The Devastating Rice Blast Airborne Pathogen Magnaporthe oryzae—A Review on Genes Studied with Mutant Analysis
Abstract
1. Introduction
2. The Features of M. oryzae Genome
2.1. Genome Sequencing
2.2. Transcriptomic and Secretome Analysis
3. Molecular Dissection of M. oryzae Biology
3.1. Genes Mainly Related to Fungal Development
3.2. Autophagy in Different Biological Processes of M. oryzae
3.3. Effector-Related Genes
3.4. Signaling Pathways in M. oryzae
3.4.1. Heterotrimeric G Protein Subunits and Regulatory Proteins
3.4.2. Components of cAMP Pathway
3.4.3. Mitogen-Activated Protein Kinase (MAPK) Cascade
3.4.4. Monomeric GTPase Modules (Ras Superfamily)
3.4.5. Target of Rapamycin (TOR) Signaling Pathway
3.4.6. Ubiquitination Cascade
3.5. Multifunctional Genes Involved in Different Aspects of M. oryzae Biology
3.5.1. Transcription Factors
3.5.2. Kinases and Phosphatases
3.5.3. Peroxisomal- and Mitochondrial-Related Genes
3.5.4. Other Important Genes in M. oryzae Biology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fairbrothers, D.E. An annotated bibliography of the floristic publications of New Jersey from 1753–1961. Bull. Torrey Bot. Club 1964, 91, 47–56. [Google Scholar] [CrossRef]
- Rossman, A.Y.; Howard, R.J.; Valent, B. Pyricularia grisea, the Correct Name for the Rice Blast Disease Fungus. Mycologia 1990, 82, 509–512. [Google Scholar] [CrossRef]
- Couch, B.C.; Kohn, L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 2002, 94, 683–693. [Google Scholar] [CrossRef] [PubMed]
- TeBeest, D.O.; Guerber, C.; Ditmore, M. Rice Blast. Available online: https://www.apsnet.org/edcenter/disandpath/fungalasco/pdlessons/Pages/RiceBlast.aspx (accessed on 15 January 2023).
- Meng, Q.; Gupta, R.; Min, C.W.; Kwon, S.W.; Wang, Y.; Je, B.I.; Kim, Y.J.; Jeon, J.S.; Agrawal, G.K.; Rakwal, R.; et al. Proteomics of Rice—Magnaporthe oryzae interaction: What have we learned so far? Front. Plant Sci. 2019, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Boddy, L. Pathogens of Autotrophs. In The Fungi; Academic Press: Cambridge, MA, USA, 2016; pp. 245–292. [Google Scholar] [CrossRef]
- Ryan, J.R. Biosecurity and Bioterrorism: Containing and Preventing Biological Threats, 2nd ed.; Butterworth-Heinemann: Oxford, UK, 2016; pp. 1–373. [Google Scholar] [CrossRef]
- Campos-Soriano, L.; Valè, G.; Lupotto, E.; Segundo, B.S. Investigation of rice blast development in susceptible and resistant rice cultivars using a gfp-expressing Magnaporthe oryzae isolate. Plant Pathol. 2013, 62, 1030–1037. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Ebbole, D.J. Magnaporthe as a Model for Understanding Host-Pathogen Interactions. Annu. Rev. Phytopathol. 2007, 45, 437–456. [Google Scholar] [CrossRef]
- Ribot, C.; Hirsch, J.; Balzergue, S.; Tharreau, D.; Nottéghem, J.-L.; Lebrun, M.-H.; Morel, J.-B. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 2008, 165, 114–124. [Google Scholar] [CrossRef]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic Fungal Cell Death Is Necessary for Infection by the Rice Blast Fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef]
- Kankanala, P.; Czymmek, K.; Valent, B. Roles for Rice Membrane Dynamics and Plasmodesmata during Biotrophic Invasion by the Blast Fungus. Plant Cell 2007, 19, 706–724. [Google Scholar] [CrossRef] [PubMed]
- Valent, B.; Khang, C.H. Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 2010, 13, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Khang, C.H.; Berruyer, R.; Giraldo, M.C.; Kankanala, P.; Park, S.-Y.; Czymmek, K.; Kang, S.; Valent, B. Translocation of Magnaporthe oryzae Effectors into Rice Cells and Their Subsequent Cell-to-Cell Movement. Plant Cell 2010, 22, 1388–1403. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef]
- Mosquera, G.; Giraldo, M.C.; Khang, C.H.; Coughlan, S.; Valent, B. Interaction Transcriptome Analysis Identifies Magnaporthe oryzae BAS1-4 as Biotrophy-Associated Secreted Proteins in Rice Blast Disease. Plant Cell 2009, 21, 1273–1290. [Google Scholar] [CrossRef]
- Chao, C.-C.T.; Ellingboe, A.H. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 1991, 69, 2130–2134. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Nunes, C.C.; Sailsbery, J.; Clay, B.; Brown, D.; John, T.; Oh, Y.; Young, N.; Fitzgerald, M.; Haas, B.J.; et al. Genome Sequences of Three Phytopathogenic Species of the Magnaporthaceae Family of Fungi. G3 Genes Genomes Genet. 2015, 5, 2539–2545. [Google Scholar] [CrossRef]
- Xue, M.; Yang, J.; Li, Z.; Hu, S.; Yao, N.; Dean, R.A.; Zhao, W.; Shen, M.; Zhang, H.; Li, C.; et al. Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus Magnaporthe oryzae. PLoS Genet. 2012, 8, e1002869. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae Genome Reveals a Fungal Effector, Which Is Able to Induce Resistance Response in Transgenic Rice Line Containing Resistance Gene, Pi54. Front. Plant Sci. 2016, 7, 1140. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Mahato, A.K.; Jain, P.; Rathour, R.; Sharma, V.; Sharma, T.R. Comparative Genomics Reveals the High Copy Number Variation of a Retro Transposon in Different Magnaporthe Isolates. Front. Microbiol. 2019, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Chen, M.; Zhong, Z.; Tang, W.; Lin, L.; Zhang, X.; Jiang, H.; Zhang, D.; Miao, C.; Tang, H.; et al. PacBio Sequencing Reveals Transposable Elements as a Key Contributor to Genomic Plasticity and Virulence Variation in Magnaporthe oryzae. Mol. Plant 2017, 10, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.L.; Eto, Y.; Nakao, T.; Tosa, Y.; Nakayashiki, H.; Mayama, S.; Leong, S.A. Analysis of the Structure of the AVR1-CO39 Avirulence Locus in Virulent Rice-Infecting Isolates of Magnaporthe grisea. Mol. Plant-Microbe Interact. 2002, 15, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lebrun, M.H.; Farrall, L.; Valent, B. Gain of Virulence Caused by Insertion of a Pot3 Transposon in a Magnaporthe grisea Avirulence Gene. Mol. Plant-Microbe Interact. 2001, 14, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Oliveira-Garcia, E.; Lin, G.; Hu, Y.; Dalby, M.; Migeon, P.; Tang, H.; Farman, M.; Cook, D.; White, F.F.; et al. Effector gene reshuffling involves dispensable mini-chromosomes in the wheat blast fungus. PLoS Genet. 2019, 15, e1008272. [Google Scholar] [CrossRef]
- Oh, Y.; Donofrio, N.; Pan, H.; Coughlan, S.; Brown, D.E.; Meng, S.; Mitchell, T.; Dean, R.A. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2008, 9, R85. [Google Scholar] [CrossRef] [PubMed]
- Mathioni, S.M.; Beló, A.; Rizzo, C.J.; Dean, R.A.; Donofrio, N.M. Transcriptome profiling of the rice blast fungus during invasive plant infection and in vitro stresses. BMC Genom. 2011, 12, 49. [Google Scholar] [CrossRef]
- Kawahara, Y.; Oono, Y.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Minami, E. Simultaneous RNA-Seq Analysis of a Mixed Transcriptome of Rice and Blast Fungus Interaction. PLoS ONE 2012, 7, e49423. [Google Scholar] [CrossRef]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Guo, X.; Zhang, X.; Chen, Y.; Liu, Y.; et al. Global Genome and Transcriptome Analyses of Magnaporthe oryzae Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution. PLoS Pathog. 2015, 11, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-H.; Jeong, S.-H.; Kim, S.H.; Singh, R.; Lee, J.-E.; Cho, Y.-S.; Agrawal, G.K.; Rakwal, R.; Jwa, N.-S. Secretome analysis of Magnaporthe oryzae using in vitro systems. Proteomics 2012, 12, 878–900. [Google Scholar] [CrossRef]
- Liu, N.; Qi, L.; Huang, M.; Chen, D.; Yin, C.; Zhang, Y.; Wang, X.; Yuan, G.; Wang, R.-J.; Yang, J.; et al. Comparative Secretome Analysis of Magnaporthe oryzae Identified Proteins Involved in Virulence and Cell Wall Integrity. Genom. Proteom. Bioinform. 2022, 20, 728–746. [Google Scholar] [CrossRef]
- Foster, A.J.; Martin-Urdiroz, M.; Yan, X.; Wright, H.S.; Soanes, D.M.; Talbot, N.J. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rowe, D.; Subedi, P.; Zhang, W.; Suelter, T.; Valent, B.; Cook, D.E. CRISPR-Cas12a induced DNA double-strand breaks are repaired by multiple pathways with different mutation profiles in Magnaporthe oryzae. Nat. Commun. 2022, 13, 7168. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Singh, P.; Chung, W.-C.; Ash, J.; Kim, T.S.; Hang, L.; Park, S. Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 2006, 43, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, G.; Lin, C.; He, C. Conidiophore Stalk-less1 Encodes a Putative Zinc-Finger Protein Involved in the Early Stage of Conidiation and Mycelial Infection in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2009, 22, 402–410. [Google Scholar] [CrossRef]
- Liu, S.; Wei, Y.; Zhang, S.-H. The C3HC type zinc-finger protein (ZFC3) interacting with Lon/MAP1 is important for mitochondrial gene regulation, infection hypha development and longevity of Magnaporthe oryzae. BMC Microbiol. 2020, 20, 23. [Google Scholar] [CrossRef]
- Saitoh, H.; Hirabuchi, A.; Fujisawa, S.; Mitsuoka, C.; Terauchi, R.; Takano, Y. MoST1 encoding a hexose transporter-like protein is involved in both conidiation and mycelial melanization of Magnaporthe oryzae. FEMS Microbiol. Lett. 2014, 352, 104–113. [Google Scholar] [CrossRef]
- Geoghegan, I.A.; Gurr, S.J. Investigating chitin deacetylation and chitosan hydrolysis during vegetative growth in Magnaporthe oryzae. Cell. Microbiol. 2017, 19, e12743. [Google Scholar] [CrossRef]
- Sangappillai, V.; Nadarajah, K. Fatty Acid Synthase Beta Dehydratase in the Lipid Biosynthesis Pathway Is Required for Conidiogenesis, Pigmentation and Appressorium Formation in Magnaporthe oryzae S6. Int. J. Mol. Sci. 2020, 21, 7224. [Google Scholar] [CrossRef]
- Saha, P.; Ghosh, S.; Roy-Barman, S. MoLAEA Regulates Secondary Metabolism in Magnaporthe oryzae. Msphere 2020, 5, e00936-19. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Wang, Y.; Li, H.-J.; Lu, J.-P.; Liu, X.-H.; Lin, F.-C. Disruption and molecular characterization of calpains-related (MoCAPN1, MoCAPN3 and MoCAPN4) genes in Magnaporthe oryzae. Microbiol. Res. 2014, 169, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, J.; Zhou, W.; Chen, X.-L.; Huang, J.-G.; Cheng, Z.-H.; Zhao, W.-S.; Zhang, Y.; Peng, Y.-L. A spindle pole antigen gene MoSPA2 is important for polar cell growth of vegetative hyphae and conidia, but is dispensable for pathogenicity in Magnaporthe oryzae. Curr. Genet. 2014, 60, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lu, J.-P.; Li, X.-D.; Liu, X.-H.; Min, H.; Lin, F.-C. Magnaporthe oryzae MTP1 gene encodes a type III transmembrane protein involved in conidiation and conidial germination. J. Zhejiang Univ. B 2008, 9, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Izawa, M.; Takekawa, O.; Arie, T.; Teraoka, T.; Yoshida, M.; Kimura, M.; Kamakura, T. Inhibition of histone deacetylase causes reduction of appressorium formation in the rice blast fungus Magnaporthe oryzae. J. Gen. Appl. Microbiol. 2009, 55, 489–498. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Wang, S.-Z.; Zhang, Z.; Hao, Z.-N.; Shi, X.-X.; Li, L.; Zhu, X.-M.; Qiu, H.-P.; Chai, R.-Y.; Wang, Y.-L.; et al. MAT Loci Play Crucial Roles in Sexual Development but Are Dispensable for Asexual Reproduction and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae. J. Fungi 2021, 7, 858. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef]
- Liu, X.-H.; Lu, J.-P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.-C. Involvement of a Magnaporthe grisea Serine/Threonine kinase gene, Mg ATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef]
- Khan, I.A.; Lu, J.-P.; Liu, X.-H.; Rehman, A.; Lin, F.-C. Multifunction of autophagy-related genes in filamentous fungi. Microbiol. Res. 2012, 167, 339–345. [Google Scholar] [CrossRef]
- Deng, Y.Z.; Naqvi, N.I. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2010, 6, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-B.; Liu, X.-H.; Lu, J.-P.; Zhang, L.; Min, H.; Lin, F.-C. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010, 6, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Zhao, Y.-H.; Zhu, X.-M.; Zeng, X.-Q.; Huang, L.-Y.; Dong, B.; Su, Z.-Z.; Wang, Y.; Lu, J.-P.; Lin, F.-C. Autophagy-related protein MoAtg14 is involved in differentiation, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2017, 7, 40018. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Liu, X.-H.; Lu, J.-P.; Zhang, F.-S.; Gao, H.-M.; Wang, H.-K.; Lin, F.-C. MgAtg9 trafficking in Magnaporthe oryzae. Autophagy 2009, 5, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-P.; Liu, X.-H.; Feng, X.-X.; Min, H.; Lin, F.-C. An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr. Genet. 2009, 55, 461–473. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, J.; Wu, X.; Liang, S.; Zhu, X.; Li, L.; Lu, J.; Liu, X.; Lin, F. MoOpy2 is essential for fungal development, pathogenicity, and autophagy in Magnaporthe oryzae. Environ. Microbiol. 2022, 24, 1653–1671. [Google Scholar] [CrossRef]
- Shi, H.-B.; Chen, N.; Zhu, X.-M.; Su, Z.-Z.; Wang, J.-Y.; Lu, J.-P.; Liu, X.-H.; Lin, F.-C. The casein kinase MoYck1 regulates development, autophagy, and virulence in the rice blast fungus. Virulence 2019, 10, 719–733. [Google Scholar] [CrossRef]
- Zhu, X.-M.; Liang, S.; Shi, H.-B.; Lu, J.-P.; Dong, B.; Liao, Q.-S.; Lin, F.-C.; Liu, X.-H. VPS9 domain-containing proteins are essential for autophagy and endocytosis in Pyricularia oryzae. Environ. Microbiol. 2018, 20, 1516–1530. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, C.; Yang, J.; Feng, W.; Liu, X.; Zuo, R.; Wang, J.; Yang, L.; Zhong, K.; Gao, C.; et al. Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoAtg3 and MoAtg9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy 2019, 15, 1234–1257. [Google Scholar] [CrossRef]
- Zhang, S.; Liang, M.; Naqvi, N.I.; Lin, C.; Qian, W.; Zhang, L.-H.; Deng, Y.Z. Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae. Autophagy 2017, 13, 1318–1330. [Google Scholar] [CrossRef]
- Zheng, W.; Zhou, J.; He, Y.; Xie, Q.; Chen, A.; Zheng, H.; Shi, L.; Zhao, X.; Zhang, C.; Huang, Q.; et al. Retromer Is Essential for Autophagy-Dependent Plant Infection by the Rice Blast Fungus. PLoS Genet. 2015, 11, e1005704. [Google Scholar] [CrossRef]
- Sun, L.; Qian, H.; Wu, M.; Zhao, W.; Liu, M.; Wei, Y.; Zhu, X.; Li, L.; Lu, J.; Lin, F.; et al. A Subunit of ESCRT-III, MoIst1, Is Involved in Fungal Development, Pathogenicity, and Autophagy in Magnaporthe oryzae. Front. Plant Sci. 2022, 13, 845139. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yin, Z.; Zhang, Z.; Liang, Y. Functional analysis of MoSnf7 in Magnaporthe oryzae. Fungal Genet. Biol. 2018, 121, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Jiang, H.; Aron, O.; Wang, M.; Wang, X.; Chen, J.; Lin, B.; Chen, X.; Zheng, Q.; Gao, X.; et al. Endoplasmic reticulum-associated degradation mediated by MoHrd1 and MoDer1 is pivotal for appressorium development and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 4953–4973. [Google Scholar] [CrossRef]
- He, M.; Xu, Y.; Chen, J.; Luo, Y.; Lv, Y.; Su, J.; Kershaw, M.J.; Li, W.; Wang, J.; Yin, J.; et al. MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 2018, 14, 1543–1561. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wu, L.; Wu, H.; Zhang, X.; Mei, J.; Zhou, X.; Wang, G.; Liu, W. Arginine methylation is required for remodelling pre- mRNA splicing and induction of autophagy in rice blast fungus. New Phytol. 2020, 225, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Shi, W.; Xu, X.-W.; Li, Z.-G.; Yin, C.-F.; Peng, J.-B.; Pan, S.; Chen, X.-L.; Zhao, W.-S.; Zhang, Y.; et al. Glutamate synthase MoGlt1-mediated glutamate homeostasis is important for autophagy, virulence and conidiation in the rice blast fungus. Mol. Plant Pathol. 2018, 19, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-M.; Li, L.; Cai, Y.-Y.; Wu, X.-Y.; Shi, H.-B.; Liang, S.; Qu, Y.-M.; Naqvi, N.I.; Del Poeta, M.; Dong, B.; et al. A VASt-domain protein regulates autophagy, membrane tension, and sterol homeostasis in rice blast fungus. Autophagy 2021, 17, 2939–2961. [Google Scholar] [CrossRef]
- Rovenich, H.; Boshoven, J.C.; Thomma, B.P. Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 2014, 20, 96–103. [Google Scholar] [CrossRef]
- de Guillen, K.; Ortiz-Vallejo, D.; Gracy, J.; Fournier, E.; Kroj, T.; Padilla, A. Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi. PLoS Pathog. 2015, 11, e1005228. [Google Scholar] [CrossRef]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the 1146 blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 1995, 8, 939–948. [Google Scholar] [CrossRef]
- Khang, C.H.; Park, S.-Y.; Lee, Y.-H.; Valent, B.; Kang, S. Genome Organization and Evolution of the AVR-Pita Avirulence Gene Family in the Magnaporthe grisea Species Complex. Mol. Plant-Microbe Interact. 2008, 21, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhang, Z.; Zhao, Q.; Feng, Q.; Zhang, H.; et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef]
- Miki, S.; Matsui, K.; Kito, H.; Otsuka, K.; Ashizawa, T.; Yasuda, N.; Fukiya, S.; Sato, J.; Hirayae, K.; Fujita, Y.; et al. Molecular cloning and characterization of the AVR-Pia locus from a Japanese field isolate of Magnaporthe oryzae. Mol. Plant Pathol. 2009, 10, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Mochizuki, S.; Ishii-Minami, N.; Fujisawa, Y.; Kawahara, Y.; Yoshida, Y.; Okada, K.; Ando, S.; Matsumura, H.; Terauchi, R.; et al. Magnaporthe oryzae Glycine-Rich Secretion Protein, Rbf1 Critically Participates in Pathogenicity through the Focal Formation of the Biotrophic Interfacial Complex. PLoS Pathog. 2016, 12, e1005921. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, C.-Y.; Park, S.-Y.; Kim, K.-T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef]
- Saitoh, H.; Fujisawa, S.; Mitsuoka, C.; Ito, A.; Hirabuchi, A.; Ikeda, K.; Irieda, H.; Yoshino, K.; Yoshida, K.; Matsumura, H.; et al. Large-Scale Gene Disruption in Magnaporthe oryzae Identifies MC69, a Secreted Protein Required for Infection by Monocot and Dicot Fungal Pathogens. PLoS Pathog. 2012, 8, e1002711. [Google Scholar] [CrossRef]
- Yi, M.; Chi, M.-H.; Khang, C.H.; Park, S.-Y.; Kang, S.; Valent, B.; Lee, Y.-H. The ER Chaperone LHS1 Is Involved in Asexual Development and Rice Infection by the Blast Fungus Magnaporthe oryzae. Plant Cell 2009, 21, 681–695. [Google Scholar] [CrossRef]
- Talbot, N.J.; Kershaw, M.J.; Wakley, G.E.; De Vries, O.M.; Wessels, J.G.; Hamer, J.E. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 1996, 8, 985–999. [Google Scholar] [CrossRef]
- Kim, S.; Ahn, I.-P.; Rho, H.-S.; Lee, Y.-H. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 2005, 57, 1224–1237. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Hydrophobins: Multipurpose Proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Mitchell, T.K.; Dean, R.A. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 2007, 273, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.-Z.; Zhang, L.; Zhuang, H.-Q.; Shi, W.-J.; Yang, X.-F.; Qiu, D.-W.; Zeng, H.-M. The Secreted Protein MoHrip1 Is Necessary for the Virulence of Magnaporthe oryzae. Int. J. Mol. Sci. 2019, 20, 1643. [Google Scholar] [CrossRef] [PubMed]
- Mogga, V.; Delventhal, R.; Weidenbach, D.; Langer, S.; Bertram, P.M.; Andresen, K.; Thines, E.; Kroj, T.; Schaffrath, U. Magnaporthe oryzae effectors MoHEG13 and MoHEG16 interfere with host infection and MoHEG13 counteracts cell death caused by Magnaporthe-NLPs in tobacco. Plant Cell Rep. 2016, 35, 1169–1185. [Google Scholar] [CrossRef]
- Oliveira-Garcia, E.; Valent, B. How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 2015, 26, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.; et al. Effector-Mediated Suppression of Chitin-Triggered Immunity by Magnaporthe oryzae Is Necessary for Rice Blast Disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Liu, M.; Wang, Y.; Zou, Y.; You, Y.; Yang, L.; Hu, J.; Zhang, H.; Zheng, X.; et al. Magnaporthe oryzae Auxiliary Activity Protein MoAa91 Functions as Chitin-Binding Protein To Induce Appressorium Formation on Artificial Inductive Surfaces and Suppress Plant Immunity. Mbio 2020, 11, e03304-19. [Google Scholar] [CrossRef]
- Dai, M.-D.; Wu, M.; Li, Y.; Su, Z.-Z.; Lin, F.-C.; Liu, X.-H. The chitin deacetylase PoCda7 is involved in the pathogenicity of Pyricularia oryzae. Microbiol. Res. 2021, 248, 126749. [Google Scholar] [CrossRef]
- Chen, X.-L.; Shi, T.; Yang, J.; Shi, W.; Gao, X.; Chen, D.; Xu, X.; Xu, J.-R.; Talbot, N.J.; Peng, Y.-L. N-Glycosylation of Effector Proteins by an α-1,3-Mannosyltransferase Is Required for the Rice Blast Fungus to Evade Host Innate Immunity. Plant Cell 2014, 26, 1360–1376. [Google Scholar] [CrossRef]
- Wei, Y.-Y.; Liang, S.; Zhang, Y.-R.; Lu, J.-P.; Lin, F.-C.; Liu, X.-H. MoSec61β, the beta subunit of Sec61, is involved in fungal development and pathogenicity, plant immunity, and ER-phagy in Magnaporthe oryzae. Virulence 2020, 11, 1685–1700. [Google Scholar] [CrossRef]
- Qi, Z.; Liu, M.; Dong, Y.; Zhu, Q.; Li, L.; Li, B.; Yang, J.; Li, Y.; Ru, Y.; Zhang, H.; et al. The syntaxin protein (MoSyn8) mediates intracellular trafficking to regulate conidiogenesis and pathogenicity of rice blast fungus. New Phytol. 2016, 209, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, S.; Yin, Z.; Liu, M.; Li, B.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoVrp1, a putative verprolin protein, is required for asexual development and infection in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2017, 7, srep41148. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.I.; Strathmann, M.P.; Gautam, N. Diversity of G Proteins in Signal Transduction. Science 1991, 252, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Dean, R.A. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 1997, 10, 1075–1086. [Google Scholar] [CrossRef]
- Nishimura, M.; Park, G.; Xu, J.-R. The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol. Microbiol. 2003, 50, 231–243. [Google Scholar] [CrossRef]
- Li, Y.; Que, Y.; Liu, Y.; Yue, X.; Meng, X.; Zhang, Z.; Wang, Z. The putative Gγ subunit gene MGG1 is required for conidiation, appressorium formation, mating and pathogenicity in Magnaporthe oryzae. Curr. Genet. 2015, 61, 641–651. [Google Scholar] [CrossRef]
- De Vries, L.; Zheng, B.; Fischer, T.; Elenko, E.; Farquhar, M.G. The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 235–271. [Google Scholar] [CrossRef]
- Siderovski, D.P.; Willard, F.S. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 2005, 1, 51–66. [Google Scholar] [CrossRef]
- Liu, H.; Suresh, A.; Willard, F.S.; Siderovski, D.P.; Lu, S.; Naqvi, N.I. Rgs1 regulates multiple Gα subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 2007, 26, 690–700. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, W.; Liu, K.; Huang, Q.; Zhang, X.; Yan, X.; Chen, Y.; Wang, J.; Qi, Z.; Wang, Z.; et al. Eight RGS and RGS-like Proteins Orchestrate Growth, Differentiation, and Pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1002450. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yan, X.; Ryder, L.S.; Cruz-Mireles, N.; Soanes, D.M.; Molinari, C.; Foster, A.J.; Talbot, N.J. Rgs1 is a regulator of effector gene expression during plant infection by the rice blast fungus Magnaporthe oryzae. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, X.; Wang, J.; Yang, L.; Feng, W.; Chen, C.; Gao, C.; Zhang, H.; Zheng, X.; Wang, P.; et al. MoMip11, a MoRgs7-interacting protein, functions as a scaffolding protein to regulate cAMP signaling and pathogenicity in the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2018, 20, 3168–3185. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Dean, R.A. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell 1993, 5, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.R.; Urban, M.; Sweigard, J.; Hamer, J.E. The CPKA gene of Magnaporthe grisea is required for appressorial function. Mol. Plant-Micro. Interact. 1997, 10, 187–194. [Google Scholar] [CrossRef]
- Selvaraj, P.; Tham, H.F.; Ramanujam, R.; Naqvi, N.I.; Fai, T.H. Subcellular compartmentation, interdependency and dynamics of the cyclic AMP-dependent PKA subunits during pathogenic differentiation in rice blast. Mol. Microbiol. 2017, 105, 484–504. [Google Scholar] [CrossRef]
- Ramanujam, R.; Naqvi, N.I. PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in Magnaporthe oryzae. PLoS Pathog. 2010, 6, e1000897. [Google Scholar] [CrossRef]
- Liu, X.; Qian, B.; Gao, C.; Huang, S.; Cai, Y.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. The Putative Protein Phosphatase MoYvh1 Functions Upstream of MoPdeH to Regulate the Development and Pathogenicity in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2016, 29, 496–507. [Google Scholar] [CrossRef]
- Choi, W.; Dean, R.A. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 1997, 9, 1973–1983. [Google Scholar]
- Zhou, X.; Zhang, H.; Li, G.; Shaw, B.; Xu, J.-R. The Cyclase-Associated Protein Cap1 Is Important for Proper Regulation of Infection-Related Morphogenesis in Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002911. [Google Scholar] [CrossRef]
- Nishimura, M.; Fukada, J.; Moriwaki, A.; Fujikawa, T.; Ohashi, M.; Hibi, T.; Hayashi, N. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci. Biotechnol. Biochem. 2009, 73, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Li, Y.; Yue, X.; Wang, C.; Que, Y.; Kong, D.; Ma, Z.; Talbot, N.J.; Wang, Z. Two Novel Transcriptional Regulators Are Essential for Infection-related Morphogenesis and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1002385. [Google Scholar] [CrossRef] [PubMed]
- Badaruddin, M.; Holcombe, L.J.; Wilson, R.A.; Wang, Z.-Y.; Kershaw, M.J.; Talbot, N.J. Glycogen Metabolic Genes Are Involved in Trehalose-6-Phosphate Synthase-Mediated Regulation of Pathogenicity by the Rice Blast Fungus Magnaporthe oryzae. PLoS Pathog. 2013, 9, e1003604. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.R.; Hamer, J.E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996, 10, 2696–2706. [Google Scholar] [CrossRef]
- Xu, J.-R.; Staiger, C.J.; Hamer, J.E. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 1998, 95, 12713–12718. [Google Scholar] [CrossRef]
- Dixon, K.P.; Xu, J.-R.; Smirnoff, N.; Talbot, N.J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 1999, 11, 2045–2058. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Q.; Guo, X.; Guo, M.; Qi, Z.; Tang, W.; Dong, Y.; Ye, W.; Zheng, X.; Wang, P.; et al. Pleiotropic Function of the Putative Zinc-Finger Protein MoMsn2 in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2014, 27, 446–460. [Google Scholar] [CrossRef]
- Zhao, X.; Kim, Y.; Park, G.; Xu, J.-R. A Mitogen-Activated Protein Kinase Cascade Regulating Infection-Related Morphogenesis in Magnaporthe grisea. Plant Cell 2005, 17, 1317–1329. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, C.; Zhang, Q.; Qi, L.; Li, C.; Xu, J.-R. Thioredoxins are involved in the activation of the PMK1 MAP kinase pathway during appressorium penetration and invasive growth in Magnaporthe oryzae. Environ. Microbiol. 2016, 18, 3768–3784. [Google Scholar] [CrossRef]
- Jeon, J.; Goh, J.; Yoo, S.; Chi, M.-H.; Choi, J.; Rho, H.-S.; Park, J.; Han, S.-S.; Kim, B.R.; Park, S.-Y.; et al. A Putative MAP Kinase Kinase Kinase, MCK1, Is Required for Cell Wall Integrity and Pathogenicity of the Rice Blast Fungus, Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2008, 21, 525–534. [Google Scholar] [CrossRef]
- Yin, Z.; Tang, W.; Wang, J.; Liu, X.; Yang, L.; Gao, C.; Zhang, J.; Zhang, H.; Zheng, X.; Wang, P.; et al. Phosphodiesterase MoPdeH targets MoM ck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2016, 17, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xue, C.; Bruno, K.; Nishimura, M.; Xu, J.-R. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact. 2004, 17, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Yin, Z.; Wu, H.; Liu, P.; Liu, X.; Liu, M.; Yu, R.; Gao, C.; Zhang, H.; Zheng, X.; et al. Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PLoS Pathog. 2021, 17, e1009080. [Google Scholar] [CrossRef] [PubMed]
- Osés-Ruiz, M.; Cruz-Mireles, N.; Martin-Urdiroz, M.; Soanes, D.M.; Eseola, A.B.; Tang, B.; Derbyshire, P.; Nielsen, M.; Cheema, J.; Were, V.; et al. Appressorium-mediated plant infection by Magnaporthe oryzae is regulated by a Pmk1-dependent hierarchical transcriptional network. Nat. Microbiol. 2021, 6, 1383–1397. [Google Scholar] [CrossRef]
- Park, G.; Xue, C.; Zheng, L.; Lam, S.; Xu, J.-R. MST12 Regulates Infectious Growth But Not Appressorium Formation in the Rice Blast Fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 2002, 15, 183–192. [Google Scholar] [CrossRef]
- Galhano, R.; Illana, A.; Ryder, L.S.; Rodríguez-Romero, J.; Demuez, M.; Badaruddin, M.; Martinez-Rocha, A.L.; Soanes, D.M.; Studholme, D.J.; Talbot, N.J.; et al. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 2017, 13, e1006516. [Google Scholar] [CrossRef]
- Li, G.; Zhou, X.; Kong, L.; Wang, Y.; Zhang, H.; Zhu, H.; Mitchell, T.K.; Dean, R.A.; Xu, J.-R. MoSfl1 Is Important for Virulence and Heat Tolerance in Magnaporthe oryzae. PLoS ONE 2011, 6, e19951. [Google Scholar] [CrossRef]
- Xue, C.; Park, G.; Choi, W.; Zheng, L.; Dean, R.A.; Xu, J.-R. Two Novel Fungal Virulence Genes Specifically Expressed in Appressoria of the Rice Blast Fungus. Plant Cell 2002, 14, 2107–2119. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, C.; Kong, L.; Li, G.; Xu, J.-R. A Pmk1-Interacting Gene Is Involved in Appressorium Differentiation and Plant Infection in Magnaporthe oryzae. Eukaryot. Cell 2011, 10, 1062–1070. [Google Scholar] [CrossRef]
- Mehrabi, R.; Ding, S.; Xu, J.-R. MADS-Box Transcription Factor Mig1 Is Required for Infectious Growth in Magnaporthe grisea. Eukaryot. Cell 2008, 7, 791–799. [Google Scholar] [CrossRef]
- Qi, Z.; Wang, Q.; Dou, X.; Wang, W.; Zhao, Q.; Lv, R.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 2012, 13, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Dagdas, Y.F.; Zhu, X.; Zheng, S.; Chen, L.; Cartwright, Z.; Talbot, N.J.; Wang, Z. The glycogen synthase kinase MoGsk1, regulated by Mps1 MAP kinase, is required for fungal development and pathogenicity in Magnaporthe oryzae. Sci. Rep. 2017, 7, 945. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, G.; Xu, J.-R.; Jiang, C. Penetration Peg Formation and Invasive Hyphae Development Require Stage-Specific Activation of MoGTI1 in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2016, 29, 36–45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papadaki, P.; Pizon, V.; Onken, B.; Chang, E.C. Two Ras Pathways in Fission Yeast Are Differentially Regulated by Two Ras Guanine Nucleotide Exchange Factors. Mol. Cell. Biol. 2002, 22, 4598–4606. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, X.; Xue, C.; Dai, Y.; Xu, J.-R. Bypassing Both Surface Attachment and Surface Recognition Requirements for Appressorium Formation by Overactive Ras Signaling in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2014, 27, 996–1004. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Basiewicz, M.; Soanes, D.M.; Yan, X.; Ryder, L.S.; Csukai, M.; Oses-Ruiz, M.; Valent, B.; Talbot, N.J. Conidial Morphogenesis and Septin-Mediated Plant Infection Require Smo1, a Ras GTPase-Activating Protein in Magnaporthe oryzae. Genetics 2019, 211, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Aboelfotoh Hendy, A.; Xing, J.; Chen, X.; Chen, X.-L. The farnesyltransferase β-subunit RAM1 regulates localization of RAS proteins and appressorium-mediated infection in Magnaporthe oryzae. Mol. Plant Pathol. 2019, 20, 1264–1278. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, J.; Huang, P.; Liu, X.; Lu, J.; Lin, F.-C. PoRal2 Is Involved in Appressorium Formation and Virulence via Pmk1 MAPK Pathways in the Rice Blast Fungus Pyricularia oryzae. Front. Plant Sci. 2021, 12, 702368. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, W.; Zheng, S.; Zhang, D.; Sang, W.; Chen, X.; Li, G.; Lu, G.; Wang, Z. Rac1 Is Required for Pathogenicity and Chm1-Dependent Conidiogenesis in Rice Fungal Pathogen Magnaporthe grisea. PLoS Pathog. 2008, 4, e1000202. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, Z.; Chen, J.; Liu, W.; Ke, H.; Zhou, J.; Lu, G.; Darvill, A.G.; Albersheim, P.; Wu, S.; et al. A Cdc42 ortholog is required for penetration and virulence of Magnaporthe grisea. Fungal Genet. Biol. 2009, 46, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Kim, J.-O.; Han, J.-H.; Gumilang, A.; Lee, Y.-H.; Kim, K.S. A Small GTPase RHO2 Plays an Important Role in Pre-infection Development in the Rice Blast Pathogen Magnaporthe oryzae. Plant Pathol. J. 2018, 34, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Chen, J.; Liu, W.; Zheng, S.; Zhou, J.; Lu, G.; Wang, Z. A Rho3 Homolog Is Essential for Appressorium Development and Pathogenicity of Magnaporthe grisea. Eukaryot. Cell 2007, 6, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Chen, X.; Zhong, Z.; Chen, M.; Shi, L.; Zheng, H.; Lin, Y.; Zhang, D.; Lu, G.; Li, G.; et al. Putative RhoGAP proteins orchestrate vegetative growth, conidiogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2014, 67, 37–50. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, T.; Chen, L.; Zheng, S.; Chen, S.; Zhang, D.; Li, G.; Wang, Z. Arf6 controls endocytosis and polarity during asexual development of Magnaporthe oryzae. FEMS Microbiol. Lett. 2016, 363, fnw248. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.; Yang, L.; Li, L.; Zhong, K.; Wang, W.; Liu, M.; Li, Y.; Liu, X.; Yu, R.; He, J.; et al. System-Wide Characterization of MoArf GTPase Family Proteins and Adaptor Protein MoGga1 Involved in the Development and Pathogenicity of Magnaporthe oryzae. mBio 2019, 10, e02398-19. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Li, L.; Yu, R.; He, J.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. The ArfGAP protein MoGlo3 regulates the development and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 3982–3996. [Google Scholar] [CrossRef]
- Liu, X.H.; Chen, S.M.; Gao, H.M.; Ning, G.A.; Shi, H.B.; Wang, Y.; Dong, B.; Qi, Y.Y.; Zhang, D.M.; Lu, G.D.; et al. The small GTP ase MoYpt 7 is required for membrane fusion in autophagy and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 4495–4510. [Google Scholar] [CrossRef]
- Wu, C.; Lin, Y.; Zheng, H.; Abubakar, Y.S.; Peng, M.; Li, J.; Yu, Z.; Wang, Z.; Naqvi, N.I.; Li, G.; et al. The retromer CSC subcomplex is recruited by MoYpt7 and sequentially sorted by MoVps17 for effective conidiation and pathogenicity of the rice blast fungus. Mol. Plant Pathol. 2021, 22, 284–298. [Google Scholar] [CrossRef]
- Marroquin-Guzman, M.; Wilson, R.A. GATA-Dependent Glutaminolysis Drives Appressorium Formation in Magnaporthe oryzae by Suppressing TOR Inhibition of cAMP/PKA Signaling. PLoS Pathog. 2015, 11, e1004851. [Google Scholar] [CrossRef]
- Qian, B.; Liu, X.; Jia, J.; Cai, Y.; Chen, C.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoPpe1 partners with MoSap1 to mediate TOR and cell wall integrity signalling in growth and pathogenicity of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2018, 20, 3964–3979. [Google Scholar] [CrossRef]
- Qian, B.; Liu, X.; Ye, Z.; Zhou, Q.; Liu, P.; Yin, Z.; Wang, W.; Zheng, X.; Zhang, H.; Zhang, Z. Phosphatase-associated protein MoTip41 interacts with the phosphatase MoPpe1 to mediate crosstalk between TOR and cell wall integrity signalling during infection by the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2021, 23, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Meng, S.; Qiu, J.; Wang, C.; Shu, Y.; Luo, C.; Kou, Y. MoWhi2 regulates appressorium formation and pathogenicity via the MoTor signalling pathway in Magnaporthe oryzae. Mol. Plant Pathol. 2021, 22, 969–983. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Elowsky, C.; Li, G.; Wilson, R.A. TOR-autophagy branch signaling via Imp1 dictates plant-microbe biotrophic interface longevity. PLoS Genet. 2018, 14, e1007814. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Franck, W.L.; Han, S.-O.; Shows, A.; Gokce, E.; Muddiman, D.C.; Dean, R.A. Polyubiquitin Is Required for Growth, Development and Pathogenicity in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2012, 7, e42868. [Google Scholar] [CrossRef]
- Shi, H.-B.; Chen, G.-Q.; Chen, Y.-P.; Dong, B.; Lu, J.-P.; Liu, X.-H.; Lin, F.-C. MoRad6-mediated ubiquitination pathways are essential for development and pathogenicity in Magnaporthe oryzae. Environ. Microbiol. 2016, 18, 4170–4187. [Google Scholar] [CrossRef] [PubMed]
- Prakash, C.; Manjrekar, J.; Chattoo, B.B. Skp1, a component of E3 ubiquitin ligase, is necessary for growth, sporulation, development and pathogenicity in rice blast fungus (Magnaporthe oryzae). Mol. Plant Pathol. 2016, 17, 903–919. [Google Scholar] [CrossRef]
- Guo, M.; Gao, F.; Zhu, X.; Nie, X.; Pan, Y.; Gao, Z. MoGrr1, a novel F-box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2015, 99, 8075–8088. [Google Scholar] [CrossRef]
- Lim, Y.-J.; Lee, Y.-H. F-box only and CUE proteins are crucial ubiquitination-associated components for conidiation and pathogenicity in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 2020, 144, 103473. [Google Scholar] [CrossRef]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2004, 1695, 189–207. [Google Scholar] [CrossRef]
- Que, Y.; Xu, Z.; Wang, C.; Lv, W.; Yue, X.; Xu, L.; Tang, S.; Dai, H.; Wang, Z. The putative deubiquitinating enzyme MoUbp4 is required for infection-related morphogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Curr. Genet. 2020, 66, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, D.; Matar, K.A.O.; Zheng, T.; Zhao, Q.; Xie, Y.; Gao, X.; Li, M.; Wang, B.; Lu, G.-D. The deubiquitinating enzyme MoUbp8 is required for infection-related development, pathogenicity, and carbon catabolite repression in Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2020, 104, 5081–5094. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Z.; Xing, J.; Yang, J.; Wang, Z.; Zhang, H.; Chen, D.; Peng, Y.; Chen, X. Global analysis of sumoylation function reveals novel insights into development and appressorium-mediated infection of the rice blast fungus. New Phytol. 2018, 219, 1031–1047. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Choi, J.; Lim, S.-E.; Lee, G.-W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.-S.; Jeon, J.; et al. Global Expression Profiling of Transcription Factor Genes Provides New Insights into Pathogenicity and Stress Responses in the Rice Blast Fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Choi, J.; Park, S.-Y.; Jeon, J.; Lee, Y.-H. Two conidiation-related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genet. Biol. 2013, 61, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-P.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic Analysis of Zn2Cys6 Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus. PLoS Pathog. 2014, 10, e1004432. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-Y.; Yu, Q.; Dong, B.; Zhang, Y.; Liu, X.-H.; Lin, F.-C.; Lian, S.g. MoLEU1, MoLEU2, and MoLEU4 regulated by MoLEU3 are involved in leucine biosynthesis, fungal development, and pathogenicity in Magnaporthe oryzae. Environ. Microbiol. Rep. 2019, 11, 784–796. [Google Scholar] [CrossRef]
- Odenbach, D.; Breth, B.; Thines, E.; Weber, R.W.S.; Anke, H.; Foster, A.J. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 2007, 64, 293–307. [Google Scholar] [CrossRef]
- Choi, J.; Kim, Y.; Kim, S.; Park, J.; Lee, Y.-H. MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 2009, 46, 243–254. [Google Scholar] [CrossRef]
- Hong, Y.; Cai, R.; Guo, J.; Zhong, Z.; Bao, J.; Wang, Z.; Chen, X.; Zhou, J.; Lu, G.-D. Carbon catabolite repressor MoCreA is required for the asexual development and pathogenicity of the rice blast fungus. Fungal Genet. Biol. 2020, 146, 103496. [Google Scholar] [CrossRef]
- Cao, H.; Huang, P.; Zhang, L.; Shi, Y.; Sun, D.; Yan, Y.; Liu, X.; Dong, B.; Chen, G.; Snyder, J.H.; et al. Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 2016, 211, 1035–1051. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Guo, W.; Chen, Y.; Dong, S.; Zhang, X.; Zhang, H.; Song, W.; Wang, W.; Wang, Q.; Lv, R.; et al. The Basic Leucine Zipper Transcription Factor Moatf1 Mediates Oxidative Stress Responses and Is Necessary for Full Virulence of the Rice Blast Fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2010, 23, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Park, S.-Y.; Lee, Y.-H. Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 1425–1443. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ru, Y.; Hong, L.; Zhu, Q.; Zuo, R.; Guo, X.; Wang, J.; Zhang, H.; Zheng, X.; Wang, P.; et al. System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 1377–1396. [Google Scholar] [CrossRef]
- Chen, Y.; Le, X.; Sun, Y.; Li, M.; Zhang, H.; Tan, X.; Zhang, D.; Liu, Y.; Zhang, Z. MoYcp4 is required for growth, conidiogenesis and pathogenicity in Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 1001–1011. [Google Scholar] [CrossRef]
- Cheung, N.; Tian, L.; Liu, X.; Li, X. The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis. Pathogens 2020, 9, 923. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.-Y.; Kim, K.S.; Rho, H.-S.; Chi, M.-H.; Choi, J.; Park, J.; Kong, S.; Park, J.; Goh, J.; et al. Homeobox Transcription Factors Are Required for Conidiation and Appressorium Development in the Rice Blast Fungus Magnaporthe oryzae. PLoS Genet. 2009, 5, e1000757. [Google Scholar] [CrossRef]
- Park, J.; Kong, S.; Kim, S.; Kang, S.; Lee, Y.-H. Roles of Forkhead-box Transcription Factors in Controlling Development, Pathogenicity, and Stress Response in Magnaporthe oryzae. Plant Pathol. J. 2014, 30, 136–150. [Google Scholar] [CrossRef]
- Cao, H.; Huang, P.; Yan, Y.; Shi, Y.; Dong, B.; Liu, X.; Ye, L.; Lin, F.; Lu, J. The basic helix-loop-helix transcription factor Crf1 is required for development and pathogenicity of the rice blast fungus by regulating carbohydrate and lipid metabolism. Environ. Microbiol. 2018, 20, 3427–3441. [Google Scholar] [CrossRef] [PubMed]
- Franck, W.L.; Gokce, E.; Randall, S.M.; Oh, Y.; Eyre, A.; Muddiman, D.C.; Dean, R.A. Phosphoproteome Analysis Links Protein Phosphorylation to Cellular Remodeling and Metabolic Adaptation during Magnaporthe oryzae Appressorium Development. J. Proteome Res. 2015, 14, 2408–2424. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; He, D.; Yin, C.; Yang, J.; Sun, J.; Wang, D.; Xue, M.; Li, Z.; Peng, Z.; Chen, D.; et al. PacC-dependent adaptation and modulation of host cellular pH controls hemibiotrophic invasive growth and disease development by the rice blast fungus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Landraud, P.; Chuzeville, S.; Billon-Grande, G.; Poussereau, N.; Bruel, C. Adaptation to pH and Role of PacC in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2013, 8, e69236. [Google Scholar] [CrossRef] [PubMed]
- Breth, B.; Odenbach, D.; Yemelin, A.; Schlinck, N.; Schröder, M.; Bode, M.; Antelo, L.; Andresen, K.; Thines, E.; Foster, A.J. The role of the Tra1p transcription factor of Magnaporthe oryzae in spore adhesion and pathogenic development. Fungal Genet. Biol. 2013, 57, 11–22. [Google Scholar] [CrossRef]
- Hunter, T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Chen, Y.; Ye, W.; Lin, Q.; Lu, G.; Ebbole, D.J.; Olsson, S.; Wang, Z. Magnaporthe oryzae CK2 Accumulates in Nuclei, Nucleoli, at Septal Pores and Forms a Large Ring Structure in Appressoria, and Is Involved in Rice Blast Pathogenesis. Front. Cell. Infect. Microbiol. 2019, 9, 113. [Google Scholar] [CrossRef]
- Wang, J.; Du, Y.; Zhang, H.; Zhou, C.; Qi, Z.; Zheng, X.; Wang, P.; Zhang, Z. The actin-regulating kinase homologue MoArk1 plays a pleiotropic function in Magnaporthe oryzae. Mol. Plant Pathol. 2013, 14, 470–482. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Yu, R.; Li, X.; Liu, M.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. Magnaporthe oryzae Abp1, a MoArk1 Kinase-Interacting Actin Binding Protein, Links Actin Cytoskeleton Regulation to Growth, Endocytosis, and Pathogenesis. Mol. Plant-Microbe Interact. 2019, 32, 437–451. [Google Scholar] [CrossRef]
- Han, J.-H.; Lee, H.-M.; Shin, J.-H.; Lee, Y.-H.; Su Kim, K. Role of the MoYAK 1 protein kinase gene in Magnaporthe oryzae development and pathogenicity. Environ. Microbiol. 2015, 17, 4672–4689. [Google Scholar]
- Yue, X.; Que, Y.; Deng, S.; Xu, L.; Oses-Ruiz, M.; Talbot, N.J.; Peng, Y.; Wang, Z. The cyclin dependent kinase subunit Cks1 is required for infection-associated development of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 3959–3981. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, X.; Li, X.; Liu, M.; Liu, X.; Wang, X.; Zhang, H.; Zheng, X.; Zhang, Z. The Atypical Guanylate Kinase MoGuk2 Plays Important Roles in Asexual/Sexual Development, Conidial Septation, and Pathogenicity in the Rice Blast Fungus. Front. Microbiol. 2017, 8, 2467. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Park, J.-H.; Ahn, J.-H.; Lee, Y.-H. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2008, 45, 1172–1181. [Google Scholar] [CrossRef]
- Zeng, X.-Q.; Chen, G.-Q.; Liu, X.; Dong, B.; Shi, H.-B.; Lu, J.-P.; Lin, F. Crosstalk between SNF1 Pathway and the Peroxisome-Mediated Lipid Metabolism in Magnaporthe oryzae. PLoS ONE 2014, 9, e103124. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Foster, A.J.; Yemelin, A.; Thines, E. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. MicrobiologyOpen 2014, 3, 668–687. [Google Scholar] [CrossRef]
- Islas-Flores, I.; Sanchez-Rodriguez, Y.; Brito-Argaez, L.; Peraza-Echeverria, L.; Rodriguez-Garcia, C.; Couoh-Uicab, Y.; James, A.; Tzec-Simá, M.; Canto-Canche, B.; Peraza-Echeverría, S. The Amazing Role of the Group III of Histidine Kinases in Plant Pathogenic Fungi, an Insight to Fungicide Resistance. Asian J. Biochem. 2011, 6, 1–14. [Google Scholar] [CrossRef][Green Version]
- Ryder, L.S.; Dagdas, Y.F.; Kershaw, M.J.; Venkataraman, C.; Madzvamuse, A.; Yan, X.; Cruz-Mireles, N.; Soanes, D.M.; Oses-Ruiz, M.; Styles, V.; et al. A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 2019, 574, 423–427. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.; Zhang, X.; Song, W.; Zhao, Q.; Dong, Y.; Guo, M.; Zheng, X.; Zhang, Z. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr. Genet. 2010, 56, 517–528. [Google Scholar] [CrossRef]
- Shin, J.-H.; Gumilang, A.; Kim, M.-J.; Han, J.-H.; Kim, K.S. A PAS-Containing Histidine Kinase is Required for Conidiation, Appressorium Formation, and Disease Development in the Rice Blast Fungus, Magnaporthe oryzae. Mycobiology 2019, 47, 473–482. [Google Scholar] [CrossRef]
- Du, Y.; Shi, Y.; Yang, J.; Chen, X.-L.; Xue, M.; Zhou, W.; Peng, Y.-L. A serine/threonine-protein phosphatase PP2A catalytic subunit is essential for asexual development and plant infection in Magnaporthe oryzae. Curr. Genet. 2013, 59, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cao, S.; Zhang, C.; Zhang, Y.; Zhang, Q.; Xu, J.; Wang, C. MoCDC14 is important for septation during conidiation and appressorium formation in Magnaporthe oryzae. Mol. Plant Pathol. 2018, 19, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Abubakar, Y.S.; Yang, C.; Wang, X.; Miao, P.; Lin, M.; Wen, Y.; Wu, Q.; Zhong, H.; Fan, Y.; et al. Trehalose Phosphate Synthase Complex-Mediated Regulation of Trehalose 6-Phosphate Homeostasis Is Critical for Development and Pathogenesis in Magnaporthe oryzae. Msystems 2021, 6, e00462-21. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; Jenkinson, J.M.; Gibson, R.P.; Littlechild, J.A.; Wang, Z.-Y.; Talbot, N.J. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 2007, 26, 3673–3685. [Google Scholar] [CrossRef]
- Sadat, A.; Jeon, J.; Mir, A.A.; Choi, J.; Choi, J.; Lee, Y.-H. Regulation of Cellular Diacylglycerol through Lipid Phosphate Phosphatases Is Required for Pathogenesis of the Rice Blast Fungus, Magnaporthe oryzae. PLoS ONE 2014, 9, e100726. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, P.; Sun, Q.; Li, R.; Qin, Z.; Sha, G.; Zhou, Y.; Bi, R.; Zhang, H.; Zheng, L.; et al. The MoPah1 phosphatidate phosphatase is involved in lipid metabolism, development, and pathogenesis in Magnaporthe oryzae. Mol. Plant Pathol. 2022, 23, 720–732. [Google Scholar] [CrossRef]
- Ramos-Pamplona, M.; Naqvi, N.I. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 2006, 61, 61–75. [Google Scholar] [CrossRef]
- Goh, J.; Jeon, J.; Kim, K.S.; Park, J.; Park, S.-Y.; Lee, Y.-H. The PEX7-Mediated Peroxisomal Import System Is Required for Fungal Development and Pathogenicity in Magnaporthe oryzae. PLoS ONE 2011, 6, e28220. [Google Scholar] [CrossRef]
- Wang, J.; Li, L.; Zhang, Z.; Qiu, H.; Li, D.; Fang, Y.; Jiang, H.; Chai, R.Y.; Mao, X.; Wang, Y.; et al. One of Three Pex11 Family Members Is Required for Peroxisomal Proliferation and Full Virulence of the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2015, 10, e0134249. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Li, L.; Chai, R.-Y.; Qiu, H.-P.; Zhang, Z.; Wang, Y.-L.; Liu, X.-H.; Lin, F.-C.; Sun, G.-C. Pex13 and Pex14, the key components of the peroxisomal docking complex, are required for peroxisome formation, host infection and pathogenicity-related morphogenesis in Magnaporthe oryzae. Virulence 2019, 10, 292–314. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Chen, H.; Chai, R.; Zhang, Z.; Mao, X.; Qiu, H.; Jiang, H.; Wang, Y.; Sun, G. Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 1238–1252. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Zhang, Z.; Wang, Y.; Liu, M.; Jiang, H.; Chai, R.; Mao, X.; Qiu, H.; Liu, F.; et al. MoPex19, which Is Essential for Maintenance of Peroxisomal Structure and Woronin Bodies, Is Required for Metabolism and Development in the Rice Blast Fungus. PLoS ONE 2014, 9, e85252. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z.; Wang, Y.; Li, L.; Chai, R.; Mao, X.; Jiang, H.; Qiu, H.; Du, X.; Lin, F.; et al. PTS1 Peroxisomal Import Pathway Plays Shared and Distinct Roles to PTS2 Pathway in Development and Pathogenicity of Magnaporthe oryzae. PLoS ONE 2013, 8, e55554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Soanes, D.M.; Kershaw, M.J.; Talbot, N.J. Functional Analysis of Lipid Metabolism in Magnaporthe grisea Reveals a Requirement for Peroxisomal Fatty Acid β-Oxidation During Appressorium-Mediated Plant Infection. Mol. Plant-Microbe Interact. 2007, 20, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-L.; Shen, M.; Yang, J.; Xing, Y.; Chen, D.; Li, Z.; Zhao, W.; Zhang, Y. Peroxisomal fission is induced during appressorium formation and is required for full virulence of the rice blast fungus. Mol. Plant Pathol. 2017, 18, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Rucktäschel, R.; Girzalsky, W.; Erdmann, R. Protein import machineries of peroxisomes. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Bhambra, G.K.; Wang, Z.-Y.; Soanes, D.M.; Wakley, G.E.; Talbot, N.J. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 2006, 61, 46–60. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.-N.; Li, X.; Gu, W.; Moeketsi, E.K.; Zhou, R.; Zheng, X.; Zhang, Z.; Zhang, H. The Peroxisomal-CoA Synthetase MoPcs60 Is Important for Fatty Acid Metabolism and Infectious Growth of the Rice Blast Fungus. Front. Plant Sci. 2022, 12, 3080. [Google Scholar] [CrossRef]
- Bhadauria, V.; Banniza, S.; Vandenberg, A.; Selvaraj, G.; Wei, Y. Peroxisomal Alanine: Glyoxylate Aminotransferase AGT1 Is Indispensable for Appressorium Function of the Rice Blast Pathogen, Magnaporthe oryzae. PLoS ONE 2012, 7, e36266. [Google Scholar] [CrossRef]
- Kunau, W.-H.; Dommes, V.; Schulz, H. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: A century of continued progress. Prog. Lipid Res. 1995, 34, 267–342. [Google Scholar] [CrossRef]
- Aliyu, S.R.; Lin, L.; Chen, X.; Abdul, W.; Lin, Y.; Otieno, F.J.; Shabbir, A.; Batool, W.; Zhang, Y.; Tang, W.; et al. Disruption of putative short-chain acyl-CoA dehydrogenases compromised free radical scavenging, conidiogenesis, and pathogenesis of Magnaporthe oryzae. Fungal Genet. Biol. 2019, 127, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Watmough, N.J.; Frerman, F.E. The electron transfer flavoprotein: Ubiquinone oxidoreductases. Biochim. Biophys. Acta (BBA)—Bioenerg. 2010, 1797, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Hu, J.; Meng, X.; Zhang, Q.; Zhu, K.; Chen, X.; Chen, X.; Li, G.; Wang, Z.; et al. Functional characterization of electron-transferring flavoprotein and its dehydrogenase required for fungal development and plant infection by the rice blast fungus. Sci. Rep. 2016, 6, 24911. [Google Scholar] [CrossRef] [PubMed]
- Patkar, R.N.; Ramos-Pamplona, M.; Gupta, A.P.; Fan, Y.; Naqvi, N.I. Mitochondrial β-oxidation regulates organellar integrity and is necessary for conidial germination and invasive growth in Magnaporthe oryzae. Mol. Microbiol. 2012, 86, 1345–1363. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, L.; Zhang, T.; Zhou, R.; Ren, Y.; Li, X.; Shu, H.; Ye, W.; Zheng, X.; Zhang, Z.; et al. Transcription factor MoMsn2 targets the putative 3-methylglutaconyl-CoA hydratase-encoding gene MoAUH1 to govern infectious growth via mitochondrial fusion/fission balance in Magnaporthe oryzae. Environ. Microbiol. 2021, 23, 774–790. [Google Scholar] [CrossRef]
- Khan, I.A.; Ning, G.; Liu, X.; Feng, X.; Lin, F.; Lu, J. Mitochondrial fission protein MoFis1 mediates conidiation and is required for full virulence of the rice blast fungus Magnaporthe oryzae. Microbiol. Res. 2015, 178, 51–58. [Google Scholar] [CrossRef]
- Zhong, K.; Li, X.; Le, X.; Kong, X.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoDnm1 Dynamin Mediating Peroxisomal and Mitochondrial Fission in Complex with MoFis1 and MoMdv1 Is Important for Development of Functional Appressorium in Magnaporthe oryzae. PLoS Pathog. 2016, 12, e1005823. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, X.; Zhu, M.; Chen, M.; Zhang, S.; He, F.; Chen, X.; Lv, J.; Pei, M.; Zhang, Y.; et al. MoIVD-Mediated Leucine Catabolism Is Required for Vegetative Growth, Conidiation and Full Virulence of the Rice Blast Fungus Magnaporthe oryzae. Front. Microbiol. 2019, 10, 444. [Google Scholar] [CrossRef]
- Zhong, Z.; Norvienyeku, J.; Yu, J.; Chen, M.; Cai, R.; Hong, Y.; Chen, L.; Zhang, D.; Wang, B.; Zhou, J.; et al. Two different subcellular-localized Acetoacetyl-CoA acetyltransferases differentiate diverse functions in Magnaporthe oryzae. Fungal Genet. Biol. 2015, 83, 58–67. [Google Scholar] [CrossRef]
- Howard, R.J.; Ferrari, M.A. Role of melanin in appressorium function. Exp. Mycol. 1989, 13, 403–418. [Google Scholar] [CrossRef]
- Chumley, F.G.; Valent, B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol. Plant-Microbe Interact. 1990, 3, 135–143. [Google Scholar] [CrossRef]
- Zhu, S.; Yan, Y.; Qu, Y.; Wang, J.; Feng, X.; Liu, X.; Lin, F.; Lu, J. Role refinement of melanin synthesis genes by gene knockout reveals their functional diversity in Pyricularia oryzae strains. Microbiol. Res. 2021, 242, 126620. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Ma, Y.; Chen, C.-Y.; Shen, L.; Sun, W.; Cui, G.; Naqvi, N.I.; Deng, Y.Z. Identification and Characterization of Auxin/IAA Biosynthesis Pathway in the Rice Blast Fungus Magnaporthe oryzae. J. Fungi 2022, 8, 208. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in Budding Yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef]
- Raman, V.; Simon, S.A.; Demirci, F.; Nakano, M.; Meyers, B.C.; Donofrio, N.M. Small RNA Functions Are Required for Growth and Development of Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2017, 30, 517–530. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Saletore, Y.; Meyer, K.; Korlach, J.; Vilfan, I.D.; Jaffrey, S.; Mason, C.E. The birth of the Epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 2012, 13, 1–12. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Wang, J.; Liu, X.; Lin, F.; Lu, J. N6-methyladenosine RNA methylation is involved in virulence of the rice blast fungus Pyricularia oryzae (syn. Magnaporthe oryzae). FEMS Microbiol. Lett. 2019, 366, fny286. [Google Scholar]
- Zhang, W.; Huang, J.; Cook, D.E. Histone modification dynamics at H3K27 are associated with altered transcription of in planta induced genes in Magnaporthe oryzae. PLoS Genet. 2021, 17, e1009376. [Google Scholar] [CrossRef]
- Pham, K.T.M.; Inoue, Y.; Van Vu, B.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.-I.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar] [CrossRef]
- Kwon, S.; Lee, J.; Jeon, J.; Kim, S.; Park, S.-Y.; Jeon, J.; Lee, Y.-H. Role of the Histone Acetyltransferase Rtt109 in Development and Pathogenicity of the Rice Blast Fungus. Mol. Plant-Microbe Interact. 2018, 31, 1200–1210. [Google Scholar] [CrossRef]
- Dubey, A.; Lee, J.; Kwon, S.; Lee, Y.; Jeon, J. A MYST family histone acetyltransferase, MoSAS3, is required for development and pathogenicity in the rice blast fungus. Mol. Plant Pathol. 2019, 20, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Cao, X.; Qu, Z.; Zhang, S.; Naqvi, N.I.; Deng, Y.Z. The Histone Deacetylases MoRpd3 and MoHst4 Regulate Growth, Conidiation, and Pathogenicity in the Rice Blast Fungus Magnaporthe oryzae. Msphere 2021, 6, e00118-21. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-L.; Liu, W.; Iliuk, A.; Ribot, C.; Vallet, J.; Tao, A.; Wang, Y.; Lebrun, M.-H.; Xu, J.-R. The Tig1 Histone Deacetylase Complex Regulates Infectious Growth in the Rice Blast Fungus Magnaporthe oryzae. Plant Cell 2010, 22, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Huh, A.; Dubey, A.; Kim, S.; Jeon, J.; Lee, Y.-H. MoJMJ1, Encoding a Histone Demethylase Containing JmjC Domain, Is Required for Pathogenic Development of the Rice Blast Fungus, Magnaporthe oryzae. Plant Pathol. J. 2017, 33, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.J.; Wang, Z.-Y.; Jones, M.A.; Smirnoff, N.; Talbot, N.J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Tan, L.; Nie, X.; Zhu, X.; Pan, Y.; Gao, Z. The Pmt2p-Mediated Protein O-Mannosylation Is Required for Morphogenesis, Adhesive Properties, Cell Wall Integrity and Full Virulence of Magnaporthe oryzae. Front. Microbiol. 2016, 7, 630. [Google Scholar] [CrossRef]
- Pan, Y.; Pan, R.; Tan, L.; Zhang, Z.; Guo, M. Pleiotropic roles of O-mannosyltransferase MoPmt4 in development and pathogenicity of Magnaporthe oryzae. Curr. Genet. 2019, 65, 223–239. [Google Scholar] [CrossRef]
- Choi, J.; Kim, K.S.; Rho, H.-S.; Lee, Y.-H. Differential roles of the phospholipase C genes in fungal development and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 2011, 48, 445–455. [Google Scholar] [CrossRef]
- Rho, H.-S.; Jeon, J.; Lee, Y.-H. Phospholipase C-mediated calcium signalling is required for fungal development and pathogenicity in Magnaporthe oryzae. Mol. Plant Pathol. 2009, 10, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-H.; Shin, J.-H.; Lee, Y.-H.; Kim, K.S. Distinct roles of the YPEL gene family in development and pathogenicity in the ascomycete fungus Magnaporthe oryzae. Sci. Rep. 2018, 8, 14461. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zhang, W.; Zhang, S.; Yang, G.; Tan, L.; Guo, M. Class I myosin mediated endocytosis and polarization growth is essential for pathogenicity of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2021, 105, 7395–7410. [Google Scholar] [CrossRef]
- Fu, T.; Park, G.-C.; Han, J.H.; Shin, J.-H.; Park, H.-H.; Kim, K.S. MoRBP9 Encoding a Ran-Binding Protein Microtubule-Organizing Center Is Required for Asexual Reproduction and Infection in the Rice Blast Pathogen Magnaporthe oryzae. Plant Pathol. J. 2019, 35, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.; Rawat, K.; Ashar, H.; Patkar, R.; Manjrekar, J. Dual role for fungal-specific outer kinetochore proteins during cell cycle and development in Magnaporthe oryzae. J. Cell Sci. 2019, 132, jcs224147. [Google Scholar] [CrossRef] [PubMed]
- Matar, K.A.O.; Chen, X.; Chen, D.; Anjago, W.M.; Norvienyeku, J.; Lin, Y.; Chen, M.; Wang, Z.; Ebbole, D.J.; Lu, G.-D. WD40-repeat protein MoCreC is essential for carbon repression and is involved in conidiation, growth and pathogenicity of Magnaporthe oryzae. Curr. Genet. 2017, 63, 685–696. [Google Scholar] [CrossRef]
- Li, Y.; Liang, S.; Yan, X.; Wang, H.; Li, D.; Soanes, D.M.; Talbot, N.J.; Wang, Z.; Wang, Z. Characterization of MoLDB1 required for vegetative growth, infection-related morphogenesis, and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2010, 23, 1260–1274. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, H.; Xie, X.; Ji, J.; Dong, Y.; Du, Y.; Tang, W.; Zheng, X.; Wang, P.; Zhang, Z. Comparative proteomic analyses reveal that the regulators of G-protein signaling proteins regulate amino acid metabolism of the rice blast fungus Magnaporthe oryzae. Proteomics 2014, 14, 2508–2522. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, H.; Hong, L.; Wang, J.; Zheng, X.; Zhang, Z. Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae. Mol. Plant Pathol. 2013, 14, 870–884. [Google Scholar] [CrossRef]
- Yang, C.D.; Dang, X.; Zheng, H.W.; Chen, X.F.; Lin, X.L.; Zhang, D.M.; Abubakar, Y.S.; Chen, X.; Lu, G.; Wang, Z.; et al. Two Rab5 Homologs Are Essential for the Development and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 620. [Google Scholar] [CrossRef]
- Zhou, T.; Qin, L.; Zhu, X.; Shen, W.; Zou, J.; Wang, Z.; Wei, Y. The D-lactate dehydrogenase MoDLD1 is essential for growth and infection-related development in Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 3938–3958. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, X.; Sun, J.; Kang, Z.; Ding, S.; Xu, J.-R.; Peng, Y.-L. A Novel Protein Com1 Is Required for Normal Conidium Morphology and Full Virulence in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2010, 23, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Wilson, R.A. Characterizing Roles for the Glutathione Reductase, Thioredoxin Reductase and Thioredoxin Peroxidase-Encoding Genes of Magnaporthe oryzae during Rice Blast Disease. PLoS ONE 2014, 9, e87300. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Guo, Z.; Xi, Y.; Yuan, M.; Lin, Y.; Wu, C.; Abubakar, Y.S.; Dou, X.; Li, G.; Wang, Z.; et al. Sorting nexin (MoVps17) is required for fungal development and plant infection by regulating endosome dynamics in the rice blast fungus. Environ. Microbiol. 2017, 19, 4301–4317. [Google Scholar] [CrossRef]
- Norvienyeku, J.; Zhong, Z.; Lin, L.; Dang, X.; Chen, M.; Lin, X.; Zhang, H.; Anjago, W.M.; Lin, L.; Abdul, W.; et al. Methylmalonate-semialdehyde dehydrogenase mediated metabolite homeostasis essentially regulate conidiation, polarized germination and pathogenesis in Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 4256–4277. [Google Scholar] [CrossRef]
- Yan, X.; Que, Y.; Wang, H.; Wang, C.; Li, Y.; Yue, X.; Ma, Z.; Talbot, N.J.; Wang, Z. The MET13 Methylenetetrahydrofolate Reductase Gene Is Essential for Infection-Related Morphogenesis in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2013, 8, e76914. [Google Scholar] [CrossRef]
- Aron, O.; Wang, M.; Mabeche, A.W.; Wajjiha, B.; Li, M.; Yang, S.; You, H.; Cai, Y.; Zhang, T.; Li, Y.; et al. MoCpa1-mediated arginine biosynthesis is crucial for fungal growth, conidiation, and plant infection of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2021, 105, 5915–5929. [Google Scholar] [CrossRef]
- Kong, L.-A.; Yang, J.; Li, G.-T.; Qi, L.-L.; Zhang, Y.-J.; Wang, C.-F.; Zhao, W.-S.; Xu, J.-R.; Peng, Y.-L. Different Chitin Synthase Genes Are Required for Various Developmental and Plant Infection Processes in the Rice Blast Fungus Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002526. [Google Scholar] [CrossRef]
- Heupel, S.; Roser, B.; Kuhn, H.; Lebrun, M.-H.; Villalba, F.; Requena, N. Erl1, a novel era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont Glomus intraradices. Mol. Plant-Microbe Interact. 2010, 23, 67–81. [Google Scholar] [CrossRef]
- Chen, Y.; Zuo, R.; Zhu, Q.; Sun, Y.; Li, M.; Dong, Y.; Ru, Y.; Zhang, H.; Zheng, X.; Zhang, Z. MoLys2 is necessary for growth, conidiogenesis, lysine biosynthesis, and pathogenicity in Magnaporthe oryzae. Fungal Genet. Biol. 2014, 67, 51–57. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Zhu, S.; Wang, J.; Liu, X.; Lin, F.; Lu, J. The Methylcitrate Cycle is Required for Development and Virulence in the Rice Blast Fungus Pyricularia oryzae. Mol. Plant-Microbe Interact. 2019, 32, 1148–1161. [Google Scholar] [CrossRef]
- Wu, M.-H.; Huang, L.-Y.; Sun, L.-X.; Qian, H.; Wei, Y.-Y.; Liang, S.; Zhu, X.-M.; Li, L.; Lu, J.-P.; Lin, F.-C.; et al. A putative D-arabinono-1, 4-lactone oxidase, MoAlo1, is required for fungal growth, conidiogenesis, and pathogenicity in Magnaporthe oryzae. J. Fungi 2022, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Cao, H.; Huang, P.; Wang, J.; Liu, X.; Lu, J.; Lin, F.-C. A kelch domain cell end protein, PoTea1, mediates cell polarization during appressorium morphogenesis in Pyricularia oryzae. Microbiol. Res. 2022, 259, 126999. [Google Scholar] [CrossRef]
- Liu, T.-B.; Chen, G.-Q.; Min, H.; Lin, F.-C. MoFLP1, encoding a novel fungal fasciclin-like protein, is involved in conidiation and pathogenicity in Magnaporthe oryzae. J. Zhejiang Univ. B 2009, 10, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Z.; Qu, Z.; Naqvi, N.I. Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development. PLoS Pathog. 2015, 11, e1004972. [Google Scholar] [CrossRef]
- Jeon, J.; Rho, H.; Kim, S.; Kim, K.S.; Lee, Y.-H. Role of MoAND1-mediated nuclear positioning in morphogenesis and pathogenicity in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 2014, 69, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Tan, L.; Nie, X.; Zhang, Z. A class-II myosin is required for growth, conidiation, cell wall integrity and pathogenicity of Magnaporthe oryzae. Virulence 2017, 8, 1335–1354. [Google Scholar] [CrossRef]
- Chen, G.; Liu, X.; Zhang, L.; Cao, H.; Lu, J.; Lin, F. Involvement of MoVMA11, a Putative Vacuolar ATPase c’ Subunit, in Vacuolar Acidification and Infection-Related Morphogenesis of Magnaporthe oryzae. PLoS ONE 2013, 8, e67804. [Google Scholar] [CrossRef]
- Liu, X.H.; Liang, S.; Wei, Y.Y.; Zhu, X.M.; Li, L.; Liu, P.P.; Zheng, Q.X.; Zhou, H.N.; Zhang, Y.; Mao, L.J.; et al. Metabolomics Analysis Identifies Sphingolipids as Key Signaling Moieties in Appressorium Morphogenesis and Function in Magnaporthe oryzae. mBio 2019, 10, e01467-19. [Google Scholar] [CrossRef]
- Fernandez, J.; Wright, J.D.; Hartline, D.; Quispe, C.F.; Madayiputhiya, N.; Wilson, R.A. Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection. PLoS Genet. 2012, 8, e1002673. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Yan, Y.; Cao, H.; Liu, X.; Lin, F.; Lu, J. Glycerol-3-Phosphate Shuttle Is Involved in Development and Virulence in the Rice Blast Fungus Pyricularia oryzae. Front. Plant Sci. 2018, 9, 687. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Xu, X.; Chen, G.; Zhang, D.; Tang, M.; Xu, F.; Liu, X.; Wang, H.; Zhou, B. Autophagy-associated alpha-arrestin signaling is required for conidiogenous cell development in Magnaporthe oryzae. Sci. Rep. 2016, 6, 30963. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; Yang, C.; Huang, J.; Chen, X.; Zhou, J.; Li, G.; Norvienyeku, J.; Wang, Z. A HOPS Protein, MoVps41, Is Crucially Important for Vacuolar Morphogenesis, Vegetative Growth, Reproduction and Virulence in Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 1091. [Google Scholar] [CrossRef] [PubMed]
- Donofrio, N.; Oh, Y.; Lundy, R.; Pan, H.; Brown, D.; Jeong, J.; Coughlan, S.; Mitchell, T.; Dean, R. Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 2006, 43, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Cao, J.; Du, A.; An, Q.; Chen, X.; Yuan, S.; Batool, W.; Shabbir, A.; Zhang, D.; Wang, Z.; et al. eIF3k Domain-Containing Protein Regulates Conidiogenesis, Appressorium Turgor, Virulence, Stress Tolerance, and Physiological and Pathogenic Development of Magnaporthe oryzae Oryzae. Front. Plant Sci. 2021, 12, 748120. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Cao, H.; Shi, Y.; Huang, P.; Dong, B.; Liu, X.; Lin, F.; Lu, J. The regulatory factor X protein MoRfx1 is required for development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 1075–1088. [Google Scholar] [CrossRef]
- Song, W.; Dou, X.; Qi, Z.; Wang, Q.; Zhang, X.; Zhang, H.; Guo, M.; Dong, S.; Zhang, Z.; Wang, P.; et al. R-SNARE Homolog MoSec22 Is Required for Conidiogenesis, Cell Wall Integrity, and Pathogenesis of Magnaporthe oryzae. PLoS ONE 2010, 5, e13193. [Google Scholar] [CrossRef]
- Sarkar, A.; Roy-Barman, S. Spray-Induced Silencing of Pathogenicity Gene MoDES1 via Exogenous Double-Stranded RNA Can Confer Partial Resistance Against Fungal Blast in Rice. Front. Plant Sci. 2021, 12, 733129. [Google Scholar] [CrossRef]
- Li, Y.; Yue, X.; Que, Y.; Yan, X.; Ma, Z.; Talbot, N.J.; Wang, Z. Characterisation of Four LIM Protein-Encoding Genes Involved in Infection-Related Development and Pathogenicity by the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2014, 9, e88246. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Li, C.; Zeng, Y.; Tan, X.; Zhang, D.; Liu, Y. MoPer1 is required for growth, conidiogenesis, and pathogenicity in Magnaporthe oryzae. Rice 2018, 11, 64. [Google Scholar] [CrossRef]
- Fernandez, J.; Lopez, V.; Kinch, L.; Pfeifer, M.A.; Gray, H.; Garcia, N.; Grishin, N.V.; Khang, C.-H.; Orth, K. Role of Two Metacaspases in Development and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. mBio 2021, 12, e03471-20. [Google Scholar] [CrossRef]
- Yan, X.; Ma, W.-B.; Li, Y.; Wang, H.; Que, Y.-W.; Ma, Z.H.; Talbot, N.J.; Wang, Z.-Y. A sterol 14α-demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2011, 48, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Fukada, F.; Kodama, S.; Nishiuchi, T.; Kajikawa, N.; Kubo, Y. Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 2019, 222, 1909–1923. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Wang, Q.; Qi, Z.; Song, W.; Wang, W.; Guo, M.; Zhang, H.; Zhang, Z.; Wang, P.; Zheng, X. MoVam7, a Conserved SNARE Involved in Vacuole Assembly, Is Required for Growth, Endocytosis, ROS Accumulation, and Pathogenesis of Magnaporthe oryzae. PLoS ONE 2011, 6, e16439. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-M.; Liu, X.; Shi, H.-B.; Lu, J.-P.; Yang, J.; Lin, F. MoMon1 is required for vacuolar assembly, conidiogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Res. Microbiol. 2013, 164, 300–309. [Google Scholar] [CrossRef]
- Abdul, W.; Aliyu, S.R.; Lin, L.; Sekete, M.; Chen, X.; Otieno, F.J.; Yang, T.; Lin, Y.; Norvienyeku, J.; Wang, Z. Family-Four Aldehyde Dehydrogenases Play an Indispensable Role in the Pathogenesis of Magnaporthe oryzae. Front. Plant Sci. 2018, 9, 980. [Google Scholar] [CrossRef]
- Kou, Y.; Tan, Y.H.; Ramanujam, R.; Naqvi, N.I. Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 2017, 214, 330–342. [Google Scholar] [CrossRef]
- DeZwaan, T.M.; Carroll, A.M.; Valent, B.; Sweigard, J.A. Magnaporthe grisea Pth11p Is a Novel Plasma Membrane Protein That Mediates Appressorium Differentiation in Response to Inductive Substrate Cues. Plant Cell 1999, 11, 2013–2030. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Chai, R.; Qiu, H.; Jiang, H.; Mao, X.; Wang, Y.; Liu, F.; Sun, G. An S-(Hydroxymethyl)Glutathione Dehydrogenase Is Involved in Conidiation and Full Virulence in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2015, 10, e0120627. [Google Scholar] [CrossRef]
- Li, X.; Gao, C.; Li, L.; Liu, M.; Yin, Z.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2017, 13, e1006449. [Google Scholar] [CrossRef]
- Patkar, R.N.; Suresh, A.; Naqvi, N.I. MoTea4-Mediated Polarized Growth Is Essential for Proper Asexual Development and Pathogenesis in Magnaporthe oryzae. Eukaryot. Cell 2010, 9, 1029–1038. [Google Scholar] [CrossRef]
- Zhu, X.; Li, L.; Wang, J.; Zhao, L.; Shi, H.; Bao, J.; Su, Z.; Liu, X.; Lin, F. Vacuolar Protein-Sorting Receptor MoVps13 Regulates Conidiation and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae. J. Fungi 2021, 7, 1084. [Google Scholar] [CrossRef]
- Chen, Y.; Zhai, S.; Zhang, H.; Zuo, R.; Wang, J.; Guo, M.; Zheng, X.; Wang, P.; Zhang, Z. Shared and distinct functions of two Gti1/P ac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2014, 16, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Park, S.-Y.; Kim, S.G.; Yoo, J.S.; Park, S.; Gupta, R.; Kang, K.Y.; Kim, S.T. Secreted Alpha-N-Arabinofuranosidase B Protein Is Required for the Full Virulence of Magnaporthe oryzae and Triggers Host Defences. PLoS ONE 2016, 11, e0165149. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhao, Q.; Liu, X.; Zhang, X.; Qi, Z.; Zhang, H.; Zheng, X.; Zhang, Z. MoMyb1 is required for asexual development and tissue-specific infection in the rice blast fungus Magnaporthe oryzae. BMC Microbiol. 2015, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Völz, R.; Song, H.; Harris, W.; Lee, Y.-H. Characterization of the MYB Genes Reveals Insights Into Their Evolutionary Conservation, Structural Diversity, and Functional Roles in Magnaporthe oryzae. Front. Microbiol. 2021, 12, 721530. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Liu, M.; Dong, Y.; Yang, J.; Zhang, H.; Zheng, X.; Zhang, Z. Orotate phosphoribosyl transferase MoPyr5 is involved in uridine 5′-phosphate synthesis and pathogenesis of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2016, 100, 3655–3666. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, V.C.; Chandarana, P.M.; Chattoo, B.B.; Patkar, R.N.; Manjrekar, J. Fungal Histidine Phosphotransferase Plays a Crucial Role in Photomorphogenesis and Pathogenesis in Magnaporthe oryzae. Front. Chem. 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Hong, L.; Tang, W.; Li, L.; Wang, X.; Ma, H.; Wang, Z.; Zhang, H.; Zheng, X.; Zhang, Z. Threonine deaminase MoIlv1 is important for conidiogenesis and pathogenesis in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2014, 73, 53–60. [Google Scholar] [CrossRef]
- Dang, Y.; Wei, Y.; Wang, Y.; Liu, S.; Julia, C.; Zhang, S. Cleavage of PrePL by Lon promotes growth and pathogenesis in Magnaporthe oryzae. Environ. Microbiol. 2021, 23, 4881–4895. [Google Scholar] [CrossRef]
- Batool, W.; Shabbir, A.; Lin, L.; Chen, X.; An, Q.; He, X.; Pan, S.; Chen, S.; Chen, Q.; Wang, Z.; et al. Translation Initiation Factor eIF4E Positively Modulates Conidiogenesis, Appressorium Formation, Host Invasion and Stress Homeostasis in the Filamentous Fungi Magnaporthe oryzae. Front. Plant Sci. 2021, 12, 646343. [Google Scholar] [CrossRef]
- Fan, G.; Zhang, K.; Huang, H.; Zhang, H.; Zhao, A.; Chen, L.; Chen, R.; Li, G.; Wang, Z.; Lu, G.-D. Multiprotein-bridging factor 1 regulates vegetative growth, osmotic stress, and virulence in Magnaporthe oryzae. Curr. Genet. 2017, 63, 293–309. [Google Scholar] [CrossRef]
- Shi, H.-B.; Chen, N.; Zhu, X.-M.; Liang, S.; Li, L.; Wang, J.-Y.; Lu, J.-P.; Lin, F.-C.; Liu, X.-H. F-box proteins MoFwd1, MoCdc4 and MoFbx15 regulate development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2019, 21, 3027–3045. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Q.; Liu, L.; Li, X.; Lin, A.; Li, C. MoMCP1, a Cytochrome P450 Gene, Is Required for Alleviating Manganese Toxin Revealed by Transcriptomics Analysis in Magnaporthe oryzae. Int. J. Mol. Sci. 2019, 20, 1590. [Google Scholar] [CrossRef]
- Li, M.; Liu, X.; Liu, Z.; Sun, Y.; Liu, M.; Wang, X.; Zhang, H.; Zheng, X.; Zhang, Z. Correction: Glycoside Hydrolase MoGls2 Controls Asexual/Sexual Development, Cell Wall Integrity and Infectious Growth in the Rice Blast Fungus. PLoS ONE 2017, 12, e0186552. [Google Scholar] [CrossRef] [PubMed]
- Thaker, A.; Mehta, K.; Patkar, R. Feruloyl esterase Fae1 is required specifically for host colonisation by the rice-blast fungus Magnaporthe oryzae. Curr. Genet. 2021, 68, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhai, S.; Sun, Y.; Li, M.; Dong, Y.; Wang, X.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoTup1 is required for growth, conidiogenesis and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 2015, 16, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Reza, H.; Shah, H.; Manjrekar, J.; Chattoo, B.B. Magnesium Uptake by CorA Transporters Is Essential for Growth, Development and Infection in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2016, 11, e0159244. [Google Scholar] [CrossRef]
- Liu, X.; Pan, X.; Chen, D.; Yin, C.; Peng, J.; Shi, W.; Qi, L.; Wang, R.; Zhao, W.; Zhang, Z.; et al. Prp19-associated splicing factor Cwf15 regulates fungal virulence and development in the rice blast fungus. Environ. Microbiol. 2021, 23, 5901–5916. [Google Scholar] [CrossRef]
- Sabnam, N.; Barman, S.R. WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. Biol. 2017, 105, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, X.; Zhang, S.; Yang, J.; Chen, D.; Liu, M.; Zhang, H.; Zheng, X.; Wang, P.; Peng, Y.; et al. MoCAP proteins regulated by MoArk1-mediated phosphorylation coordinate endocytosis and actin dynamics to govern development and virulence of Magnaporthe oryzae. PLoS Genet. 2017, 13, e1006814. [Google Scholar] [CrossRef] [PubMed]
- Sadat, A.; Han, J.-H.; Kim, S.; Lee, Y.-H.; Kim, K.S.; Choi, J. The Membrane-Bound Protein, MoAfo1, Is Involved in Sensing Diverse Signals from Different Surfaces in the Rice Blast Fungus. Plant Pathol. J. 2021, 37, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kong, L.; Chen, X.; Wang, D.; Qi, L.; Zhao, W.; Zhang, Y.; Liu, X.; Peng, Y.-L. A carnitine–acylcarnitine carrier protein, MoCrc1, is essential for pathogenicity in Magnaporthe oryzae. Curr. Genet. 2012, 58, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, X.; Wei, Y.; Liu, J.; Lin, F.; Zhang, S.-H. An ATP-dependent protease homolog ensures basic standards of survival and pathogenicity for Magnaporthe oryzae. Eur. J. Plant Pathol. 2015, 141, 703–716. [Google Scholar] [CrossRef]
- Goh, J.; Kim, K.S.; Park, J.; Jeon, J.; Park, S.-Y.; Lee, Y.-H. The cell cycle gene MoCDC15 regulates hyphal growth, asexual development and plant infection in the rice blast pathogen Magnaporthe oryzae. Fungal Genet. Biol. 2011, 48, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, J.; Zhu, X.; Dong, B.; Liu, X.; Lu, J.; Lin, F. The P5-type ATPase Spf1 is required for development and virulence of the rice blast fungus Pyricularia oryzae. Curr. Genet. 2020, 66, 385–395. [Google Scholar] [CrossRef]
- Li, Y.; Yan, X.; Wang, H.; Liang, S.; Ma, W.-B.; Fang, M.-Y.; Talbot, N.J.; Wang, Z.-Y. MoRic8 Is a Novel Component of G-Protein Signaling During Plant Infection by the Rice Blast Fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2010, 23, 317–331. [Google Scholar] [CrossRef]
- Liu, X.-H.; Zhuang, F.-L.; Lu, J.-P.; Lin, F.-C. Identification and molecular cloning Moplaa gene, a homologue of Homo sapiens PLAA, in Magnaporthe oryzae. Microbiol. Res. 2011, 167, 8–13. [Google Scholar] [CrossRef]
- Aron, O.; Wang, M.; Lin, L.; Batool, W.; Lin, B.; Shabbir, A.; Wang, Z.; Tang, W. MoGLN2 is important for vegetative growth, conidiogenesis, maintenance of cell wall integrity and pathogenesis of Magnaporthe oryzae. J. Fungi 2021, 7, 463. [Google Scholar] [CrossRef]
- Dang, Y.; Wei, Y.; Zhang, P.; Liu, X.; Li, X.; Wang, S.; Liang, H.; Zhang, S.-H. The Bicarbonate Transporter (MoAE4) Localized on Both Cytomembrane and Tonoplast Promotes Pathogenesis in Magnaporthe oryzae. J. Fungi 2021, 7, 955. [Google Scholar] [CrossRef]
- Goh, J.; Jeon, J.; Lee, Y.-H. ER retention receptor, MoERR1 is required for fungal development and pathogenicity in the rice blast fungus, Magnaporthe oryzae. Sci. Rep. 2017, 7, 1259. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, X.-M.; Shi, H.-B.; Feng, X.-X.; Liu, X.-H.; Lin, F.-C. MoFap7, a ribosome assembly factor, is required for fungal development and plant colonization of Magnaporthe oryzae. Virulence 2019, 10, 1047–1063. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Gao, C.; Wang, J.; Yin, Z.; Zhang, J.; Ji, J.; Zhang, H.; Zheng, X.; Zhang, Z.; Wang, P. Disruption of actin motor function due to MoMyo5 mutation impairs host penetration and pathogenicity in Magnaporthe oryzae. Mol. Plant Pathol. 2018, 19, 689–699. [Google Scholar] [CrossRef] [PubMed]
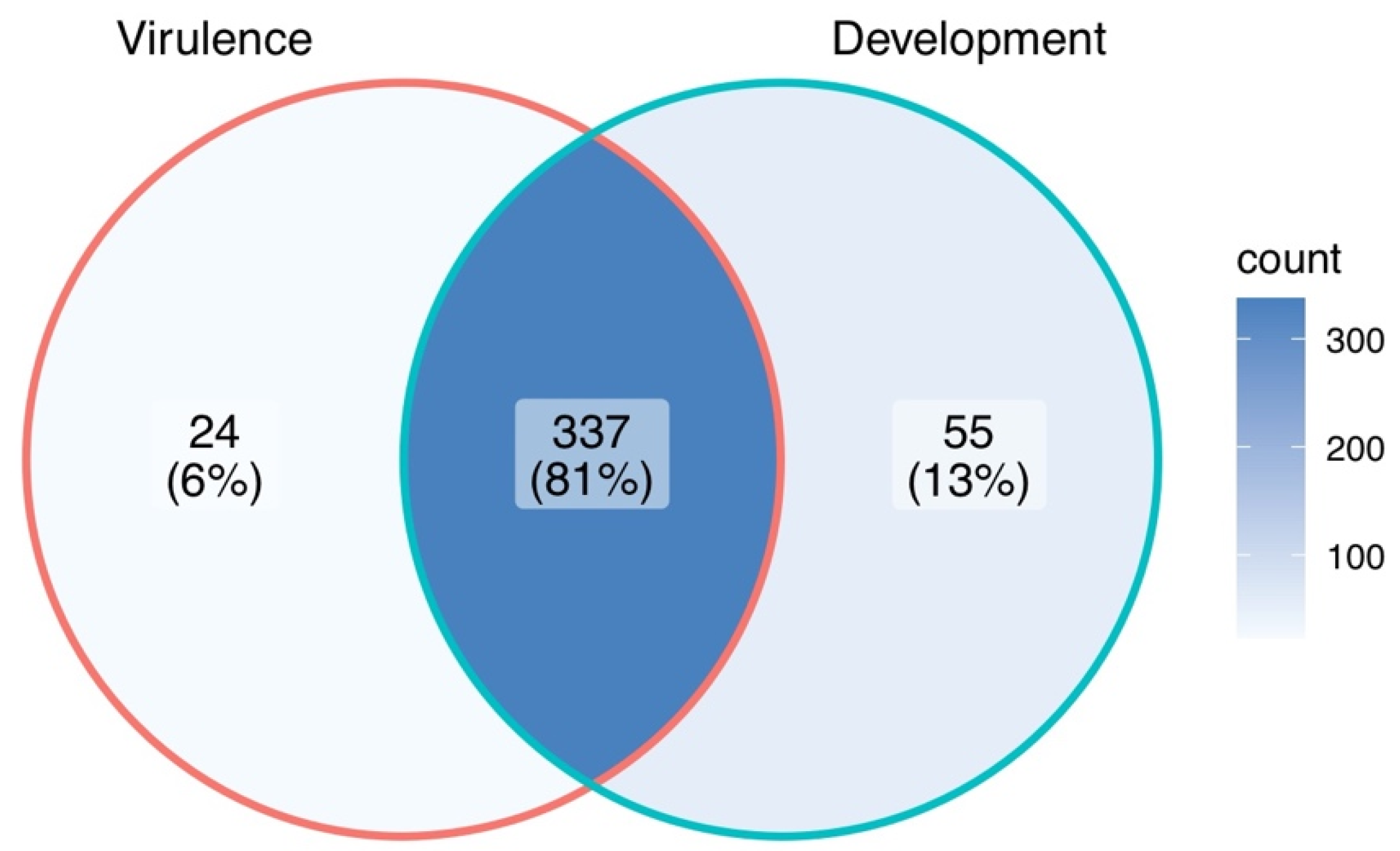
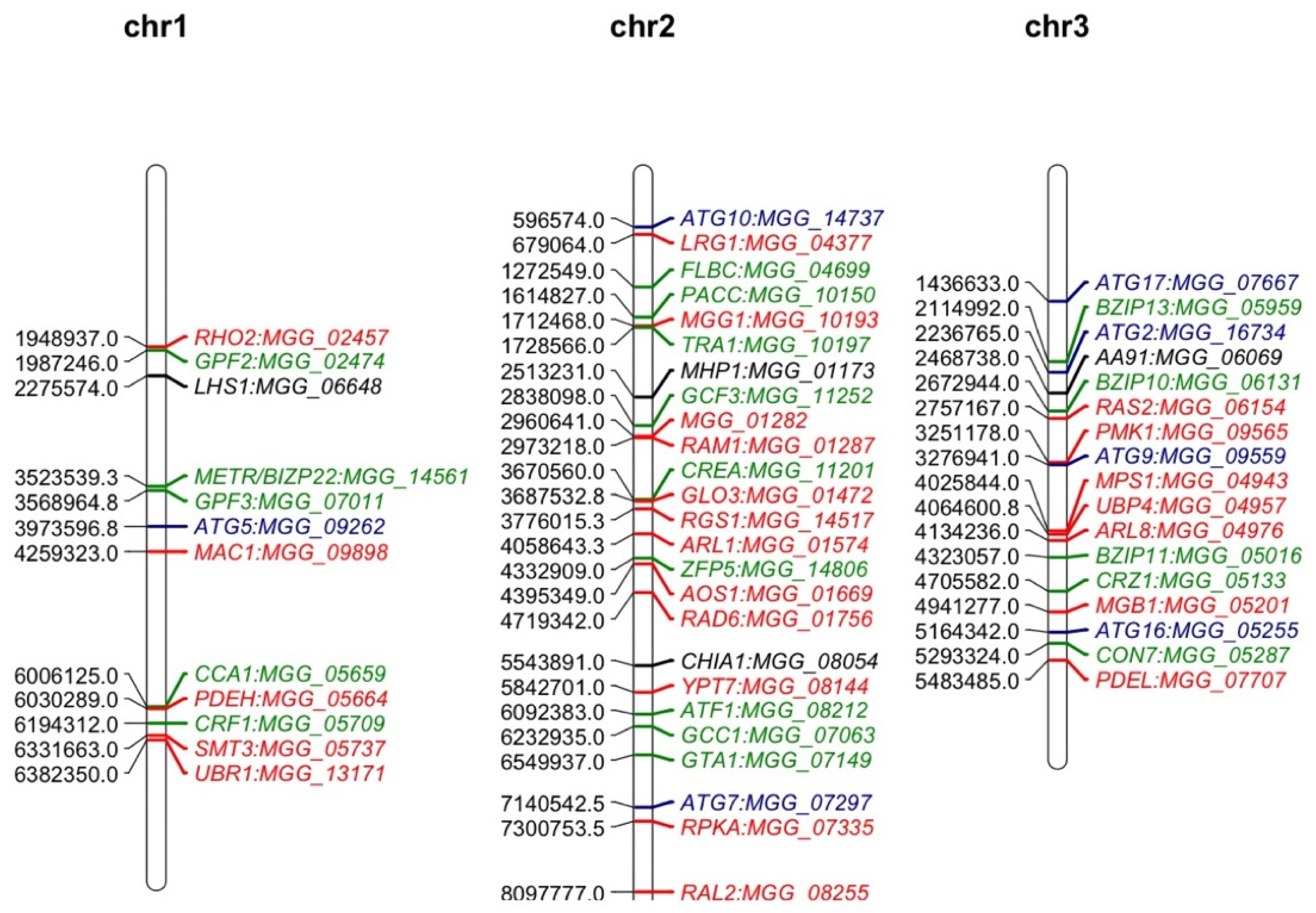
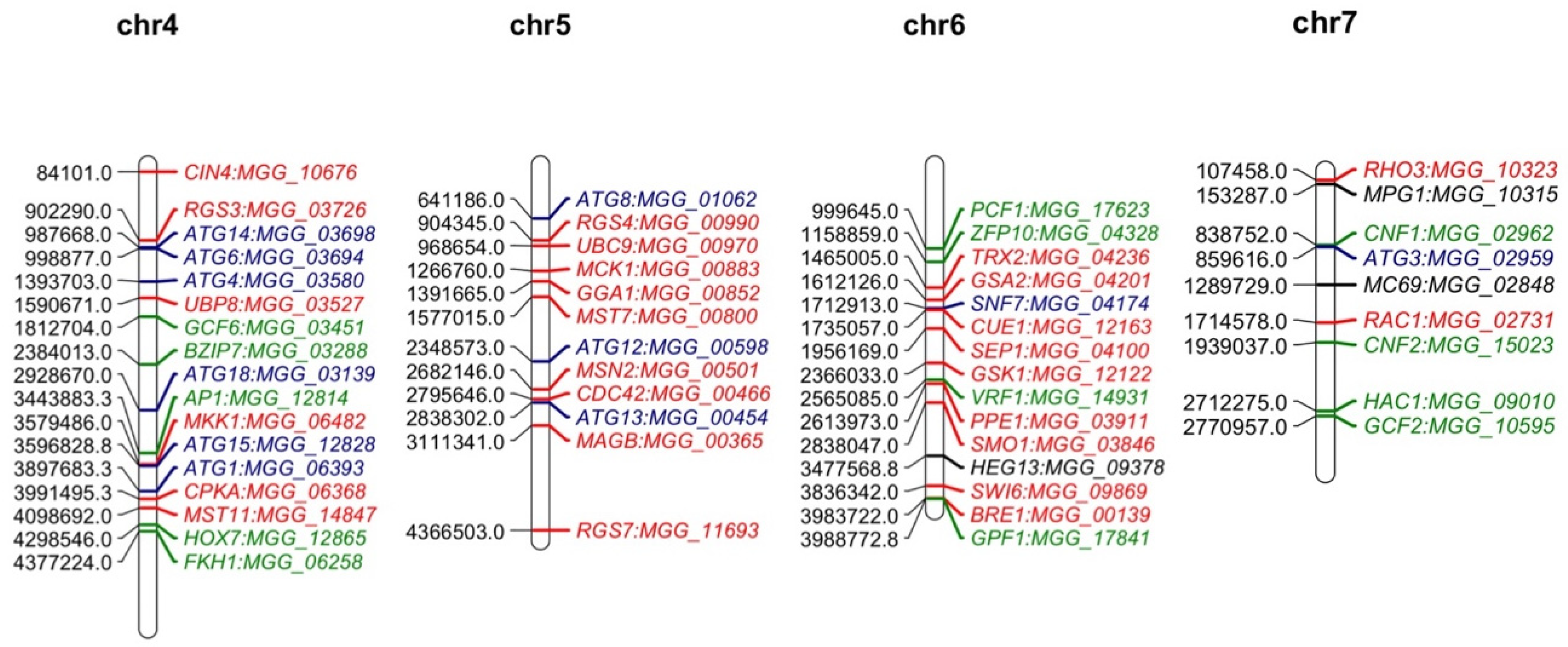

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, J.; Zhao, H.; Li, J.; Gong, Y.; Li, X. The Devastating Rice Blast Airborne Pathogen Magnaporthe oryzae—A Review on Genes Studied with Mutant Analysis. Pathogens 2023, 12, 379. https://doi.org/10.3390/pathogens12030379
Tan J, Zhao H, Li J, Gong Y, Li X. The Devastating Rice Blast Airborne Pathogen Magnaporthe oryzae—A Review on Genes Studied with Mutant Analysis. Pathogens. 2023; 12(3):379. https://doi.org/10.3390/pathogens12030379
Chicago/Turabian StyleTan, Jinyi, Haikun Zhao, Josh Li, Yihan Gong, and Xin Li. 2023. "The Devastating Rice Blast Airborne Pathogen Magnaporthe oryzae—A Review on Genes Studied with Mutant Analysis" Pathogens 12, no. 3: 379. https://doi.org/10.3390/pathogens12030379
APA StyleTan, J., Zhao, H., Li, J., Gong, Y., & Li, X. (2023). The Devastating Rice Blast Airborne Pathogen Magnaporthe oryzae—A Review on Genes Studied with Mutant Analysis. Pathogens, 12(3), 379. https://doi.org/10.3390/pathogens12030379






