Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence
Abstract
1. Introduction
1.1. S. suis Metabolism
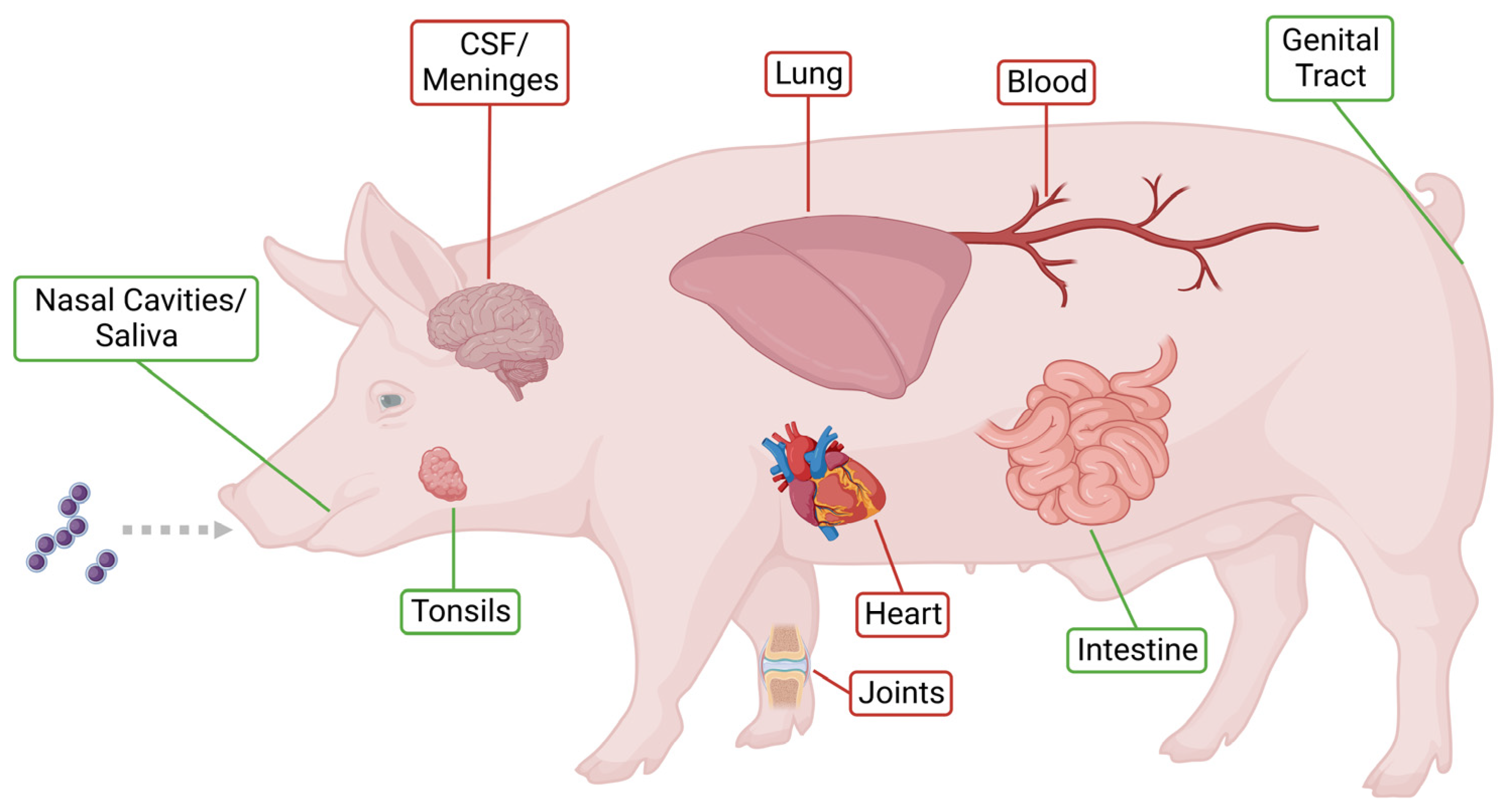
1.2. The Respiratory Habitat of S. suis
2. Transcriptomic Response of S. suis to Host Environments
2.1. Transcriptional Response of S. suis after In Vivo and In Vitro Infection
2.2. Transcriptomic Analysis of S. suis in Blood and Cerebrospinal Fluid
3. S. suis Metabolism, Biological Fitness and Virulence
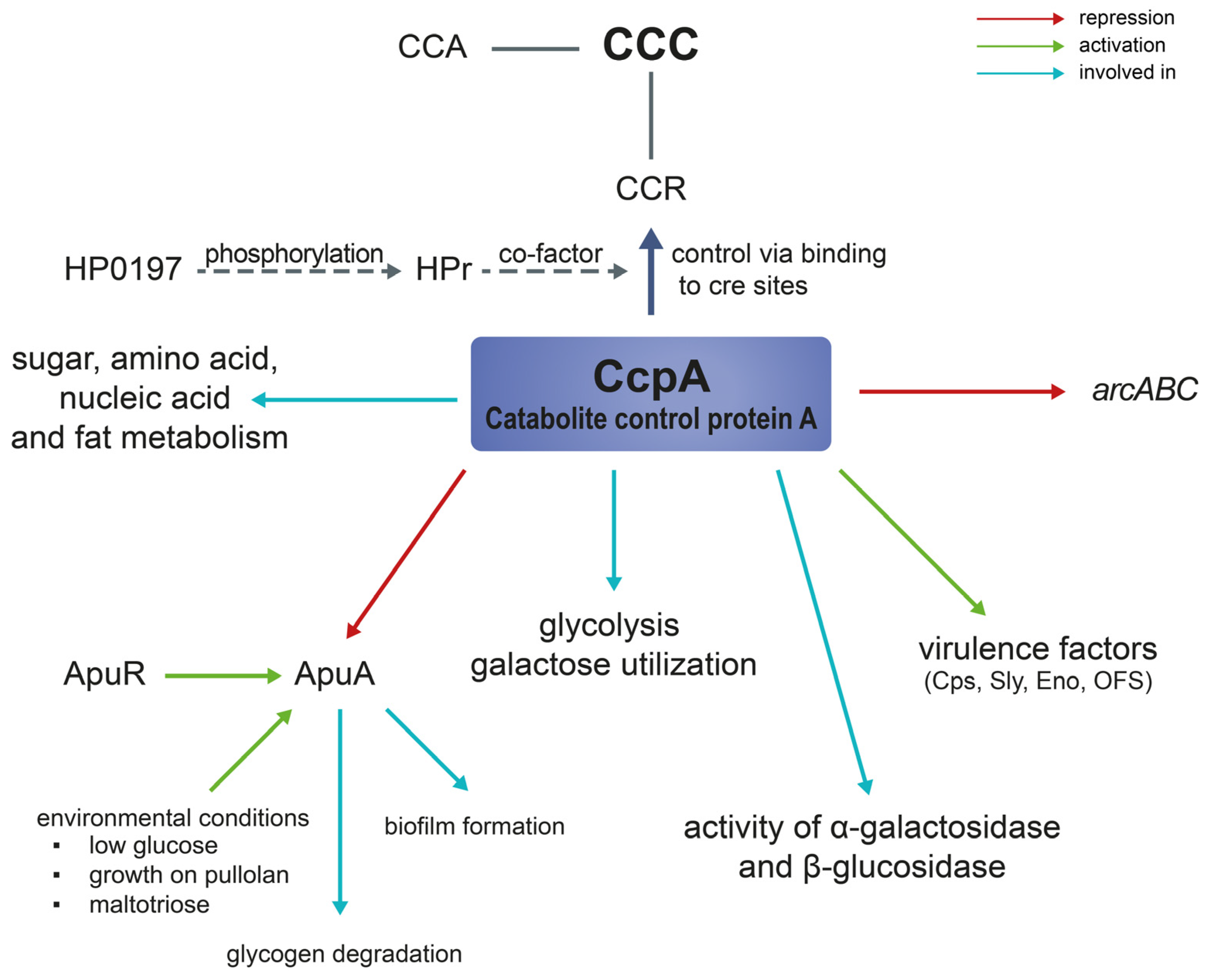
3.1. Catabolite Control Protein A (CcpA)
3.2. Amylopullulanase (apuA)
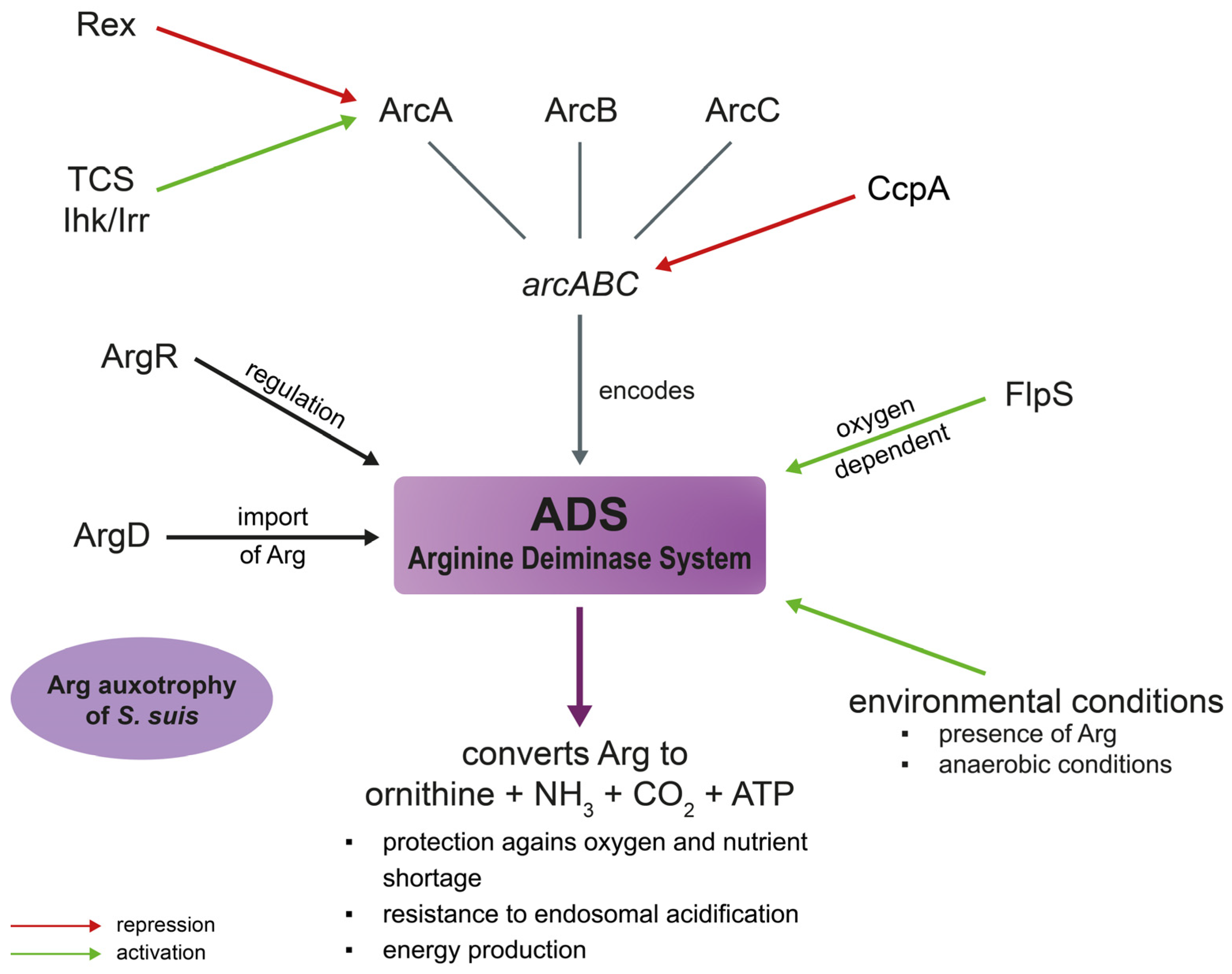
3.3. Arginine Deiminase System (ADS)
3.4. Amino Acid Metabolism
4. S. suis Metabolism and Co-Infections
5. S. suis Metabolism and Antibiotic Resistance
6. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Segura, M.; Calzas, C.; Grenier, D.; Gottschalk, M. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: Fighting against nonspecific defenses. FEBS Lett. 2016, 590, 3772–3799. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes. Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef]
- Votsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Dekker, N.; Bouma, A.; Daemen, I.; Klinkenberg, D.; van Leengoed, L.; Wagenaar, J.A.; Stegeman, A. Effect of spatial separation of pigs on spread of Streptococcus suis serotype 9. PLoS ONE 2013, 8, e61339. [Google Scholar] [CrossRef]
- Tang, J.; Wang, C.; Feng, Y.; Yang, W.; Song, H.; Chen, Z.; Yu, H.; Pan, X.; Zhou, X.; Wang, H.; et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006, 3, e151. [Google Scholar] [CrossRef]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef]
- Gottschalk, M.; Segura, M. The pathogenesis of the meningitis caused by Streptococcus suis: The unresolved questions. Vet. Microbiol. 2000, 76, 259–272. [Google Scholar] [CrossRef]
- Madsen, L.W.; Bak, H.; Nielsen, B.; Jensen, H.E.; Aalbaek, B.; Riising, H.J. Bacterial colonization and invasion in pigs experimentally exposed to Streptococcus suis serotype 2 in aerosol. J. Vet. Med. Infect. Dis. Vet. Public Health 2002, 49, 211–215. [Google Scholar] [CrossRef]
- Arends, J.P.; Hartwig, N.; Rudolphy, M.; Zanen, H.C. Carrier rate of Streptococcus suis capsular type 2 in palatine tonsils of slaughtered pigs. J. Clin. Microbiol. 1984, 20, 945–947. [Google Scholar] [CrossRef]
- Rohmer, L.; Hocquet, D.; Miller, S.I. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011, 19, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta Biomembr. 2018, 1860, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; Fulde, M.; de Greeff, A.; Rohde, M.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiol. (Read. Engl.) 2011, 157, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Hondorp, E.R.; McIver, K.S. The Mga virulence regulon: Infection where the grass is greener. Mol. Microbiol. 2007, 66, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.R.; Virtaneva, K.; Porcella, S.F.; Barry, W.T.; Gowen, B.B.; Johnson, C.R.; Wright, F.A.; Musser, J.M. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 2005, 166, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.P.; Choi, S.C.; Pavinski Bitar, P.D.; Gurjar, A.A.; Stanhope, M.J. Transcriptomic and genomic evidence for Streptococcus agalactiae adaptation to the bovine environment. BMC Genom. 2013, 14, 920. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Watanabe, T.; Arai, S.; Kim, H.; Tohya, M.; Ishida-Kuroki, K.; Võ, T.H.; Nguyễn, T.P.B.; Nakagawa, I.; Osawa, R.; et al. Characterization of pig saliva as the major natural habitat of Streptococcus suis by analyzing oral, fecal, vaginal, and environmental microbiota. PLoS ONE 2019, 14, e0215983. [Google Scholar] [CrossRef]
- Koczula, A.; Jarek, M.; Visscher, C.; Valentin-Weigand, P.; Goethe, R.; Willenborg, J. Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid. Pathogens 2017, 6, 7. [Google Scholar] [CrossRef]
- Van der Putten, B.C.L.; Roodsant, T.J.; Haagmans, M.A.; Schultsz, C.; van der Ark, K.C.H. Five Complete Genome Sequences Spanning the Dutch Streptococcus suis Serotype 2 and Serotype 9 Populations. Microbiol. Resour. Announc. 2020, 9, e01439-19. [Google Scholar] [CrossRef]
- Willenborg, J.; Huber, C.; Koczula, A.; Lange, B.; Eisenreich, W.; Valentin-Weigand, P.; Goethe, R. Characterization of the pivotal carbon metabolism of Streptococcus suis serotype 2 under ex vivo and chemically defined in vitro conditions by isotopologue profiling. J. Biol. Chem. 2015, 290, 5840–5854. [Google Scholar] [CrossRef]
- Hoskins, J.; Alborn, W.E., Jr.; Arnold, J.; Blaszczak, L.C.; Burgett, S.; DeHoff, B.S.; Estrem, S.T.; Fritz, L.; Fu, D.J.; Fuller, W.; et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 2001, 183, 5709–5717. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, M.L.; van Baarlen, P.; Orrù, G.; Piga, R.; Bongers, R.S.; Wels, M.; De Greeff, A.; Smith, H.E.; Wells, J.M. Carbohydrate availability regulates virulence gene expression in Streptococcus suis. PLoS ONE 2014, 9, e89334. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Tan, J.; Zhang, F.F.; Li, H.Q.; Ji, H.Y.; Fang, S.P.; Wu, C.C.; Rao, Y.L.; Zeng, Y.B.; Yang, Q. Exogenous glycogen utilization effects the transcriptome and pathogenicity of Streptococcus suis serotype 2. Front. Cell. Infect. Microbiol. 2022, 12, 938286. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; de Greeff, A.; Jarek, M.; Valentin-Weigand, P.; Goethe, R. The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol. Microbiol. 2014, 92, 61–83. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Ferrando, M.L.; Fuentes, S.; de Greeff, A.; Smith, H.; Wells, J.M. ApuA, a multifunctional alpha-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiol. (Read. Engl.) 2010, 156, 2818–2828. [Google Scholar] [CrossRef]
- Willenborg, J.; Goethe, R. Metabolic traits of pathogenic streptococci. FEBS Lett. 2016, 590, 3905–3919. [Google Scholar] [CrossRef]
- Bidossi, A.; Mulas, L.; Decorosi, F.; Colomba, L.; Ricci, S.; Pozzi, G.; Deutscher, J.; Viti, C.; Oggioni, M.R. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 2012, 7, e33320. [Google Scholar] [CrossRef]
- Juliao, P.C.; Marrs, C.F.; Xie, J.; Gilsdorf, J.R. Histidine auxotrophy in commensal and disease-causing nontypeable Haemophilus influenzae. J. Bacteriol. 2007, 189, 4994–5001. [Google Scholar] [CrossRef]
- Fittipaldi, N.; Harel, J.; D’Amours, B.; Lacouture, S.; Kobisch, M.; Gottschalk, M. Potential use of an unencapsulated and aromatic amino acid-auxotrophic Streptococcus suis mutant as a live attenuated vaccine in swine. Vaccine 2007, 25, 3524–3535. [Google Scholar] [CrossRef]
- Holden, M.T.; Hauser, H.; Sanders, M.; Ngo, T.H.; Cherevach, I.; Cronin, A.; Goodhead, I.; Mungall, K.; Quail, M.A.; Price, C.; et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 2009, 4, e6072. [Google Scholar] [CrossRef]
- Arndt, E.R.; Farzan, A.; Slavic, D.; MacInnes, J.I.; Friendship, R.M. An epidemiological study of Streptococcus suis serotypes of pigs in Ontario determined by a multiplex polymerase chain reaction. Can. Vet. J. 2018, 59, 997–1000. [Google Scholar] [PubMed]
- Pirolo, M.; Espinosa-Gongora, C.; Alberdi, A.; Eisenhofer, R.; Soverini, M.; Eriksen, E.; Pedersen, K.S.; Guardabassi, L. Bacterial topography of the upper and lower respiratory tract in pigs. Anim. Microbiome 2023, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Fredriksen, S.; Guan, X.; Boekhorst, J.; Molist, F.; van Baarlen, P.; Wells, J.M. Environmental and maternal factors shaping tonsillar microbiota development in piglets. BMC Microbiol. 2022, 22, 224. [Google Scholar] [CrossRef] [PubMed]
- Pirolo, M.; Espinosa-Gongora, C.; Bogaert, D.; Guardabassi, L. The porcine respiratory microbiome: Recent insights and future challenges. Anim. Microbiome 2021, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cai, R.; Huang, A.; Wang, X.; Qu, W.; Shi, L.; Li, C.; Yan, H. Comparison of Oropharyngeal Microbiota in Healthy Piglets and Piglets with Respiratory Disease. Front. Microbiol. 2018, 9, 3218. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S.; Yassin, S.A.; Rickard, A.H. Community interactions of oral streptococci. Adv. Appl. Microbiol. 2014, 87, 43–110. [Google Scholar] [CrossRef]
- Nobbs, A.H.; Jenkinson, H.F.; Everett, D.B. Generic determinants of Streptococcus colonization and infection. Infect. Genet. Evol. 2015, 33, 361–370. [Google Scholar] [CrossRef]
- Nitsche-Schmitz, D.P.; Rohde, M.; Chhatwal, G.S. Invasion mechanisms of Gram-positive pathogenic cocci. Thromb. Haemost. 2007, 98, 488–496. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, H.; Bian, C.; Chen, H.; Shen, Y.; Gao, X.; Ma, J.; Yao, H.; Wang, L.; Wu, Z. The antimicrobial systems of Streptococcus suis promote niche competition in pig tonsils. Virulence 2022, 13, 781–793. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Jack, R.W.; Tagg, J.R.; Ray, B. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 1995, 59, 171–200. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Kawada-Matsuo, M.; Le, M.N.; Hisatsune, J.; Oogai, Y.; Nakano, Y.; Nakata, M.; Miyawaki, S.; Sugai, M.; Komatsuzawa, H. Comprehensive analysis of bacteriocins in Streptococcus mutans. Sci. Rep. 2021, 11, 12963. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Giménez-Lirola, L.G.; Halbur, P.G. Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 2011, 12, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; et al. Coinfections and their molecular consequences in the porcine respiratory tract. Vet. Res. 2020, 51, 80. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, S.; Dong, X.; Li, J.; Grenier, D.; Yi, L. In vitro Mixed Biofilm of Streptococcus suis and Actinobacillus pleuropneumoniae Impacts Antibiotic Susceptibility and Modulates Virulence Factor Gene Expression. Front. Microbiol. 2020, 11, 507. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, C.; Shao, J.; Zhu, Z.; Wang, W.; Zhang, W.; Tang, M.; Pei, N.; Fan, H.; Li, J.; et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 2014, 20, 882–898. [Google Scholar] [CrossRef]
- Arenas, J.; Bossers-de Vries, R.; Harders-Westerveen, J.; Buys, H.; Ruuls-van Stalle, L.M.F.; Stockhofe-Zurwieden, N.; Zaccaria, E.; Tommassen, J.; Wells, J.M.; Smith, H.E.; et al. In vivo transcriptomes of Streptococcus suis reveal genes required for niche-specific adaptation and pathogenesis. Virulence 2019, 10, 334–351. [Google Scholar] [CrossRef]
- Schwerk, C.; Adam, R.; Borkowski, J.; Schneider, H.; Klenk, M.; Zink, S.; Quednau, N.; Schmidt, N.; Stump, C.; Sagar, A.; et al. In vitro transcriptome analysis of porcine choroid plexus epithelial cells in response to Streptococcus suis: Release of pro-inflammatory cytokines and chemokines. Microbes. Infect. 2011, 13, 953–962. [Google Scholar] [CrossRef] [PubMed]
- De Greeff, A.; Benga, L.; Wichgers Schreur, P.J.; Valentin-Weigand, P.; Rebel, J.M.; Smith, H.E. Involvement of NF-kappaB and MAP-kinases in the transcriptional response of alveolar macrophages to Streptococcus suis. Vet. Microbiol. 2010, 141, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Tan, C.; Fang, L.; Xiao, S.; Chen, H. Microarray analyses of THP-1 cells infected with Streptococcus suis serotype 2. Vet. Microbiol. 2011, 150, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, P.; Sun, H.; Wu, Z.; Gottschalk, M.; Qi, K.; Zheng, H. Comparative transcriptomic analysis reveal genes involved in the pathogenicity increase of Streptococcus suis epidemic strains. Virulence 2022, 13, 1455–1470. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zheng, H.; Zhang, J.; Jing, H.; Wang, L.; Xiong, Y.; Wang, W.; Zhou, Z.; Sun, Q.; Luo, X.; et al. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 2009, 199, 97–107. [Google Scholar] [CrossRef]
- Feng, L.; Zhu, J.; Chang, H.; Gao, X.; Gao, C.; Wei, X.; Yuan, F.; Bei, W. The CodY regulator is essential for virulence in Streptococcus suis serotype 2. Sci. Rep. 2016, 6, 21241. [Google Scholar] [CrossRef]
- Li, J.; Tan, C.; Zhou, Y.; Fu, S.; Hu, L.; Hu, J.; Chen, H.; Bei, W. The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet. Microbiol. 2011, 148, 99–104. [Google Scholar] [CrossRef]
- Federle, M.J.; McIver, K.S.; Scott, J.R. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 1999, 181, 3649–3657. [Google Scholar] [CrossRef]
- Lee, S.F.; Delaney, G.D.; Elkhateeb, M. A two-component covRS regulatory system regulates expression of fructosyltransferase and a novel extracellular carbohydrate in Streptococcus mutans. Infect. Immun. 2004, 72, 3968–3973. [Google Scholar] [CrossRef]
- Zheng, C.; Xu, J.; Li, J.; Hu, L.; Xia, J.; Fan, J.; Guo, W.; Chen, H.; Bei, W. Two Spx regulators modulate stress tolerance and virulence in Streptococcus suis serotype 2. PLoS ONE 2014, 9, e108197. [Google Scholar] [CrossRef]
- Neila-Ibanez, C.; Brogaard, L.; Pailler-Garcia, L.; Martinez, J.; Segales, J.; Segura, M.; Heegaard, P.M.H.; Aragon, V. Piglet innate immune response to Streptococcus suis colonization is modulated by the virulence of the strain. Vet. Res. 2021, 52, 145. [Google Scholar] [CrossRef] [PubMed]
- Sanford, S.E. Gross and histopathological findings in unusual lesions caused by Streptococcus suis in pigs. II. Central nervous system lesions. Can. J. Vet. Res. = Rev. Can. De Rech. Vet. 1987, 51, 486–489. [Google Scholar]
- Williams, A.E.; Blakemore, W.F. Pathogenesis of meningitis caused by Streptococcus suis type 2. J. Infect. Dis. 1990, 162, 474–481. [Google Scholar] [CrossRef]
- Beineke, A.; Bennecke, K.; Neis, C.; Schroder, C.; Waldmann, K.H.; Baumgartner, W.; Valentin-Weigand, P.; Baums, C.G. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet. Microbiol. 2008, 128, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Lauer, A.N.; Scholtysik, R.; Beineke, A.; Baums, C.G.; Klose, K.; Valentin-Weigand, P.; Ishikawa, H.; Schroten, H.; Klein-Hitpass, L.; Schwerk, C. A Comparative Transcriptome Analysis of Human and Porcine Choroid Plexus Cells in Response to Streptococcus suis Serotype 2 Infection Points to a Role of Hypoxia. Front. Cell. Infect. Microbiol. 2021, 11, 639620. [Google Scholar] [CrossRef]
- Tenenbaum, T.; Matalon, D.; Adam, R.; Seibt, A.; Wewer, C.; Schwerk, C.; Galla, H.J.; Schroten, H. Dexamethasone prevents alteration of tight junction-associated proteins and barrier function in porcine choroid plexus epithelial cells after infection with Streptococcus suis in vitro. Brain Res. 2008, 1229, 1–17. [Google Scholar] [CrossRef]
- Schwerk, C.; Papandreou, T.; Schuhmann, D.; Nickol, L.; Borkowski, J.; Steinmann, U.; Quednau, N.; Stump, C.; Weiss, C.; Berger, J.; et al. Polar invasion and translocation of Neisseria meningitidis and Streptococcus suis in a novel human model of the blood-cerebrospinal fluid barrier. PLoS ONE 2012, 7, e30069. [Google Scholar] [CrossRef]
- Tesorero, R.A.; Yu, N.; Wright, J.O.; Svencionis, J.P.; Cheng, Q.; Kim, J.H.; Cho, K.H. Novel regulatory small RNAs in Streptococcus pyogenes. PLoS ONE 2013, 8, e64021. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Mann, B.; van Opijnen, T.; Wang, J.; Obert, C.; Wang, Y.D.; Carter, R.; McGoldrick, D.J.; Ridout, G.; Camilli, A.; Tuomanen, E.I.; et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012, 8, e1002788. [Google Scholar] [CrossRef]
- Kumar, R.; Shah, P.; Swiatlo, E.; Burgess, S.C.; Lawrence, M.L.; Nanduri, B. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genom. 2010, 11, 350. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hong, S.H. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 2012, 326, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Pichon, C.; du Merle, L.; Caliot, M.E.; Trieu-Cuot, P.; Le Bouguenec, C. An in silico model for identification of small RNAs in whole bacterial genomes: Characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Res. 2012, 40, 2846–2861. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.G.R.; Charlesworth, J.; Miller, E.L.; Casey, M.J.; Lloyd, C.T.; Gottschalk, M.; Tucker, A.W.D.; Welch, J.J.; Weinert, L.A. Genome Reduction Is Associated with Bacterial Pathogenicity across Different Scales of Temporal and Ecological Divergence. Mol. Biol. Evol. 2021, 38, 1570–1579. [Google Scholar] [CrossRef]
- Moran, N.A. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 2002, 108, 583–586. [Google Scholar] [CrossRef]
- Zientz, E.; Dandekar, T.; Gross, R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 2004, 68, 745–770. [Google Scholar] [CrossRef]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Shelburne, S.A.; Davenport, M.T.; Keith, D.B.; Musser, J.M. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008, 16, 318–325. [Google Scholar] [CrossRef]
- Poncet, S.; Milohanic, E.; Mazé, A.; Abdallah, J.N.; Aké, F.; Larribe, M.; Deghmane, A.E.; Taha, M.K.; Dozot, M.; De Bolle, X.; et al. Correlations between carbon metabolism and virulence in bacteria. Contrib. Microbiol. 2009, 16, 88–102. [Google Scholar] [CrossRef]
- Nair, A.; Sarma, S.J. The impact of carbon and nitrogen catabolite repression in microorganisms. Microbiol. Res. 2021, 251, 126831. [Google Scholar] [CrossRef]
- Moreno, M.S.; Schneider, B.L.; Maile, R.R.; Weyler, W.; Saier, M.H., Jr. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: Novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 2001, 39, 1366–1381. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J.; Francke, C.; Postma, P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Baliga, N.S.; Camilli, A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 2005, 187, 8340–8349. [Google Scholar] [CrossRef]
- Tang, Y.; Wu, W.; Zhang, X.; Lu, Z.; Chen, J.; Fang, W. Catabolite control protein A of Streptococcus suis type 2 contributes to sugar metabolism and virulence. J. Microbiol. 2012, 50, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Lang, X.; Wan, Z.; Bu, Z.; Wang, X.; Wang, X.; Zhu, L.; Wan, J.; Sun, Y.; Wang, X. Catabolite control protein A is an important regulator of metabolism in Streptococcus suis type 2. Biomed. Rep. 2014, 2, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Lang, X.; Wan, Z.; Pan, Y.; Bu, Z.; Wang, X.; Wang, X.; Ji, X.; Zhu, L.; Wan, J.; Sun, Y.; et al. Catabolite control protein A has an important role in the metabolic regulation of Streptococcus suis type 2 according to iTRAQ-based quantitative proteomic analysis. Mol. Med. Rep. 2015, 12, 5967–5972. [Google Scholar] [CrossRef]
- Lang, X.; Wan, Z.; Pan, Y.; Wang, X.; Wang, X.; Bu, Z.; Qian, J.; Zeng, H.; Wang, X. Investigation into the role of catabolite control protein A in the metabolic regulation of Streptococcus suis serotype 2 using gene expression profile analysis. Exp. Ther. Med. 2015, 10, 127–132. [Google Scholar] [CrossRef]
- Richardson, A.R.; Somerville, G.A.; Sonenshein, A.L. Regulating the Intersection of Metabolism and Pathogenesis in Gram-positive Bacteria. Microbiol. Spectr. 2015, 3, 129–165. [Google Scholar] [CrossRef]
- Gruening, P.; Fulde, M.; Valentin-Weigand, P.; Goethe, R. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 2006, 188, 361–369. [Google Scholar] [CrossRef]
- Winterhoff, N.; Goethe, R.; Gruening, P.; Rohde, M.; Kalisz, H.; Smith, H.E.; Valentin-Weigand, P. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the Arginine Deiminase system of Streptococcus pyogenes. J. Bacteriol. 2002, 184, 6768–6776. [Google Scholar] [CrossRef]
- Benga, L.; Fulde, M.; Neis, C.; Goethe, R.; Valentin-Weigand, P. Polysaccharide capsule and suilysin contribute to extracellular survival of Streptococcus suis co-cultivated with primary porcine phagocytes. Vet. Microbiol. 2008, 132, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.E.; Damman, M.; van der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Seitz, M.; Baums, C.G.; Neis, C.; Benga, L.; Fulde, M.; Rohde, M.; Goethe, R.; Valentin-Weigand, P. Subcytolytic effects of suilysin on interaction of Streptococcus suis with epithelial cells. Vet. Microbiol. 2013, 167, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Brinsmade, S.R. CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr. Genet. 2017, 63, 417–425. [Google Scholar] [CrossRef]
- Zhang, A.; Chen, B.; Yuan, Z.; Li, R.; Liu, C.; Zhou, H.; Chen, H.; Jin, M. HP0197 contributes to CPS synthesis and the virulence of Streptococcus suis via CcpA. PLoS ONE 2012, 7, e50987. [Google Scholar] [CrossRef]
- Zhang, A.; Xie, C.; Chen, H.; Jin, M. Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics 2008, 8, 3506–3515. [Google Scholar] [CrossRef]
- Zhang, A.; Chen, B.; Li, R.; Mu, X.; Han, L.; Zhou, H.; Chen, H.; Meilin, J. Identification of a surface protective antigen, HP0197 of Streptococcus suis serotype 2. Vaccine 2009, 27, 5209–5213. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Allen, G.S.; Diel, M.; Seidel, G.; Hillen, W.; Brennan, R.G. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell 2004, 118, 731–741. [Google Scholar] [CrossRef]
- Shelburne, S.A., 3rd; Keith, D.; Horstmann, N.; Sumby, P.; Davenport, M.T.; Graviss, E.A.; Brennan, R.G.; Musser, J.M. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 2008, 105, 1698–1703. [Google Scholar] [CrossRef]
- Wen, Z.T.; Burne, R.A. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Env. Microbiol. 2002, 68, 1196–1203. [Google Scholar] [CrossRef]
- Almengor, A.C.; Kinkel, T.L.; Day, S.J.; McIver, K.S. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A streptococcus. J. Bacteriol. 2007, 189, 8405–8416. [Google Scholar] [CrossRef] [PubMed]
- Grenier, D.; Grignon, L.; Gottschalk, M. Characterisation of biofilm formation by a Streptococcus suis meningitis isolate. Vet. J. 2009, 179, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Wu, Z.; Lu, C. Reduced virulence is an important characteristic of biofilm infection of Streptococcus suis. FEMS Microbiol. Lett. 2011, 316, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Bulock, L.L.; Ahn, J.; Shinde, D.; Pandey, S.; Sarmiento, C.; Thomas, V.C.; Guda, C.; Bayles, K.W.; Sadykov, M.R. Interplay of CodY and CcpA in Regulating Central Metabolism and Biofilm Formation in Staphylococcus aureus. J. Bacteriol. 2022, 204, e0061721. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Chen, Z.; Itzek, A.; Herzberg, M.C.; Kreth, J. CcpA regulates biofilm formation and competence in Streptococcus gordonii. Mol. Oral Microbiol. 2012, 27, 83–94. [Google Scholar] [CrossRef]
- Gough, H.; Luke, G.A.; Beeley, J.A.; Geddes, D.A. Human salivary glucose analysis by high-performance ion-exchange chromatography and pulsed amperometric detection. Arch. Oral Biol. 1996, 41, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Navarro, D.; Abelilla, J.J.; Stein, H.H. Structures and characteristics of carbohydrates in diets fed to pigs: A review. J. Anim. Sci. Biotechnol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Tenenbaum, T.; Asmat, T.M.; Seitz, M.; Schroten, H.; Schwerk, C. Biological activities of suilysin: Role in Streptococcus suis pathogenesis. Future Microbiol. 2016, 11, 941–954. [Google Scholar] [CrossRef]
- Jacobs, A.A.; Loeffen, P.L.; van den Berg, A.J.; Storm, P.K. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 1994, 62, 1742–1748. [Google Scholar] [CrossRef]
- Takeuchi, D.; Akeda, Y.; Nakayama, T.; Kerdsin, A.; Sano, Y.; Kanda, T.; Hamada, S.; Dejsirilert, S.; Oishi, K. The contribution of suilysin to the pathogenesis of Streptococcus suis meningitis. J. Infect. Dis. 2014, 209, 1509–1519. [Google Scholar] [CrossRef]
- Lalonde, M.; Segura, M.; Lacouture, S.; Gottschalk, M. Interactions between Streptococcus suis serotype 2 and different epithelial cell lines. Microbiol. (Read. Engl.) 2000, 146 Pt 8, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Norton, P.M.; Rolph, C.; Ward, P.N.; Bentley, R.W.; Leigh, J.A. Epithelial invasion and cell lysis by virulent strains of Streptococcus suis is enhanced by the presence of suilysin. FEMS Immunol. Med. Microbiol. 1999, 26, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Charland, N.; Nizet, V.; Rubens, C.E.; Kim, K.S.; Lacouture, S.; Gottschalk, M. Streptococcus suis serotype 2 interactions with human brain microvascular endothelial cells. Infect. Immun. 2000, 68, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Wu, N.H.; Nerlich, A.; Herrler, G.; Valentin-Weigand, P.; Seitz, M. Dynamic Virus-Bacterium Interactions in a Porcine Precision-Cut Lung Slice Coinfection Model: Swine Influenza Virus Paves the Way for Streptococcus suis Infection in a Two-Step Process. Infect. Immun. 2015, 83, 2806–2815. [Google Scholar] [CrossRef]
- Meng, F.; Wu, N.H.; Seitz, M.; Herrler, G.; Valentin-Weigand, P. Efficient suilysin-mediated invasion and apoptosis in porcine respiratory epithelial cells after streptococcal infection under air-liquid interface conditions. Sci. Rep. 2016, 6, 26748. [Google Scholar] [CrossRef]
- Fulde, M.; Willenborg, J.; de Greeff, A.; Benga, L.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiol. (Read. Engl.) 2011, 157, 572–582. [Google Scholar] [CrossRef]
- Zúñiga, M.; Pérez, G.; González-Candelas, F. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 2002, 25, 429–444. [Google Scholar] [CrossRef]
- Champomier Vergès, M.C.; Zuñiga, M.; Morel-Deville, F.; Pérez-Martínez, G.; Zagorec, M.; Ehrlich, S.D. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 1999, 180, 297–304. [Google Scholar] [CrossRef]
- Fulde, M.; Willenborg, J.; Huber, C.; Hitzmann, A.; Willms, D.; Seitz, M.; Eisenreich, W.; Valentin-Weigand, P.; Goethe, R. The arginine-ornithine antiporter ArcD contributes to biological fitness of Streptococcus suis. Front. Cell. Infect. Microbiol. 2014, 4, 107. [Google Scholar] [CrossRef]
- Hashim, S.; Kwon, D.H.; Abdelal, A.; Lu, C.D. The arginine regulatory protein mediates repression by arginine of the operons encoding glutamate synthase and anabolic glutamate dehydrogenase in Pseudomonas aeruginosa. J. Bacteriol. 2004, 186, 3848–3854. [Google Scholar] [CrossRef]
- Larsen, R.; Kok, J.; Kuipers, O.P. Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J. Biol. Chem. 2005, 280, 19319–19330. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Lu, C.D.; Abdelal, A.T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1997, 179, 5300–5308. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; Koczula, A.; Fulde, M.; de Greeff, A.; Beineke, A.; Eisenreich, W.; Huber, C.; Seitz, M.; Valentin-Weigand, P.; Goethe, R. FlpS, the FNR-Like Protein of Streptococcus suis Is an Essential, Oxygen-Sensing Activator of the Arginine Deiminase System. Pathogens 2016, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Liu, C.; Wang, Q.; Xuan, C.; Zheng, B.; Tang, J.; Yan, J.; Zhang, J.; Li, M.; Cheng, H.; et al. The two-component system Ihk/Irr contributes to the virulence of Streptococcus suis serotype 2 strain 05ZYH33 through alteration of the bacterial cell metabolism. Microbiol. (Read. Engl.) 2012, 158, 1852–1866. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, Y.; Ni, Y.; Zhou, J.; Han, L.; Yu, Z.; Mao, A.; Wang, D.; Fan, H.; He, K. The Redox-Sensing Regulator Rex Contributes to the Virulence and Oxidative Stress Response of Streptococcus suis Serotype 2. Front. Cell. Infect. Microbiol. 2018, 8, 317. [Google Scholar] [CrossRef]
- Neis, E.P.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [PubMed]
- Dresen, M.; Schaaf, D.; Arenas, J.; de Greeff, A.; Valentin-Weigand, P.; Nerlich, A. Streptococcus suis TrpX is part of a tryptophan uptake system, and its expression is regulated by a T-box regulatory element. Sci. Rep. 2022, 12, 13920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gagnon, C.A.; Savard, C.; Music, N.; Srednik, M.; Segura, M.; Lachance, C.; Bellehumeur, C.; Gottschalk, M. Capsular sialic acid of Streptococcus suis serotype 2 binds to swine influenza virus and enhances bacterial interactions with virus-infected tracheal epithelial cells. Infect. Immun. 2013, 81, 4498–4508. [Google Scholar] [CrossRef]
- Dang, Y.; Lachance, C.; Wang, Y.; Gagnon, C.A.; Savard, C.; Segura, M.; Grenier, D.; Gottschalk, M. Transcriptional approach to study porcine tracheal epithelial cells individually or dually infected with swine influenza virus and Streptococcus suis. BMC Vet. Res. 2014, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Auray, G.; Lachance, C.; Wang, Y.; Gagnon, C.A.; Segura, M.; Gottschalk, M. Transcriptional Analysis of PRRSV-Infected Porcine Dendritic Cell Response to Streptococcus suis Infection Reveals Up-Regulation of Inflammatory-Related Genes Expression. PLoS ONE 2016, 11, e0156019. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Liu, Y.; Yang, J.; Guo, L.; Ren, S.; Chen, Z.; Liu, Z.; Zhang, Y.; Qiu, W.; et al. Porcine reproductive and respiratory syndrome virus NADC30-like strain accelerates Streptococcus suis serotype 2 infection in vivo and in vitro. Transbound Emerg. Dis. 2019, 66, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, H.; Hao, Q.; Li, M.; Liu, J.; Fan, H. Coinfection with porcine circovirus type 2 and Streptococcus suis serotype 2 enhances pathogenicity by dysregulation of the immune responses in piglets. Vet. Microbiol. 2020, 243, 108653. [Google Scholar] [CrossRef]
- Wu, N.H.; Meng, F.; Seitz, M.; Valentin-Weigand, P.; Herrler, G. Sialic acid-dependent interactions between influenza viruses and Streptococcus suis affect the infection of porcine tracheal cells. J. Gen. Virol. 2015, 96, 2557–2568. [Google Scholar] [CrossRef] [PubMed]
- Vötsch, D.; Willenborg, M.; Baumgärtner, W.; Rohde, M.; Valentin-Weigand, P. Bordetella bronchiseptica promotes adherence, colonization, and cytotoxicity of Streptococcus suis in a porcine precision-cut lung slice model. Virulence 2021, 12, 84–95. [Google Scholar] [CrossRef]
- Mathieu-Denoncourt, A.; Letendre, C.; Auger, J.P.; Segura, M.; Aragon, V.; Lacouture, S.; Gottschalk, M. Limited Interactions between Streptococcus Suis and Haemophilus Parasuis in In Vitro Co-Infection Studies. Pathogens 2018, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, H.; Fan, H.; Wang, X. Coinfection with Porcine Circovirus Type 2 (PCV2) and Streptococcus suis Serotype 2 (SS2) Enhances the Survival of SS2 in Swine Tracheal Epithelial Cells by Decreasing Reactive Oxygen Species Production. Infect. Immun. 2020, 88, e00537-20. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.H. Aging: A revisited theory based on free radicals generated by NOX family NADPH oxidases. Exp. Gerontol. 2007, 42, 256–262. [Google Scholar] [CrossRef]
- Brieger, K.; Schiavone, S.; Miller, F.J., Jr.; Krause, K.H. Reactive oxygen species: From health to disease. Swiss Med. Wkly 2012, 142, w13659. [Google Scholar] [CrossRef]
- Palmieri, C.; Varaldo, P.E.; Facinelli, B. Streptococcus suis, an Emerging Drug-Resistant Animal and Human Pathogen. Front. Microbiol. 2011, 2, 235. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2021, 130, 797–806. [Google Scholar] [CrossRef]
- Uruén, C.; García, C.; Fraile, L.; Tommassen, J.; Arenas, J. How Streptococcus suis escapes antibiotic treatments. Vet. Res. 2022, 53, 91. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Bont, L.; Engelhard, D.; Moore, E.; Fernández, D.; Kreisberg-Greenblatt, R.; Oved, K.; Eden, E.; Hays, J.P. A multifaceted ‘omics’ approach for addressing the challenge of antimicrobial resistance. Future Microbiol. 2015, 10, 365–376. [Google Scholar] [CrossRef]
- Kuang, S.F.; Chen, Y.T.; Chen, J.J.; Peng, X.X.; Chen, Z.G.; Li, H. Synergy of alanine and gentamicin to reduce nitric oxide for elevating killing efficacy to antibiotic-resistant Vibrio alginolyticus. Virulence 2021, 12, 1737–1753. [Google Scholar] [CrossRef]
- Wu, T.; Wang, X.; Dong, Y.; Xing, C.; Chen, X.; Li, L.; Dong, C.; Li, Y. Effects of l-Serine on Macrolide Resistance in Streptococcus suis. Microbiol. Spectr. 2022, 10, e0068922. [Google Scholar] [CrossRef]
- Wang, Q.; Lv, Y.; Pang, J.; Li, X.; Lu, X.; Wang, X.; Hu, X.; Nie, T.; Yang, X.; Xiong, Y.Q.; et al. In vitro and in vivo activity of d-serine in combination with β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. Acta Pharm Sin. 2019, 9, 496–504. [Google Scholar] [CrossRef]
- Peng, B.; Su, Y.B.; Li, H.; Han, Y.; Guo, C.; Tian, Y.M.; Peng, X.X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–262. [Google Scholar] [CrossRef]
- Escudero, J.A.; San Millan, A.; Gutierrez, B.; Hidalgo, L.; La Ragione, R.M.; AbuOun, M.; Galimand, M.; Ferrándiz, M.J.; Domínguez, L.; de la Campa, A.G.; et al. Fluoroquinolone efflux in Streptococcus suis is mediated by SatAB and not by SmrA. Antimicrob. Agents Chemother. 2011, 55, 5850–5860. [Google Scholar] [CrossRef]
- Bizzini, A.; Entenza, J.M.; Moreillon, P. Loss of penicillin tolerance by inactivating the carbon catabolite repression determinant CcpA in Streptococcus gordonii. J. Antimicrob. Chemother. 2007, 59, 607–615. [Google Scholar] [CrossRef]
- Seidl, K.; Stucki, M.; Ruegg, M.; Goerke, C.; Wolz, C.; Harris, L.; Berger-Bächi, B.; Bischoff, M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006, 50, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Collins, J.J. Boosting bacterial metabolism to combat antibiotic resistance. Cell Metab. 2015, 21, 154–155. [Google Scholar] [CrossRef]
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, M.R.; Segura, M.; Segalés, J.; Gottschalk, M. Review of the speculative role of co-infections in Streptococcus suis-associated diseases in pigs. Vet. Res. 2021, 52, 49. [Google Scholar] [CrossRef]
- Antimicrobial Resistance in G7 Countries and beyond: Economic Issues, Policies and Options for Action. Available online: https://www.oecd.org/els/health-systems/Antimicrobial-Resistance-in-G7-Countries-and-Beyond.pdf (accessed on 9 February 2023).
- Amato, S.M.; Fazen, C.H.; Henry, T.C.; Mok, W.W.; Orman, M.A.; Sandvik, E.L.; Volzing, K.G.; Brynildsen, M.P. The role of metabolism in bacterial persistence. Front. Microbiol. 2014, 5, 70. [Google Scholar] [CrossRef]
- Walsh, C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003, 1, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rock, C.O. Bacterial fatty acid metabolism in modern antibiotic discovery. Biochim. Biophys Acta Mol. Cell. Biol. Lipids 2017, 1862, 1300–1309. [Google Scholar] [CrossRef]
- Ballouche, M.; Cornelis, P.; Baysse, C. Iron metabolism: A promising target for antibacterial strategies. Recent Pat. Anti-Infect. Drug Discov. 2009, 4, 190–205. [Google Scholar] [CrossRef]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef]
- Haag, N.L.; Velk, K.K.; Wu, C. Potential Antibacterial Targets in Bacterial Central Metabolism. Int. J. Adv. Life Sci. 2012, 4, 21–32. [Google Scholar] [PubMed]
- Garmory, H.S.; Titball, R.W. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004, 72, 6757–6763. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh Aghdam, E.; Hejazi, M.S.; Barzegar, A. Riboswitches: From living biosensors to novel targets of antibiotics. Gene 2016, 592, 244–259. [Google Scholar] [CrossRef]
- Anupam, R.; Nayek, A.; Green, N.J.; Grundy, F.J.; Henkin, T.M.; Means, J.A.; Bergmeier, S.C.; Hines, J.V. 4,5-Disubstituted oxazolidinones: High affinity molecular effectors of RNA function. Bioorg. Med. Chem. Lett. 2008, 18, 3541–3544. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dresen, M.; Valentin-Weigand, P.; Berhanu Weldearegay, Y. Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence. Pathogens 2023, 12, 541. https://doi.org/10.3390/pathogens12040541
Dresen M, Valentin-Weigand P, Berhanu Weldearegay Y. Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence. Pathogens. 2023; 12(4):541. https://doi.org/10.3390/pathogens12040541
Chicago/Turabian StyleDresen, Muriel, Peter Valentin-Weigand, and Yenehiwot Berhanu Weldearegay. 2023. "Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence" Pathogens 12, no. 4: 541. https://doi.org/10.3390/pathogens12040541
APA StyleDresen, M., Valentin-Weigand, P., & Berhanu Weldearegay, Y. (2023). Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence. Pathogens, 12(4), 541. https://doi.org/10.3390/pathogens12040541






