Correlation of Lymphocyte Subpopulations, Clinical Features and Inflammatory Markers during Severe COVID-19 Onset
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Flow Cytometry Analysis
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAP | community acquired pneumonia |
| COVID-19 | Coronavirus disease |
| CRP | C-reactive protein |
| CT | computed tomography |
| FDR | False density rate |
| IL-6 | interleukin-6 |
| NK | natural killer |
| NLR | Neutrophil to Lymphocyte ratio |
| OR | odds ratio |
| PO2/FiO2 ratio | partial O2 pressure/fraction of inspired O2 |
| RT-PCR | reverse transcriptase–polymerase chain reaction |
| SD | standard deviation |
References
- Li, C.K.; Wu, H.; Yan, H.; Ma, S.; Wang, L.; Zhang, M.; Tang, X.; Temperton, N.J.; Weiss, R.A.; Brenchley, J.M.; et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008, 181, 5490–5500. [Google Scholar] [CrossRef]
- Zelba, H.; Worbs, D.; Harter, J.; Pieper, N.; Kyzirakos-Feger, C.; Kayser, S.; Seibold, M.; Bartsch, O.; Kodding, J.; Biskup, S. A Highly Specific Assay for the Detection of SARS-CoV-2-Reactive CD4(+) and CD8(+) T Cells in COVID-19 Patients. J. Immunol. 2021, 206, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e973. [Google Scholar] [CrossRef] [PubMed]
- Fafi-Kremer, S.; Bruel, T.; Madec, Y.; Grant, R.; Tondeur, L.; Grzelak, L.; Staropoli, I.; Anna, F.; Souque, P.; Fernandes-Pellerin, S.; et al. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine 2020, 59, 102915. [Google Scholar] [CrossRef]
- Liu, S.T.H.; Lin, H.M.; Baine, I.; Wajnberg, A.; Gumprecht, J.P.; Rahman, F.; Rodriguez, D.; Tandon, P.; Bassily-Marcus, A.; Bander, J.; et al. Convalescent plasma treatment of severe COVID-19: A propensity score-matched control study. Nat. Med. 2020, 26, 1708–1713. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef]
- Yin, Q.; Fu, Z.; Xie, J.; Yang, J.; Li, F.; Zhu, W.; Yu, Y.; Zhang, J. Analysis of Risk Factors of Severe COVID-19 Patients. 2020. Available online: https://www.researchsquare.com/article/rs-23272/v1 (accessed on 20 October 2022).
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Chien, J.Y.; Hsueh, P.R.; Cheng, W.C.; Yu, C.J.; Yang, P.C. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology 2006, 11, 715–722. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fontanet, A.; Zhang, P.H.; Zhan, L.; Xin, Z.T.; Baril, L.; Tang, F.; Lv, H.; Cao, W.C. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006, 193, 792–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Yi, Q.; Fan, S.; Lv, J.; Zhang, X.; Guo, L.; Lang, C.; Xiao, Q.; Xiao, K.; Yi, Z.; et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv 2020. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef]
- Chung, M.; Bernheim, A. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Duni, A.; Markopoulos, G.S.; Mallioras, I.; Pappas, H.; Pappas, E.; Koutlas, V.; Tzalavra, E.; Baxevanos, G.; Priska, S.; Gartzonika, K.; et al. The Humoral Immune Response to BNT162b2 Vaccine Is Associated with Circulating CD19+ B Lymphocytes and the Naïve CD45RA to Memory CD45RO CD4+ T Helper Cells Ratio in Hemodialysis Patients and Kidney Transplant Recipients. Front. Immunol. 2021, 12, 760249. [Google Scholar] [CrossRef]
- Liu, K.; Chen, Y.; Lin, R.; Han, K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J. Infect. 2020, 80, e14–e18. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Li, J.; Li, S.; Yang, C. Leucocyte Subsets Effectively Predict the Clinical Outcome of Patients With COVID-19 Pneumonia: A Retrospective Case-Control Study. Front. Public Health 2020, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Nong, S.; Jiang, L.; Chi, X. Correlations of disease severity and age with hematology parameter variations in patients with COVID-19 pre- and post-treatment. J. Clin. Lab. Anal. 2021, 35, e23609. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, F.; Nematollahi, P.; Salmasi, M.; Hedayat, A.; Amra, B. Association of lymphocyte subsets with mortality in severe COVID-19 pneumonia patients. J. Clin. Lab. Anal. 2021, 35, e24046. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Paul, W.E. CD4 T cells: Fates, functions, and faults. Blood J. Am. Soc. Hematol. 2008, 112, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Huang, W.; Berube, J.; McNamara, M.; Saksena, S.; Hartman, M.; Arshad, T.; Bornheimer, S.J.; O’Gorman, M. Lymphocyte Subset Counts in COVID-19 Patients: A Meta-Analysis. Cytom. Part A J. Int. Soc. Anal. Cytol. 2020, 97, 772–776. [Google Scholar] [CrossRef]
- Deng, X.; Terunuma, H.; Nieda, M. Exploring the Utility of NK Cells in COVID-19. Biomedicines 2022, 10, 1002. [Google Scholar] [CrossRef]
- Jeyaraman, M.; Muthu, S.; Bapat, A.; Jain, R.; Sushmitha, E.S.; Gulati, A.; Channaiah Anudeep, T.; Dilip, S.J.; Jha, N.K.; Kumar, D.; et al. Bracing NK cell based therapy to relegate pulmonary inflammation in COVID-19. Heliyon 2021, 7, e07635. [Google Scholar] [CrossRef]
- Varchetta, S.; Mele, D.; Oliviero, B.; Mantovani, S.; Ludovisi, S.; Cerino, A.; Bruno, R.; Castelli, A.; Mosconi, M.; Vecchia, M.; et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2021, 18, 604–612. [Google Scholar] [CrossRef]
- Jiang, M.; Guo, Y.; Luo, Q.; Huang, Z.; Zhao, R.; Liu, S.; Le, A.; Li, J.; Wan, L. T-Cell Subset Counts in Peripheral Blood Can Be Used as Discriminatory Biomarkers for Diagnosis and Severity Prediction of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 198–202. [Google Scholar] [CrossRef]
- Iannetta, M.; Buccisano, F.; Fraboni, D.; Malagnino, V.; Campogiani, L.; Teti, E.; Spalliera, I.; Rossi, B.; Di Lorenzo, A.; Palmieri, R.; et al. Baseline T-lymphocyte subset absolute counts can predict both outcome and severity in SARS-CoV-2 infected patients: A single center study. Sci. Rep. 2021, 11, 12762. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Jiang, X.; Zeng, X.; Pan, Y. Analysis of Lymphocyte Subpopulations and Cytokines in COVID-19-Associated Pneumonia and Community-Acquired Pneumonia. J. Immunol. Res. 2021, 2021, 6657894. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e1019. [Google Scholar] [CrossRef] [PubMed]
- Rha, M.S.; Shin, E.C. Activation or exhaustion of CD8(+) T cells in patients with COVID-19. Cell. Mol. Immunol. 2021, 18, 2325–2333. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine storm in COVID-19: The current evidence and treatment strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Tabrizi, R.; Lankarani, K.B.; Aria, H.; Vakili, S.; Asadian, F.; Noroozi, S.; Keshavarz, P.; Faramarz, S. The role of cytokine profile and lymphocyte subsets in the severity of coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis. Life Sci. 2020, 258, 118167. [Google Scholar] [CrossRef] [PubMed]
- Peisajovich, A.; Marnell, L.; Mold, C.; Du Clos, T.W. C-reactive protein at the interface between innate immunity and inflammation. Expert Rev. Clin. Immunol. 2008, 4, 379–390. [Google Scholar] [CrossRef]
- Acar, E.; Demir, A. The role of hemogram parameters and C-reactive protein in predicting mortality in COVID-19 infection. Int. J. Clin. Pract. 2021, 75, e14256. [Google Scholar] [CrossRef]
- Wang, L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020, 50, 332–334. [Google Scholar] [CrossRef]
- Urra, J.M.; Cabrera, C.M.; Porras, L.; Ródenas, I. Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin. Immunol. 2020, 217, 108486. [Google Scholar] [CrossRef] [PubMed]
- Mangano, C.; Oliva, B.M. Relationship between lymphocyte subsets values and C-reactive protein in COVID-19 patients. Cytom. Part A J. Int. Soc. Anal. Cytol. 2021, 99, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.; Endeman, H.; van den Akker, J.P.; Molenkamp, R.; Koopmans, M.P.; van Gorp, E.C. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020, 182, 1419–1440.e1423. [Google Scholar] [CrossRef]
- Rendeiro, A.F.; Casano, J.; Vorkas, C.K.; Singh, H.; Morales, A.; DeSimone, R.A.; Ellsworth, G.B.; Soave, R.; Kapadia, S.N.; Saito, K.; et al. Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression. Life Sci. Alliance 2021, 4, e202000955. [Google Scholar] [CrossRef]
| Characteristics | N | Mean (SD) or Frequency | CT Burden of Disease | |
|---|---|---|---|---|
| ≤50% (n = 19) | >50% (n = 15) | |||
| Age, years, mean (SD) | 42 | 55.90 (20.30) | 54.63 (16.69) | 60.13 (19.16) |
| Sex, n (%) | 42 | |||
| Female | 29 (69.05) | 15 (78.95) | 9 (60.00) | |
| Male | 13 (30.95) | 4 (21.05) | 6 (40.00) | |
| Duration of symptoms, days, mean (SD) | 38 | 6.29 (3.69) | 6.83 (4.00) | 6.35 (3.69) |
| Comorbidities, n (%) | 42 | |||
| Arterial Hypertension | 21 (50.00) | 12 (63.15) | 8 (53.30) | |
| Diabetes Mellitus | 7 (16.70) | 3 (15.80) | 3 (20.00) | |
| Coronary Artery Disease | 6 (14.30) | 2 (10.50) | 2 (13.30) | |
| Stroke | 1 (2.40) | 0 | 1 (6.60) | |
| Cancer (non-active) | 2 (4.80) | 0 | 2 (13.30) | |
| Obesity | 14 (33.30) | 9 (47.40) | 4 (26.60) | |
| Chronic Obstructive Pulmonary Disease | 1 (2.40) | 1 (5.30) | 0 | |
| Smoking (Active) | 2 (4.80) | 2 (10.50) | 0 | |
| Dyslipidemia | 7 (16.70) | 3 (15.80) | 2 (13.30) | |
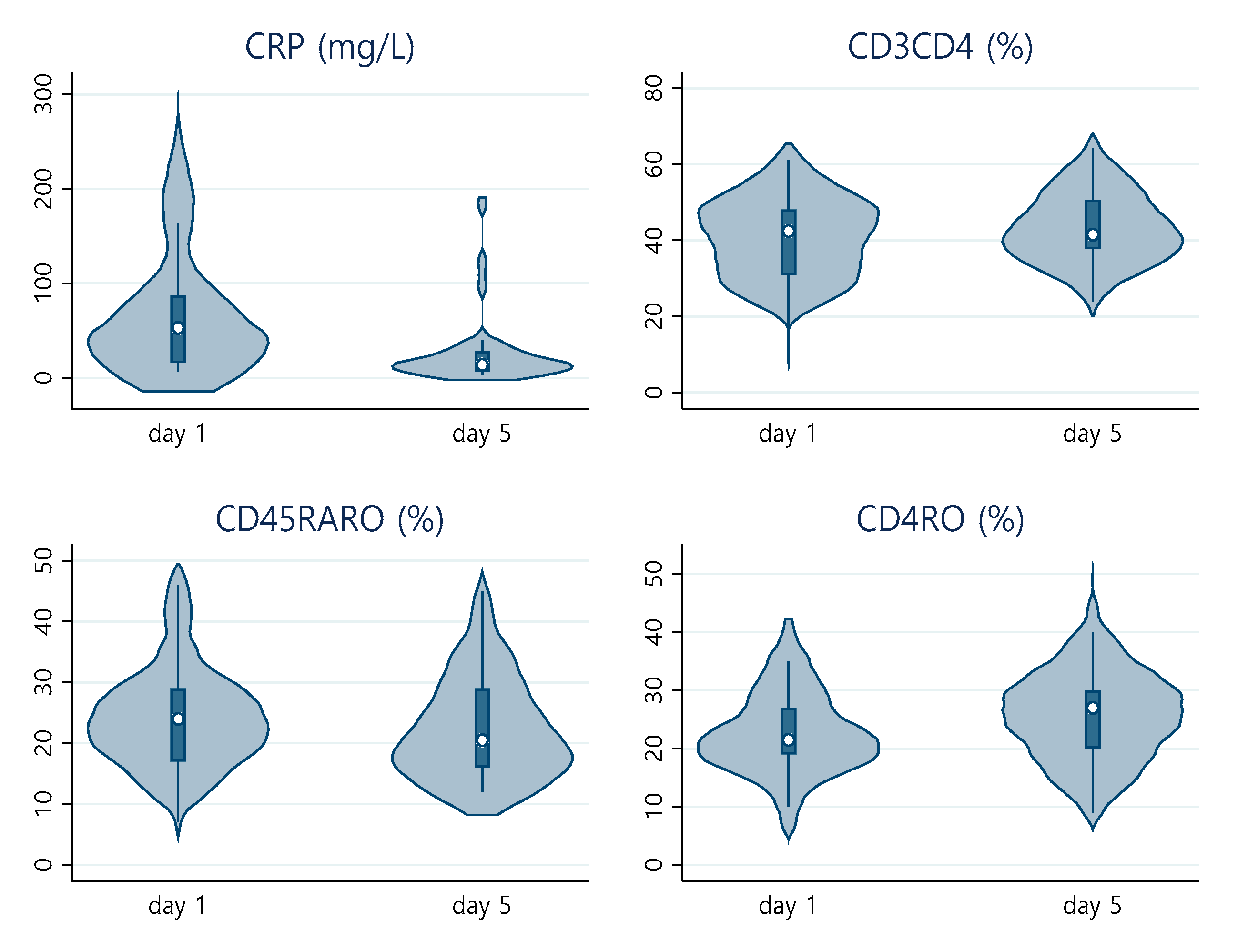
| Day 1 | ||||
| IL-6, IU/mL, mean (SD) | 32 | 39.46 (52.48) | 27.43 (21.27) | 60.73 (79.73) |
| C-reactive protein, mg/L, mean (SD) | 42 | 72.90 (70.98) | 70.05 (67.10) | 85.73 (85.06) |
| PO2/FiO2, mean (SD) | 41 | 294.59 (142.52) | 342.67 (138.76) | 244.15 (117.46) |
| CD45, mean absolute number | 42 | 1259 (811.88) | 1248.21 (712.66) | 923 (538.01) |
| CD3 %, mean (SD) | 42 | 66.69 (11.69) | 65.04 (10.82) | 64.86 (10.56) |
| CD3CD8 %, mean (SD) | 42 | 25.04 (10.38) | 26.46 (10.83) | 19.28 (6.22) |
| CD3CD4 %, mean (SD) | 42 | 40.39 (10.85) | 37.65 (10.84) | 45.03 (10.46) |
| CD3CD4CD8 %, mean (SD) | 42 | 1.17 (1.12) | 1.25 (1.45) | 1.14 (0.89) |
| CD16+56 %, mean (SD) | 42 | 19.33 (9.20) | 17.87 (8.86) | 21.54 (8.03) |
| CD19 %, mean (SD) | 42 | 13.09 (7.72) | 15.97 (7.33) | 12.82 (8.18) |
| CD45RA %, mean (SD) | 42 | 45.02 (12.17) | 46.42 (10.75) | 42.20 (10.37) |
| CD45RO %, mean (SD) | 42 | 30.29 (10.97) | 31.52 (11.56) | 31.40 (10.84) |
| CD45RA+RO+ %, mean (SD) | 42 | 24.40 (8.44) | 22 (6.70) | 25.73 (7.98) |
| CD4RA %, mean (SD) | 42 | 16.98 (11.14) | 13.78 (9.46) | 20.26 (12.45) |
| CD4RO %, mean (SD) | 42 | 22.74 (7.62) | 22.73 (8.59) | 24.86 (6.85) |
| Day 5 | ||||
| C-reactive protein, mg/L, mean (SD) | 29 | 27.93 (40.09) | 44.16 (58.83) | 15.84 (10.48) |
| PO2/FiO2, mean (SD) | 19 | 209.29 (135.93) | 260.98 (142.70) | 203.27 (138.08) |
| CD45, mean absolute number | 34 | 1745 (918.89) | 2042.68 (890.23) | 1527.78 (778.17) |
| CD3 %, mean (SD) | 34 | 67.07 (11.58) | 66.10 (10.14) | 68.61 (12.26) |
| CD3CD8 %, mean (SD) | 34 | 22.76 (7.49) | 23.66 (5.96) | 20.28 (7.83) |
| CD3CD4 %, mean (SD) | 34 | 43.28 (9.53) | 42.16 (8.56) | 47.07 (9.38) |
| CD3CD4CD8 %, mean (SD) | 34 | 1.36 (1.48) | 1.56 (1.93) | 1.24 (1.06) |
| CD16+56 %, mean (SD) | 34 | 12.65 (7.92) | 12.20 (7.35) | 11.48 (6.09) |
| CD19 %, mean (SD) | 34 | 18.88 (10.66) | 20.46 (8.81) | 18.17 (12.01) |
| CD45RA %, mean (SD) | 34 | 43.38 (14.55) | 43.87 (13.98) | 42.71 (13.32) |
| CD45RO %, mean (SD) | 34 | 33.35 (11.70) | 3287 (11.38) | 34 (10.66) |
| CD45RA+RO+ %, mean (SD) | 34 | 23.26 (8.93) | 23.25 (8.51) | 23.28 (8.37) |
| CD4RA %, mean (SD) | 34 | 16.26 (8.08) | 14.87 (7.35) | 18.07 (9.61) |
| CD4RO %, mean (SD) | 34 | 26.03 (8.52) | 25.18 (6.15) | 29.07 (9.12) |
| CT Burden of Disease | IL-6 | ||||||
|---|---|---|---|---|---|---|---|
| Cells | OR (95% CIs) | p-Value | FDR | Obs | Beta (95% CIs) | p-Value | Obs |
| CD3 | 0.957 (0.877, 1.044) | 0.327 | 0.546 | 32 | 0.208 (−1.584, 2.001) | 0.813 | 31 |
| CD3CD8 | 0.855 (0.741, 0.986) | 0.032 | 0.176 | 32 | 0.109 (−1.546, 1.764) | 0.893 | 31 |
| CD3CD4 | 1.059 (0.971, 1.155) | 0.191 | 0.499 | 32 | 0.095 (−1.647, 1.837) | 0.911 | 31 |
| CD3CD4CD8 | 0.902 (0.474, 1.717) | 0.755 | 0.755 | 32 | −3.263(−17.455, 10.929) | 0.640 | 31 |
| CD16+56 | 1.193 (1.019, 1.397) | 0.028 | 0.176 | 32 | −0.528 (−2.638, 1.581) | 0.611 | 31 |
| CD19 | 0.951 (0.857, 1.055) | 0.348 | 0.546 | 32 | 0.380 (−2.037, 2.799) | 0.749 | 31 |
| CD45RA | 0.974 (0.901, 1.053) | 0.518 | 0.633 | 32 | −0.725 (−2.541, 1.091) | 0.419 | 31 |
| CD45RO | 0.988 (0.920, 1.061) | 0.744 | 0.755 | 32 | 0.337 (−1.587, 2.262) | 0.721 | 31 |
| CD45RA+RO+ | 1.072 (0.961, 1.195) | 0.211 | 0.499 | 32 | 0.547 (−1.576, 2.670) | 0.601 | 31 |
| CD4RA | 1.057 (0.965, 1.157) | 0.227 | 0.499 | 32 | −0.561 (−2.278, 1.155) | 0.507 | 31 |
| CD4RO | 1.022 (0.925, 1.129) | 0.430 | 0.591 | 32 | 0.960 (−1.517, 3.439) | 0.433 | 31 |
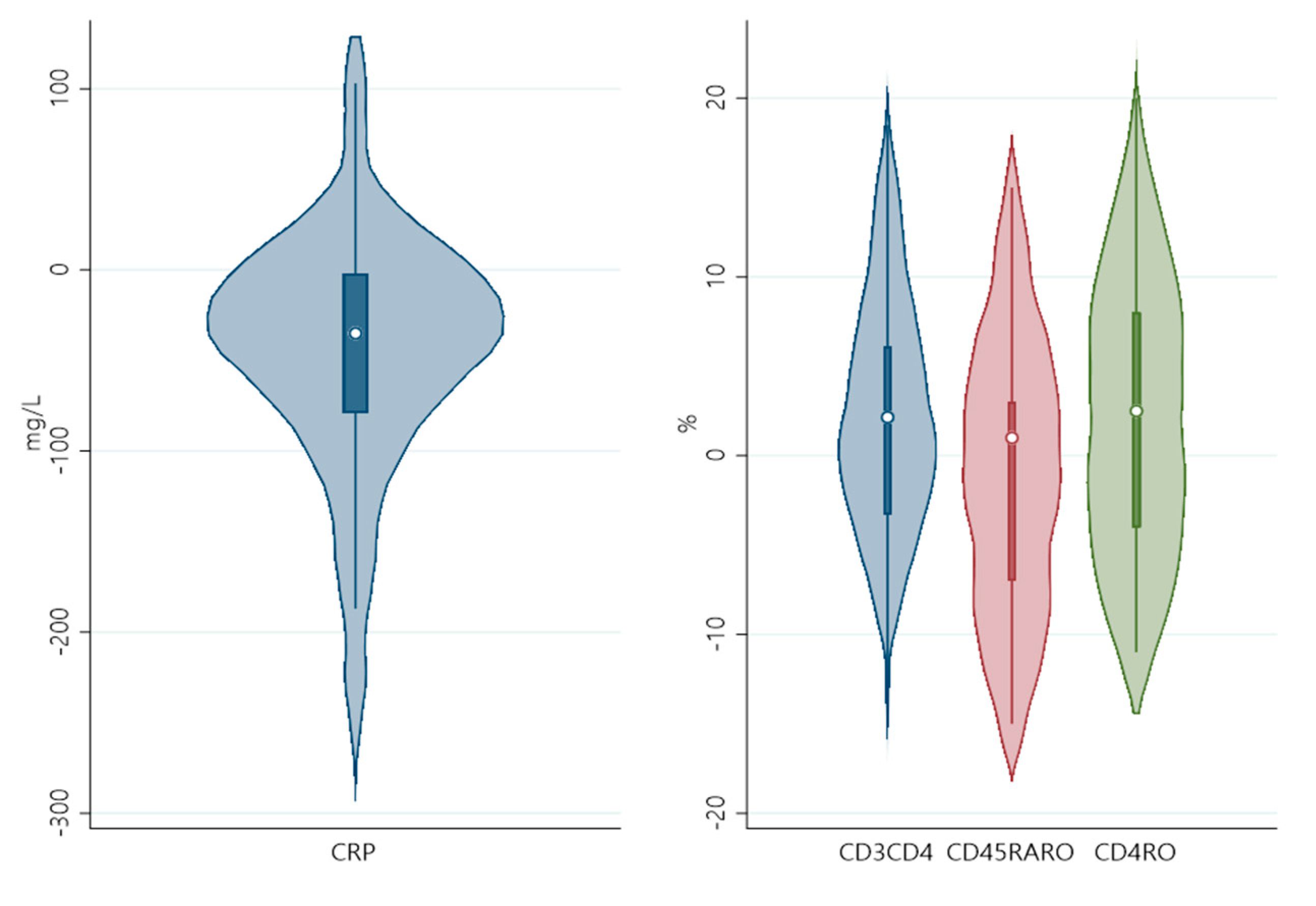
| Day5-Day1 | Delta CRP | Delta PO2/FiO2 | |||||
|---|---|---|---|---|---|---|---|
| DeltaCells | Beta (95% CIs) | p-Value | FDR | Obs | Beta (95% CIs) | p-Value | Obs |
| CD3 | −3.357(−7.720, 1.007) | 0.124 | 0.273 | 25 | −5.199(−13.540, 3.141) | 0.197 | 16 |
| CD3CD8 | 2.738(−5.057, 10.534) | 0.472 | 0.577 | 25 | −3.799(−16.438, 8.839) | 0.522 | 16 |
| CD3CD4 | −5.227(−10.353,−0.099) | 0.046 | 0.169 | 25 | −0.734(−11.779, 10.311) | 0.886 | 16 |
| CD3CD4CD8 | 13.895(−8.698, 36.488) | 0.214 | 0.392 | 25 | 17.552(−18.688, 53.793) | 0.309 | 16 |
| CD16+56 | 3.117(−0.747, 6.981) | 0.108 | 0.273 | 25 | 5.115(−1.558, 11.789) | 0.120 | 16 |
| CD19 | −0.898(−6.699, 4.902) | 0.750 | 0.825 | 25 | −2.639(−12.847, 7.569) | 0.581 | 16 |
| CD45RA | −1.241(−4.525, 2.044) | 0.440 | 0.577 | 25 | 0.210(−7.014, 7.436) | 0.950 | 16 |
| CD45RO | −2.001(−6.110, 2.108) | 0.322 | 0.506 | 25 | −3.652(−11.566, 4.262) | 0.332 | 16 |
| CD45RA+RO+ | 4.661(0.631, 8.689) | 0.026 | 0.143 | 25 | 3.827(−4.612, 12.267) | 0.340 | 16 |
| CD4RA | −0.464(−5.611, 4.683) | 0.853 | 0.853 | 25 | 6.252(−2.542, 15.048) | 0.146 | 16 |
| CD4RO | −5.327(−9.715, −0.938) | 0.020 | 0.143 | 25 | −1.157(−10.605, 8.291) | 0.793 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liontos, A.; Asimakopoulos, A.-G.; Markopoulos, G.S.; Biros, D.; Athanasiou, L.; Tsourlos, S.; Dova, L.; Rapti, I.-C.; Tsiakas, I.; Ntzani, E.; et al. Correlation of Lymphocyte Subpopulations, Clinical Features and Inflammatory Markers during Severe COVID-19 Onset. Pathogens 2023, 12, 414. https://doi.org/10.3390/pathogens12030414
Liontos A, Asimakopoulos A-G, Markopoulos GS, Biros D, Athanasiou L, Tsourlos S, Dova L, Rapti I-C, Tsiakas I, Ntzani E, et al. Correlation of Lymphocyte Subpopulations, Clinical Features and Inflammatory Markers during Severe COVID-19 Onset. Pathogens. 2023; 12(3):414. https://doi.org/10.3390/pathogens12030414
Chicago/Turabian StyleLiontos, Angelos, Alexandros-George Asimakopoulos, Georgios S. Markopoulos, Dimitrios Biros, Lazaros Athanasiou, Stavros Tsourlos, Leukothea Dova, Iro-Chrisavgi Rapti, Ilias Tsiakas, Evangelia Ntzani, and et al. 2023. "Correlation of Lymphocyte Subpopulations, Clinical Features and Inflammatory Markers during Severe COVID-19 Onset" Pathogens 12, no. 3: 414. https://doi.org/10.3390/pathogens12030414
APA StyleLiontos, A., Asimakopoulos, A.-G., Markopoulos, G. S., Biros, D., Athanasiou, L., Tsourlos, S., Dova, L., Rapti, I.-C., Tsiakas, I., Ntzani, E., Evangelou, E., Tzoulaki, I., Tsilidis, K., Vartholomatos, G., Dounousi, E., Milionis, H., & Christaki, E. (2023). Correlation of Lymphocyte Subpopulations, Clinical Features and Inflammatory Markers during Severe COVID-19 Onset. Pathogens, 12(3), 414. https://doi.org/10.3390/pathogens12030414










