Abrusci, P.; Johnson, S.; Gault, J.; Deme, J.; Caesar, J.; Dietsche, T.; Mebrhatu, M.T.; Ganief, T.; Macek, B.; et al. Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 2018, 25, 583–590. [Google Scholar] [CrossRef]
- Wagner, S.; Diepold, A. A unified nomenclature for injectisome-type type III secretion systems. In Bacterial Type III Protein Secretion Systems; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–10. [Google Scholar]
- Deng, W.; Puente, J.L.; Gruenheid, S.; Li, Y.; Vallance, B.A.; Vázquez, A.; Barba, J.; Ibarra, J.A.; O’Donnell, P.; Metalnikov, P.; et al. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 2004, 101, 3597–3602. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Ohno, R.; Sekiya, K.; Kuwae, A.; Matsuzawa, T.; Nonaka, T.; Fukuda, H.; Imajoh-Ohmi, S.; Abe, A. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 2006, 188, 2801–2811. [Google Scholar] [PubMed]
- Tseytin, I.; Dagan, A.; Oren, S.; Sal-Man, N. The role of EscD in supporting EscC polymerization in the type III secretion system of enteropathogenic Escherichia coli. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 384–395. [Google Scholar] [CrossRef]
- Zakaria, D.; Matsuda, S.; Iida, T.; Hayashi, T.; Arita, M. Genome Analysis Identifies a Novel Type III Secretion System (T3SS) Category in Vibrio Species. Microorganisms 2023, 11, 290. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Katz, A.; Porte, L.; Weitzel, T.; Varela, C.; Muñoz-Rehbein, C.; Ugalde, J.A.; Grim, C.; González-Escalona, N.; Blondel, C.J.; Bravo, V. Whole-genome sequencing reveals changes in genomic diversity and distinctive repertoires of T3SS and T6SS effector candidates in Chilean clinical Campylobacter strains. Front. Cell. Infect. Microbiol. 2023, 13, 1208825. [Google Scholar] [CrossRef]
- Malesa, R.; Pierneef, R.; Magwedere, K.; Mafuna, T.; Matle, I. Genomic characterisation of generic Escherichia coli from food-producing animals and products of animal origin in South Africa. Front. Bacteriol. 2024, 3, 1432292. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G.J. The jalview java alignment editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef] [PubMed]
- Hotinger, J.; Pendergrass, H.A.; May, A.E. Molecular targets and strategies for inhibition of the bacterial type III secretion system (T3SS); inhibitors directly binding to T3SS components. Biomolecules 2021, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Bolukaoto, J.Y.; Singh, A.; Alfinete, N.; Barnard, T.G. Occurrence of hybrid diarrhoeagenic Escherichia coli associated with multidrug resistance in environmental water, Johannesburg, South Africa. Microorganisms 2021, 9, 2163. [Google Scholar] [CrossRef]
- Whelan, R.; McVicker, G.; Leo, J.C. Staying out or going in? The interplay between type 3 and type 5 secretion systems in adhesion and invasion of enterobacterial pathogens. Int. J. Mol. Sci. 2020, 21, 4102. [Google Scholar] [CrossRef]
- Akinlabi, O.C.; Dada, R.A.; Olayinka, A.A.; Oginni-Falajiki, I.O.; Bejide, O.S.; Adewole, P.D.; Thomson, N.R.; Aboderin, A.O.; Okeke, I.N. Genomic epidemiology of enteropathogenic Escherichia coli in southwestern Nigeria. PLoS Neglected Trop. Dis. 2025, 19, e0013442. [Google Scholar] [CrossRef]
- Ochieng, J.B.; Powell, H.; Sugerman, C.E.; Omore, R.; Ogwel, B.; Juma, J.; Awuor, A.O.; Sow, S.O.; Sanogo, D.; Onwuchekwa, U.; et al. Epidemiology of Enteroaggregative, Enteropathogenic, and Shiga Toxin–Producing Escherichia coli among children aged <5 years in 3 countries in Africa, 2015–2018: Vaccine impact on Diarrhea in Africa (VIDA) Study. Clin. Infect. Dis. 2023, 76 (Supp. S1), S77–S86. [Google Scholar] [CrossRef]
- Saeed, A.; Abd, H.; Sandstrom, G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J. Med. Microbiol. 2015, 64, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Webale, M.K.; Guyah, B.; Wanjala, C.; Nyanga, P.L.; Webale, S.K.; Abonyo, C.; Kitungulu, N.; Kiboi, N.; Bowen, N. Phenotypic and genotypic antibiotic resistant diarrheagenic Escherichia coli pathotypes isolated from children with diarrhea in Nairobi City, Kenya. Ethiop. J. Health Sci. 2020, 30, 881–890. [Google Scholar]
- Mwape, K.; Bosomprah, S.; Chibesa, K.; Silwamba, S.; Luchen, C.C.; Sukwa, N.; Mubanga, C.; Phiri, B.; Chibuye, M.; Liswaniso, F.; et al. Prevalence of diarrhoeagenic Escherichia coli among children aged between 0–36 months in peri-urban areas of Lusaka. Microorganisms 2023, 11, 2790. [Google Scholar] [CrossRef]
- Hounkpe, E.C.; Sessou, P.; Farougou, S.; Daube, G.; Delcenserie, V.; Azokpota, P.; Korsak, N. Prevalence, antibiotic resistance, and virulence gene profile of Escherichia coli strains shared between food and other sources in Africa: A systematic review. Vet. World 2023, 16, 2016. [Google Scholar] [CrossRef]
- Barker, C.S.; Inoue, T.; Meshcheryakova, I.V.; Kitanobo, S.; Samatey, F.A. Function of the conserved FHIPEP domain of the flagellar type III export apparatus, protein FlhA. Mol. Microbiol. 2016, 100, 278–288. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Liao, J.; Pang, H.; Lu, Y.; Ayiku, S.; Sakyi, M.E. Molecular cloning and bioinformatics analysis of t3ss inner membrane ring hrpq from Vibrio harveyi. Genom. Appl. Biol. 2018, 9, 40–47. [Google Scholar] [CrossRef]
- Gershberg, J.; Braverman, D.; Sal-Man, N. Transmembrane domains of type III-secreted proteins affect bacterial-host interactions in enteropathogenic E. coli. Virulence 2021, 12, 902–917. [Google Scholar] [CrossRef] [PubMed]
- Mainil, J. Escherichia coli virulence factors. Vet. Immunol. Immunopathol. 2013, 152, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Bai, X.; Jin, Y.; Hu, B.; Wang, H.; Sun, H.; Fan, R.; Fu, S.; Xiong, Y. High prevalence of virulence genes in specific genotypes of atypical enteropathogenic Escherichia coli. Front. Cell. Infect. Microbiol. 2017, 7, 109. [Google Scholar] [CrossRef]
- Worrall, L.J.; Bergeron, J.R.; Strynadka, N.C. Type 3 secretion systems. In Escherichia coli; Academic Press: Cambridge, MA, USA, 2013; pp. 417–450. [Google Scholar]
- Sirisaengtaksin, N.; O’Donoghue, E.J.; Jabbari, S.; Roe, A.J.; Krachler, A.M. Bacterial outer membrane vesicles provide an alternative pathway for trafficking of Escherichia coli O157 type III secreted effectors to epithelial cells. Msphere 2023, 8, e00520-23. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Yarabbi, H.; Mortazavi, S.A.; Yavarmanesh, M.; Javadmanesh, A. In silico study of different signal peptides to express recombinant glutamate decarboxylase in the outer membrane of Escherichia coli. Int. J. Pept. Res. Ther. 2020, 26, 1879–1891. [Google Scholar] [CrossRef]
- Wagner, S.; Königsmaier, L.; Lara-Tejero, M.; Lefebre, M.; Marlovits, T.C.; Galán, J.E. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. USA 2010, 107, 17745–17750. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, S.; Gargari, S.L.M.; Rasooli, I.; Amani, J.; Bagheri, S.; Alerasool, M. An in silico chimeric multi subunit vaccine targeting virulence factors of enterotoxigenic Escherichia coli (ETEC) with its bacterial inbuilt adjuvant. J. Microbiol. Methods 2012, 90, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.H.; Candido, E.S.; Chan, L.Y.; Der Torossian Torres, M.; Oshiro, K.G.; Rezende, S.B.; Porto, W.F.; Lu, T.K.; de la Fuente-Nunez, C.; Craik, D.J.; et al. A computationally designed peptide derived from Escherichia coli as a potential drug template for antibacterial and antibiofilm therapies. ACS Infect. Dis. 2018, 4, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Angamuthu, K.; Piramanayagam, S. Evaluation of in silico protein secondary structure prediction methods by employing statistical techniques. Biomed. Biotechnol. Res. J. (BBRJ) 2017, 1, 29–36. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Felise, H.B.; Nguyen, H.V.; Pfuetzner, R.A.; Barry, K.C.; Jackson, S.R.; Blanc, M.P.; Bronstein, P.A.; Kline, T.; Miller, S.I. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 2008, 4, 325–336. [Google Scholar] [CrossRef]
- Blasey, N.; Rehrmann, D.; Riebisch, A.K.; Mühlen, S. Targeting bacterial pathogenesis by inhibiting virulence-associated Type III and Type IV secretion systems. Front. Cell. Infect. Microbiol. 2023, 12, 1065561. [Google Scholar] [CrossRef]
- Zheng, W.; Peña, A.; Ilangovan, A.; Baghshomali, Y.N.; Frankel, G.; Egelman, E.H.; Costa, T.R. Cryoelectron-microscopy structure of the enteropathogenic Escherichia coli type III secretion system EspA filament. Proc. Natl. Acad. Sci. USA 2021, 118, e2022826118. [Google Scholar] [CrossRef]

| Accession No. | Geographic Location | Region | Source of Isolation |
|---|---|---|---|
| SAMN41920845 | North America | North America | Raw poultry |
| SAMN41920846 | South Africa | Mpumalanga | Raw poultry |
| SAMN41920847 | Netherlands | Europe | Raw poultry |
| SAMN41920848 | South Africa | Free State | Raw poultry |
| SAMN41920849 | South Africa | Free State | Raw poultry |
| SAMN41920851 | South Africa | Free State | Raw poultry |
| SAMN41920852 | South Africa | Free State | Raw poultry |
| SAMN41920853 | South Africa | Free State | Raw poultry |
| SAMN41920854 | South Africa | Free State | Raw poultry |
| SAMN41920855 | South Africa | Free State | Processed pork |
| SAMN41920856 | South Africa | Free State | Raw lamb |
| SAMN41920858 | South Africa | Free State | Raw pork |
| SAMN41920860 | South Africa | Free State | Raw poultry |
| SAMN41920861 | South Africa | Gauteng | Digestive system |
| SAMN41920862 | South Africa | Gauteng | Processed beef |
| SAMN41920863 | South Africa | Gauteng | Digestive system |
| SAMN41920865 | South Africa | Gauteng | Digestive system |
| SAMN41920867 | South Africa | Gauteng | Digestive system |
| SAMN41920868 | South Africa | Gauteng | Digestive system |
| SAMN41920869 | South Africa | Gauteng | Digestive system |
| SAMN41920870 | Europe | Europe | Raw poultry |
| SAMN41920871 | South Africa | Gauteng | Digestive system |
| SAMN41920872 | South Africa | Gauteng | Digestive system |
| SAMN41920873 | South Africa | Gauteng | Digestive system |
| SAMN41920876 | South Africa | Gauteng | Water |
| SAMN41920877 | South Africa | Limpopo | RTE beef |
| SAMN41920878 | South Africa | Gauteng | Digestive system |
| Proteins | Protein Domain | Position | Function of Each Protein |
|---|---|---|---|
| LEE | |||
| Structural | |||
| escV | Pfam:FHIPEP | 26 to 663 | It is important for translocation of effector proteins |
| escD | Pfam:Yop-YscD_ppl | 157 to 404 | It is involved in assembly and function of the T3SS |
| escU | Pfam:Bac_export_2 | 2 to 241 | It is important for regulation and stabilization of the apparatus |
| escN | AAA | 169 to 349 | It hydrolyses ATP to generate energy required for operation |
| Pathogenicity | |||
| eae | LysM Pfam: IAT_beta BID_1 BID_1 BID_2 Pfam: Intimin_C | 64 to 113 166 to 442 559 to 648 659 to 746 757 to 835 838 to 939 | It is important for mediation of attachment of E. coli to the intestinal epithelium |
| Core effectors | |||
| espA | Pfam: EspA | 4 to 186 | It is important for formation of pilus-like structures that facilitate movement of effectors |
| espD | Pfam: SseC | 117 to 216 | It forms the pore in the host membrane that allows effectors to move from bacteria into the host |
| espG | Pfam: EspG | 15 to 397 | It disrupts the host cell processes |
| non-LEE | |||
| cif | Pfam: CIF | 81 to 215 | Interferes with the host cell cycle |
| nleE | NleE_fam_methyl | 13–168 | Alters immunological responses and host cell signalling |
| nleB1 | None | Alters proteins in host cells to affect immune responses |
| Proteins | Query Coverage | E-Value | Percentage Identity | Reference. Accession No. |
|---|---|---|---|---|
| LEE | ||||
| escV | 100% | 0.0 | 100% | WP_001037814.1 |
| escD | 100% | 0.0 | 99.8% | ELP0616342.1 |
| escU | 100% | 5 × 10−102 | 99.4% | WP_063856070.1 |
| escN | 100% | 0.0 | 99.8% | WP_000622546.1 |
| Eae | 100% | 0.0 | 100% | WP_000627895.1 |
| espA | 100% | 2 × 10−132 | 100% | WP_000381555.1 |
| espD | 100% | 0.0 | 100% | WP_000935768.1 |
| espG | 100% | 0.0 | 100% | AAC31534.1 |
| non-LEE | ||||
| Cif | 100% | 0.0 | 100% | WP_000652080.1 |
| nleE | 100% | 1 × 10−119 | 100% | WP_000609738.1 |
| nleB1 | 100% | 0.0 | 100% | WP_012578998.1 |
| Ramachandran Plot Analysis | escV | espA |
|---|---|---|
| MolProbity Score | 1.38 | 1.35 |
| Clash Score | 2.41 | 1.40 |
| Ramachandran Favoured | 96.14% | 92.36% |
| Ramachandran Outliers | 1.19% | 0.64% |
| Rotamer Outliers | 1.35% | 0.01% |
| C-Beta Deviations | 0 | 177 |
| Bad Bonds | 0/5384 | 4/66,450 |
| Bad Angles | 24/7285 | 350/89,750 |
| Twisted Non-Proline | 5/652 | 50/8550 |
| escV | espA | |||
|---|---|---|---|---|
| Structure | No. of Residues | Percentage (%) | No. of Residues | Percentage (%) |
| Alpha helix (Hh) | 355 | 52.59 | 132 | 68.75 |
| 310 helix (Gg) | 0 | 0.00 | 0 | 0.00 |
| Pi helix (Ii) | 0 | 0.00 | 0 | 0.00 |
| Beta bridge (Bb) | 0 | 0.00 | 0 | 0.00 |
| Extended strand (Ee) | 118 | 17.48 | 4 | 2.08 |
| Beta turn (Tt) | 37 | 5.48 | 0 | 0.00 |
| Bend region (Ss) | 0 | 0.00 | 0 | 0.00 |
| Random coil (Cc) | 165 | 24.44 | 56 | 29.17 |
| Ambiguous states | 0 | 0.00 | 0 | 0.00 |
| Other states | 0 | 0.00 | 0 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malesa, R.; Pierneef, R.; Mafuna, T.; Magwedere, K.; Seakamela, E.; Matle, I. Genome-Based In Silico Analysis of the Structural and Functional Characteristics of the Type Three Secretion System (T3SS) and Core Effector Proteins in Enteropathogenic Escherichia coli (EPEC) Strains Isolated from Food-Producing Animals and Products of Animal Origin. Pathogens 2025, 14, 1099. https://doi.org/10.3390/pathogens14111099
Malesa R, Pierneef R, Mafuna T, Magwedere K, Seakamela E, Matle I. Genome-Based In Silico Analysis of the Structural and Functional Characteristics of the Type Three Secretion System (T3SS) and Core Effector Proteins in Enteropathogenic Escherichia coli (EPEC) Strains Isolated from Food-Producing Animals and Products of Animal Origin. Pathogens. 2025; 14(11):1099. https://doi.org/10.3390/pathogens14111099
Chicago/Turabian StyleMalesa, Refiloe, Rian Pierneef, Thendo Mafuna, Kudakwashe Magwedere, Emmanuel Seakamela, and Itumeleng Matle. 2025. "Genome-Based In Silico Analysis of the Structural and Functional Characteristics of the Type Three Secretion System (T3SS) and Core Effector Proteins in Enteropathogenic Escherichia coli (EPEC) Strains Isolated from Food-Producing Animals and Products of Animal Origin" Pathogens 14, no. 11: 1099. https://doi.org/10.3390/pathogens14111099
APA StyleMalesa, R., Pierneef, R., Mafuna, T., Magwedere, K., Seakamela, E., & Matle, I. (2025). Genome-Based In Silico Analysis of the Structural and Functional Characteristics of the Type Three Secretion System (T3SS) and Core Effector Proteins in Enteropathogenic Escherichia coli (EPEC) Strains Isolated from Food-Producing Animals and Products of Animal Origin. Pathogens, 14(11), 1099. https://doi.org/10.3390/pathogens14111099






