The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Obtention of ΔFTF2 Mutants in the Weakly Virulent Strain FOP-SP4 of F. oxysporum f. sp. phaseoli
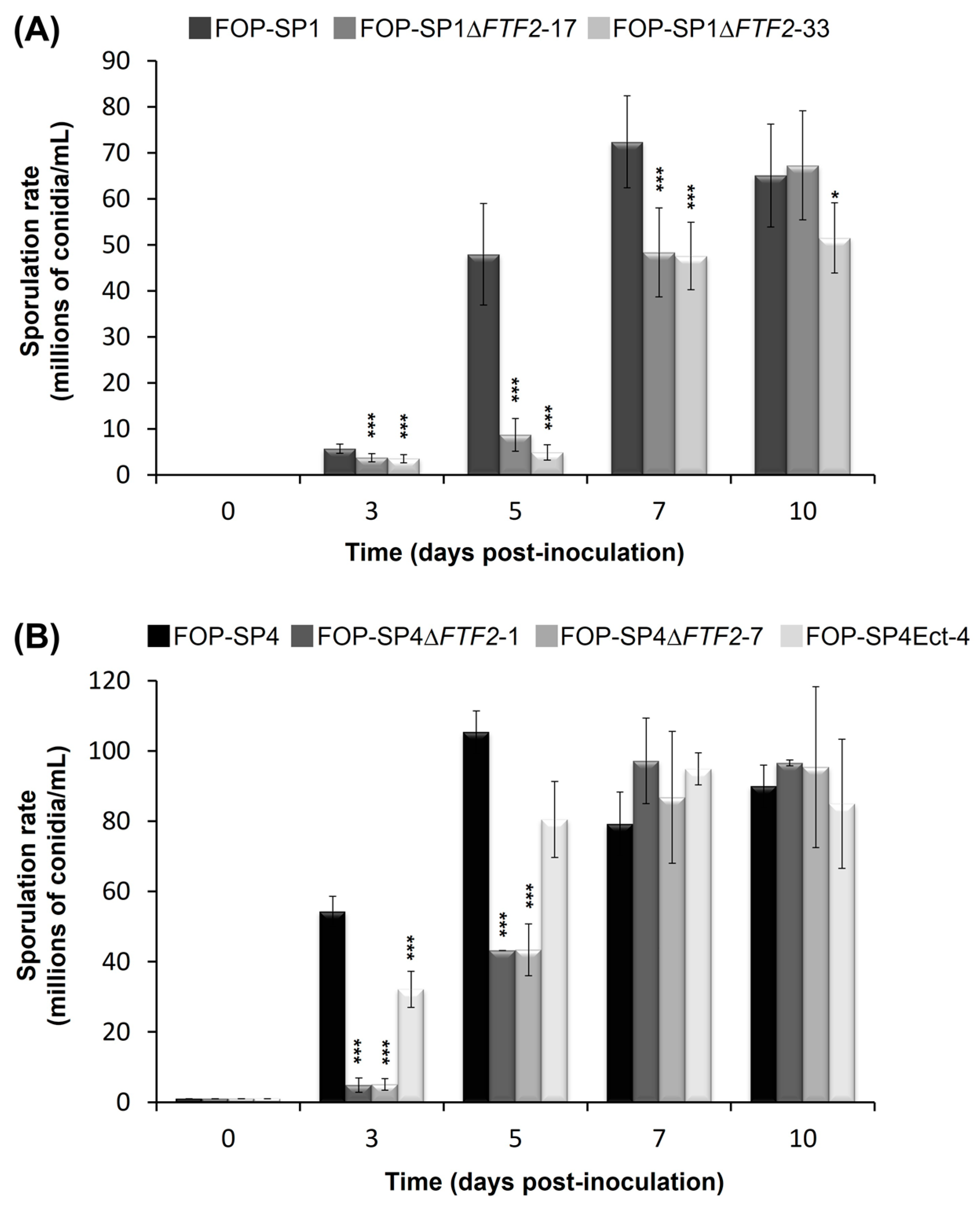
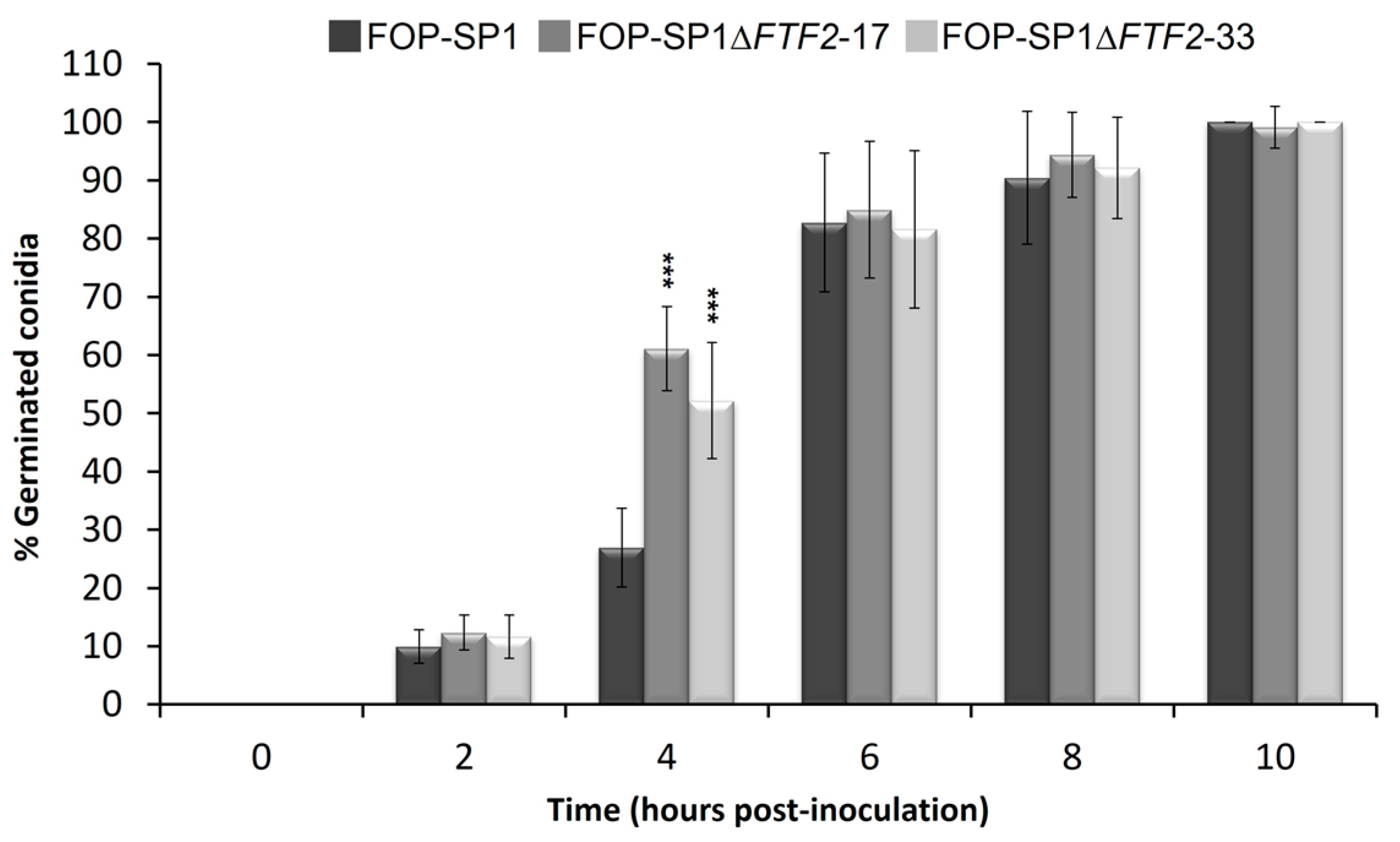
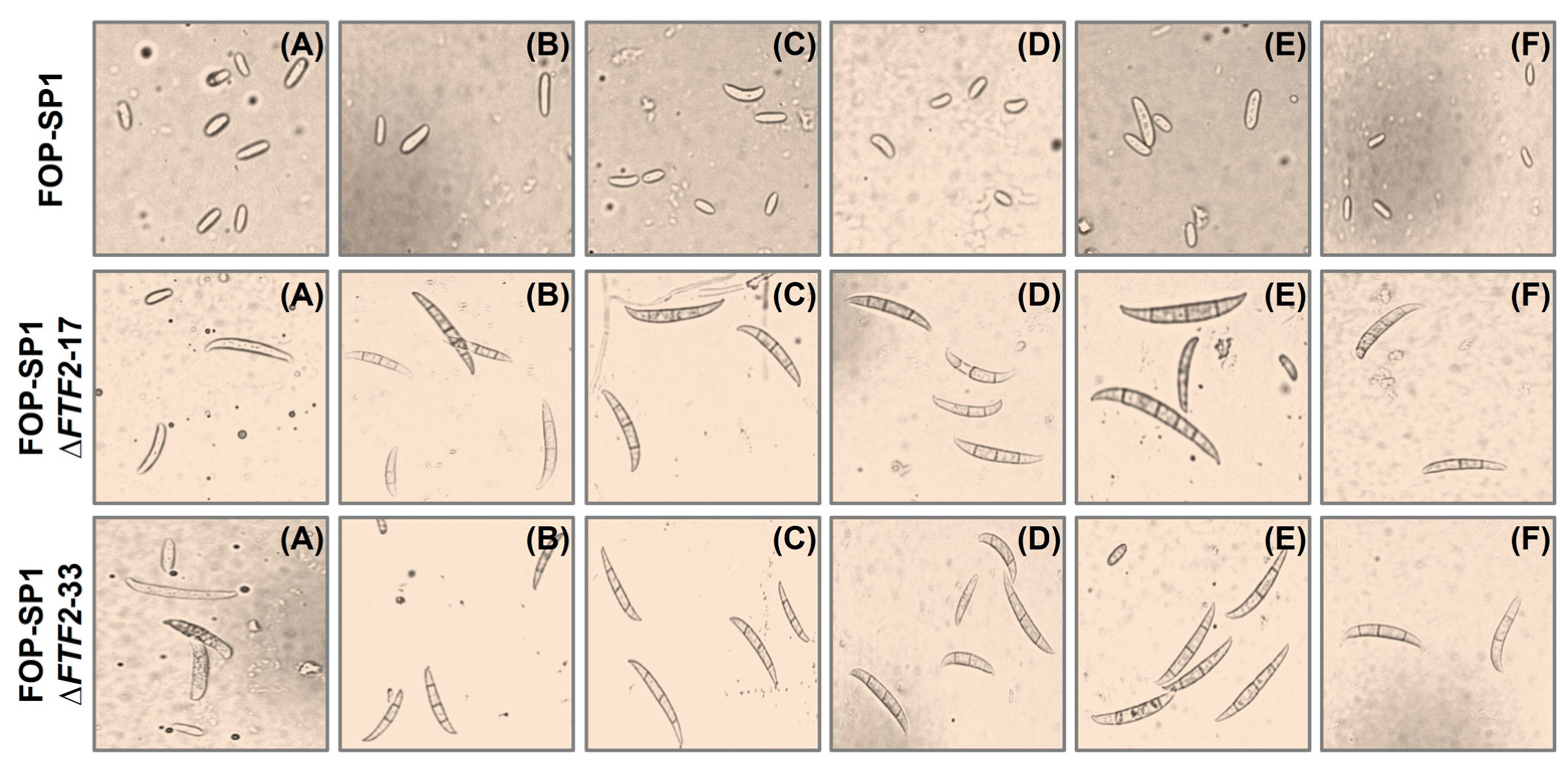
2.3. Sporulation, Germination, and Saprophytic Growth Assays
2.4. Inoculation of Common Bean Plants
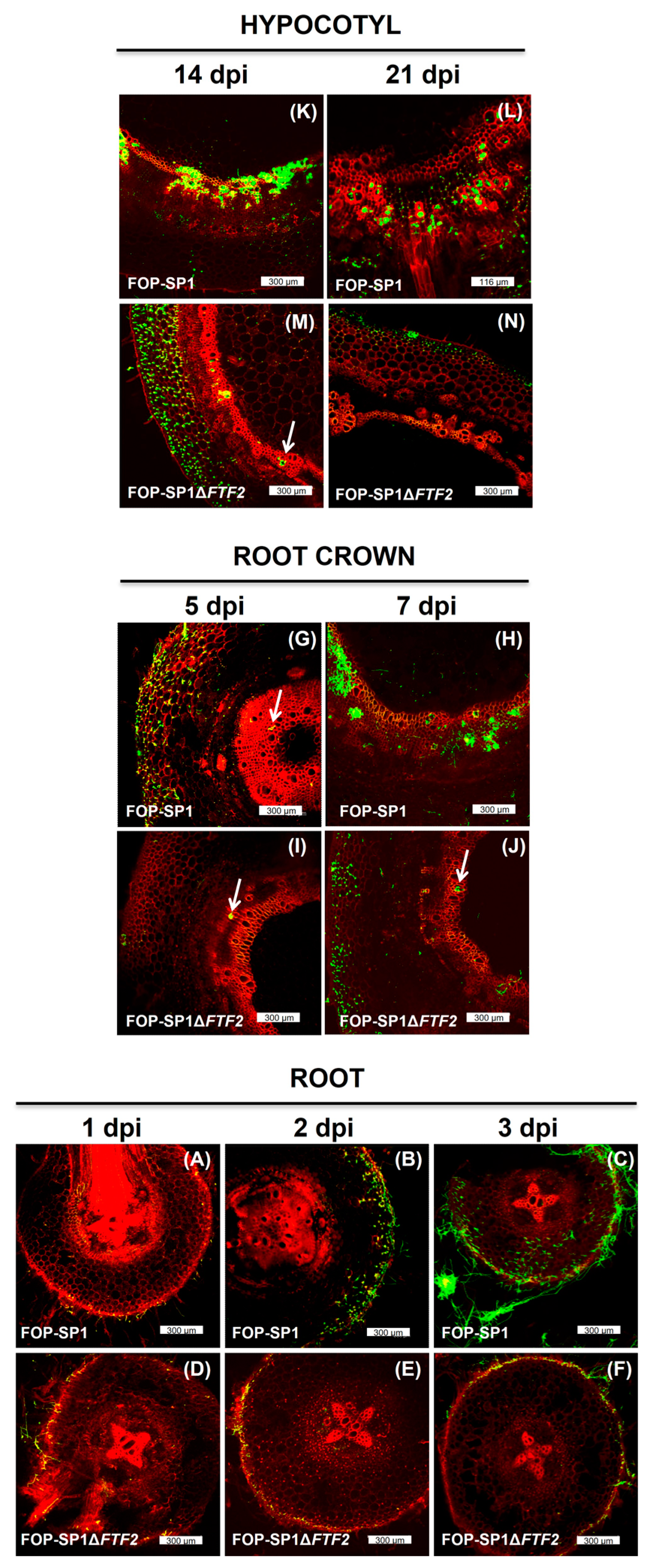
2.5. Confocal Laser Microscopy
2.6. Nucleic Acid Extraction and Purification
2.7. Analysis of Gene Expression and Fungal Biomass Quantification
3. Results
3.1. Obtention of ΔFTF2 Mutants by Gene Replacement
3.2. Functional Domains of the FTF Transcription Factors
3.3. Phenoypic Characterization of ΔFTF2 Mutants
3.4. Host Plant Colonization by ΔFTF2 Mutants
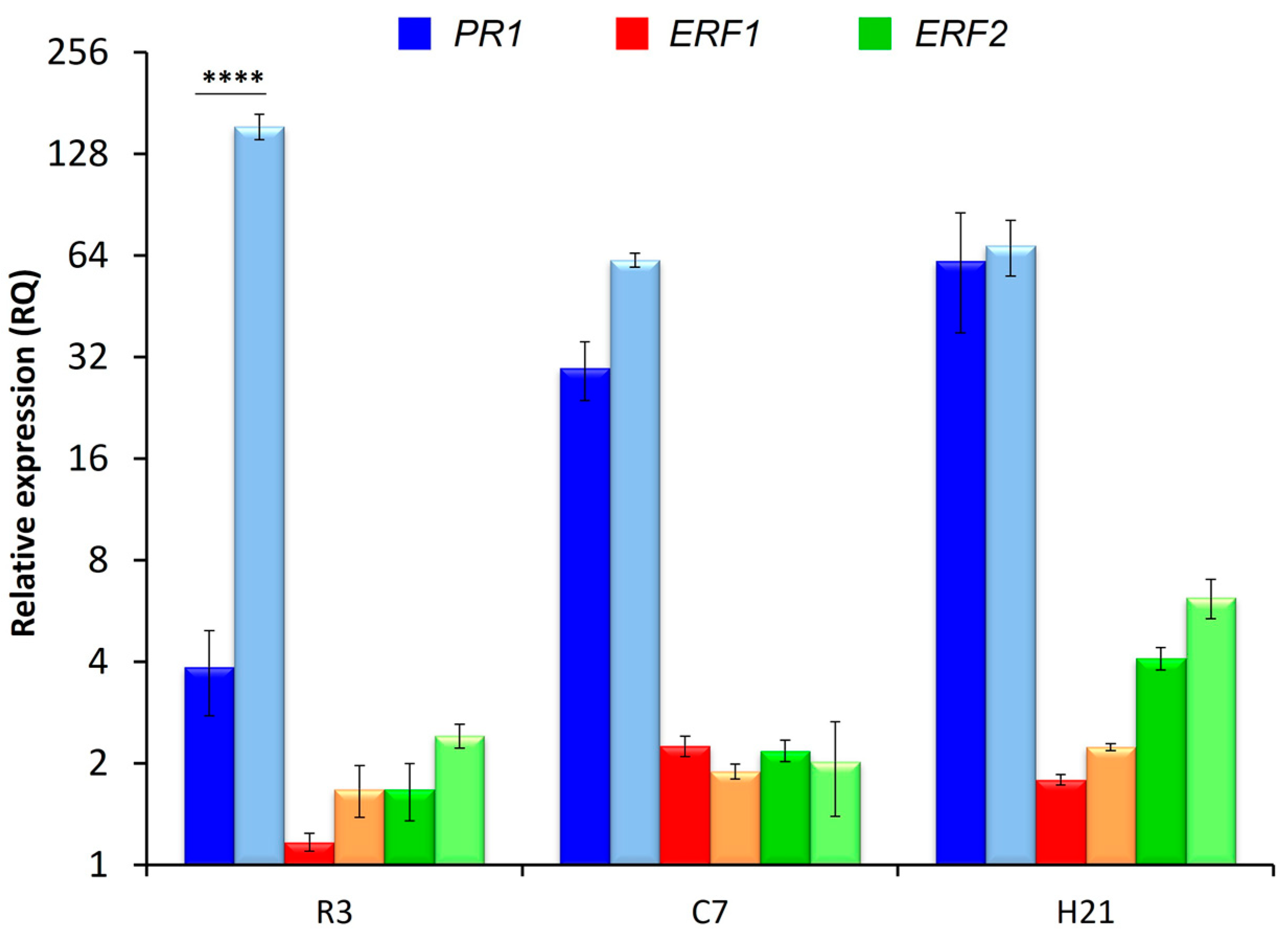
3.5. Plant Defensive Response
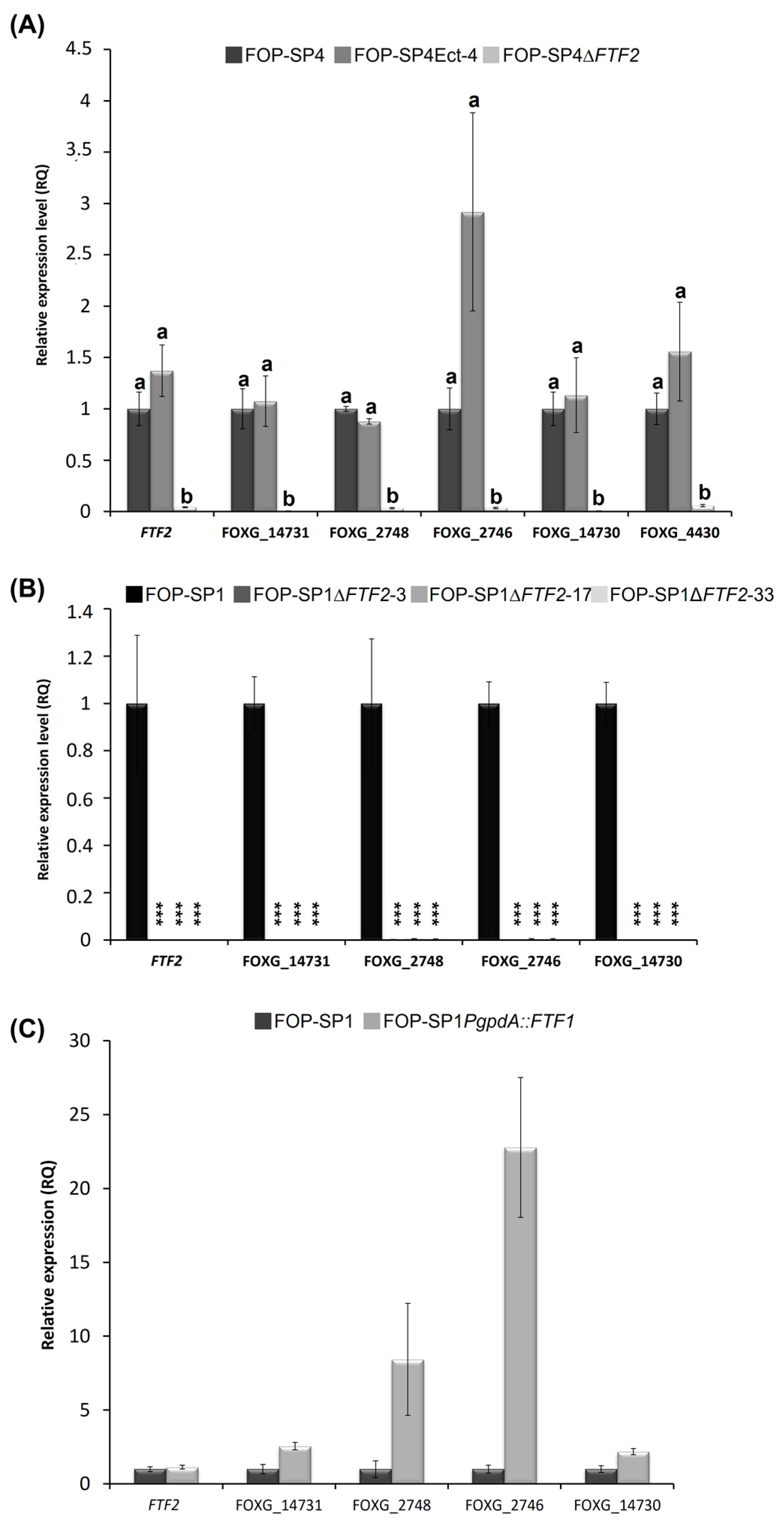
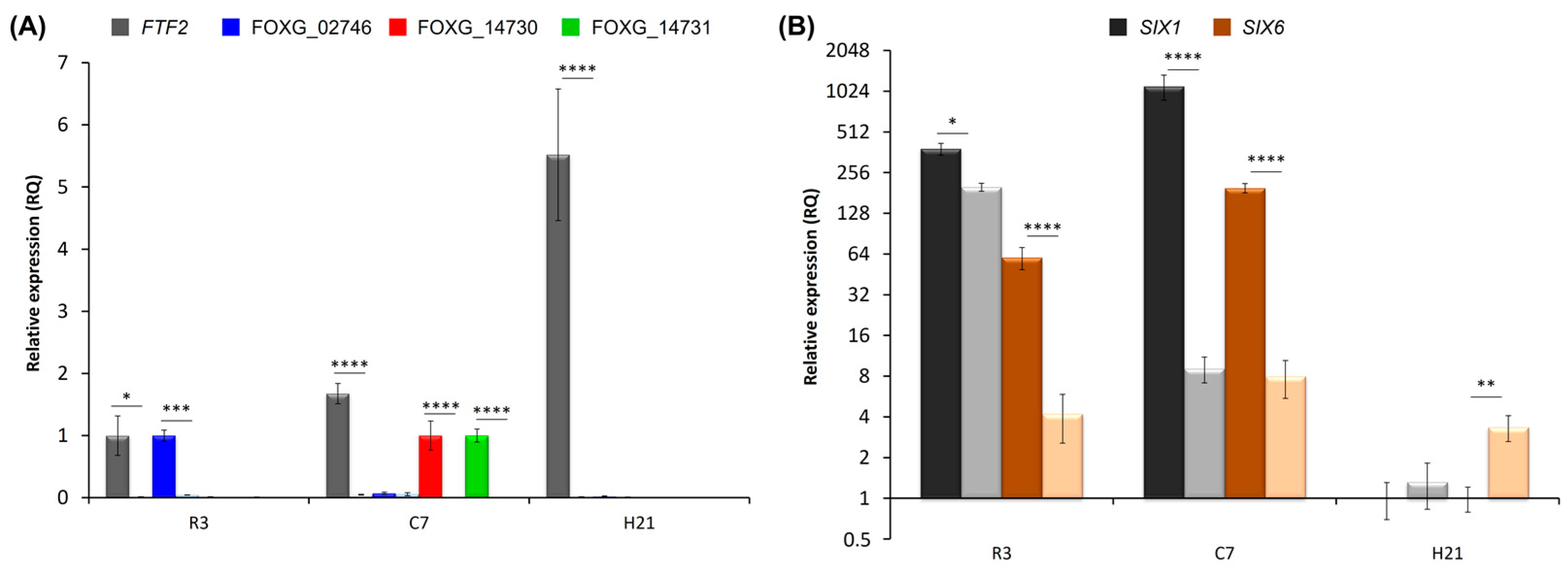
3.6. Analysis of FTF2-Responsive Genes
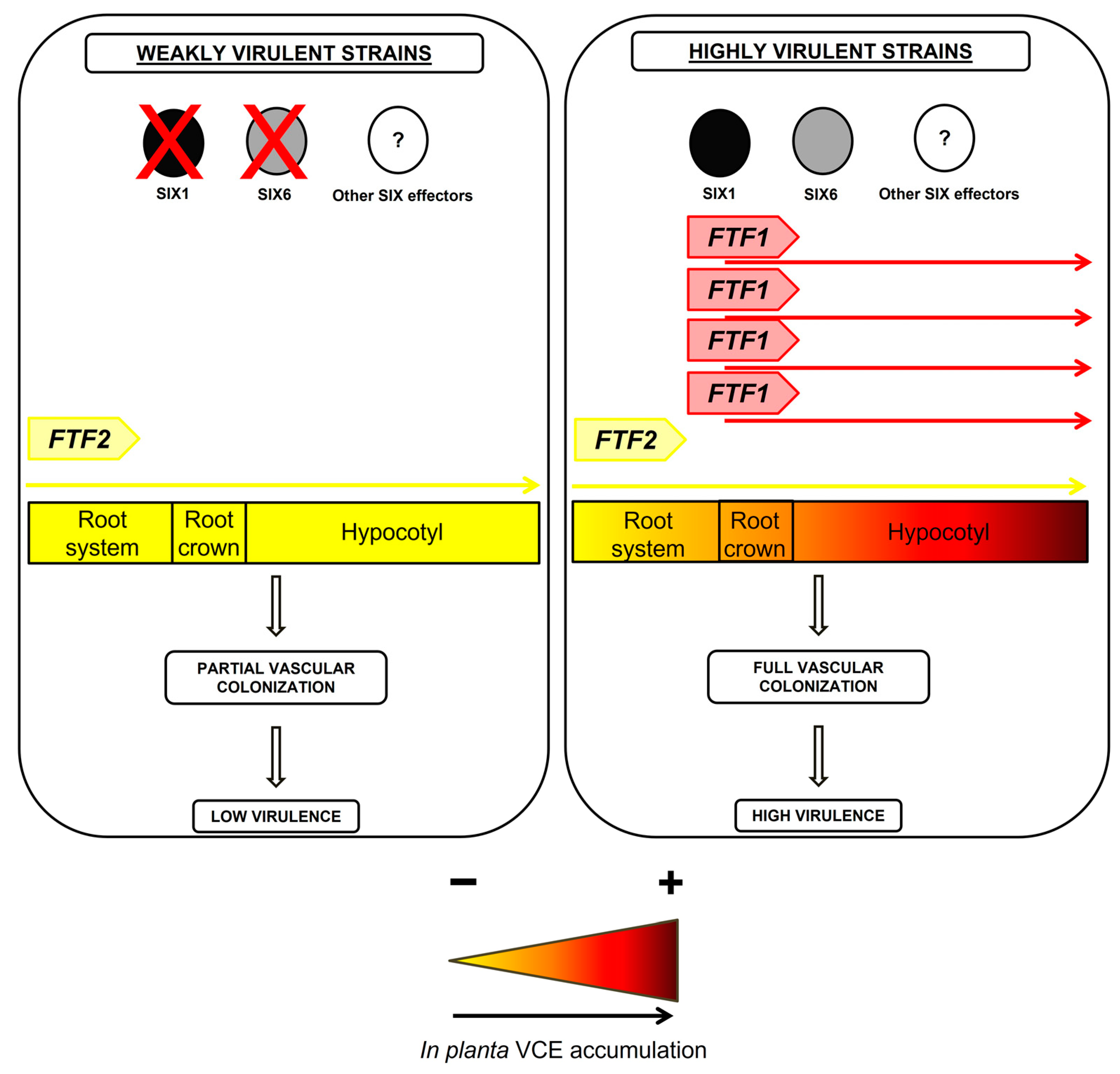
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Armstrong, G.M.; Armstrong, J.K. Fusarium: Diseases, Biology and Taxonomy; Pennsilvania State University Press: University Park, PA, USA, 1981; pp. 391–400. [Google Scholar]
- Edel-Hermann, V.; Lecomte, C. Current Status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Nag, P.; Aggarwal, P.R.; Ghosh, S.; Narula, K.; Tayal, R.; Maheshwari, N.; Chakraborty, N.; Chakraborty, S. Interplay of neuronal and non-neuronal genes regulates intestinal DAF-16-mediated immune response during Fusarium infection of Caenorhabditis elegans. Cell Death Discov. 2017, 3, 17073. [Google Scholar] [CrossRef]
- Nag, P.; Paul, S.; Shriti, S.; Das, S. Defence response in plants and animals against a common fungal pathogen, Fusarium oxysporum. Curr. Res. Microb. Sci. 2022, 3, 100135. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ayhan, D.H.; Martínez-Soto, D.; Cochavi, S.M.; Ma, L.-J. Accessory chromosomes of the Fusarium oxysporum species complex and their contribution to host niche adaptation. In Plant Relationships: Fungal-Plant Interactions; Scott, B., Mesarich, C., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 371–388. ISBN 978-3-031-16503-0. [Google Scholar]
- Ma, L.-J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Henry, P.M.; Pincot, D.D.A.; Jenner, B.N.; Borrero, C.; Aviles, M.; Nam, M.; Epstein, L.; Knapp, S.J.; Gordon, T.R. Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporum f. sp. fragariae. New Phytol. 2021, 230, 327–340. [Google Scholar] [CrossRef]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, e1000061-6. [Google Scholar] [CrossRef]
- Niño-Sánchez, J.; Casado-del Castillo, V.; Tello, V.; de Vega-Bartol, J.J.; Ramos, B.; Sukno, S.A.; Díaz-Mínguez, J.M. The FTF gene family regulates virulence and expression of SIX effectors in Fusarium oxysporum. Mol. Plant Pathol. 2016, 17, 1124–1139. [Google Scholar] [CrossRef]
- Ramos, B.; Alves-Santos, F.M.; García-Sánchez, M.A.; Martín-Rodrigues, N.; Eslava, A.P.; Díaz-Mínguez, J.M. The gene coding for a new transcription factor (Ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet. Biol. 2007, 44, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Alves-Santos, F.M.; Benito, E.P.; Eslava, A.P.; Diaz-Minguez, J.M. Genetic diversity of Fusarium oxysporum strains from common bean fields in Spain. Appl. Environ. Microbiol. 1999, 65, 3335–3340. [Google Scholar] [CrossRef]
- Alves-Santos, F.; Cordeiro-Rodrigues, L.; Sayagués, J.M.; Martín-Dominguez, R.; García-Benavides, P.; Crespo, M.C.; Díaz-Mínguez, J.M.; Eslava, A.P. Pathogenicity and race characterization of Fusarium oxysporum f. sp. phaseoli isolates from Spain and Greece. Plant Pathol. 2002, 51, 605–611. [Google Scholar]
- de Vega-Bartol, J.J.; Martín-Dominguez, R.; Ramos, B.; García-Sánchez, M.-A.; Díaz-Mínguez, J.M. New virulence groups in Fusarium oxysporum f. sp. phaseoli: The expression of the gene coding for the transcription factor Ftf1 correlates with virulence. Phytopathology 2011, 101, 470–479. [Google Scholar] [CrossRef]
- Mullins, E.; Chen, X.; Romaine, P.; Raina, R.; Geiser, D.; Kang, S. Agrobacterium-mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology 2001, 91, 173–180. [Google Scholar] [CrossRef]
- Correll, J.C.; Klittich, C.J.R.; Leslie, J.F. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 1987, 77, 1640–1646. [Google Scholar] [CrossRef]
- Castillo, V.C.-D.; Benito, E.P.; Díaz-Mínguez, J.M. In planta gene expression analysis and colonization of Fusarium oxysporum. Methods Mol. Biol. 2022, 2391, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Alves-Santos, F.M.; Ramos, B.; García-Sánchez, M.A.; Eslava, A.P.; Díaz-Mínguez, J.M. A DNA-based procedure for in planta detection of Fusarium oxysporum f. sp. phaseoli. Phytopathology 2002, 92, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Afanador, L.K.; Haley, S.D.; Kelly, J.D.; Beebe, S. Adoption of a “mini-prep” DNA extraction method for RAPD marker analysis in common bean (Phaseolus vulgaris L.). Bean Improv. Coop. Annu. Rep. 1993, 36, 10–11. [Google Scholar]
- Niño-Sánchez, J.; Tello, V.; Casado-del Castillo, V.; Thon, M.R.; Benito, E.P.; Díaz-Mínguez, J.M. Gene expression patterns and dynamics of the colonization of common bean (Phaseolus vulgaris L.) by highly virulent and eeakly virulent strains of Fusarium oxysporum. Front. Microbiol. 2015, 6, 605–614. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Coleman, J. GAL4 transcription factor Is Not a “zinc finger” but forms a Zn(II)2 Cys6 binuclear cluster. Proc. Natl. Acad. Sci. USA 1990, 87, 2077–2081. [Google Scholar] [CrossRef]
- Schjerling, P.; Holmberg, S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996, 24, 4599–4607. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- van der Does, H.C.; Fokkens, L.; Yang, A.; Schmidt, S.M.; Langereis, L.; Lukasiewicz, J.M.; Hughes, T.R.; Rep, M. Transcription factors encoded on core and accessory chromosomes of Fusarium oxysporum induce expression of effector genes. PLoS Genet. 2016, 12, e1006401-38. [Google Scholar] [CrossRef]
- Croll, D.; McDonald, B.A. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 2012, 8, e1002608. [Google Scholar] [CrossRef] [PubMed]
- Casado-del Castillo, V. The Roles of FTF2 and the Pathogenicity Chromosome of Fusarium oxysporum. Ph.D. Thesis, University of Salamanca, Salamanca, Spain, 2017. [Google Scholar]
- Macpherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef]
- Bellizzi, J.J.; Sorger, P.K.; Harrison, S.C. Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure 2007, 15, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Purvis, A.; Singleton, M.R. Insights into kinetochore–DNA interactions from the structure of Cep3Δ. EMBO Rep. 2008, 9, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Ohara, T.; Inoue, I.; Namiki, F.; Kunoh, H.; Tsuge, T. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics 2004, 166, 113–124. [Google Scholar] [CrossRef]
- Ohara, T.; Tsuge, T. FoSTUA, encoding a basic helix-loop-h protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Euk. Cell 2004, 3, 1412–1422. [Google Scholar] [CrossRef]
- López-Berges, M.S.; Hera, C.; Sulyok, M.; Schäfer, K.; Capilla, J.; Guarro, J.; Di Pietro, A. The Velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Microbiol. 2012, 87, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Wessels, J.G.H. Hydrophobins, unique fungal proteins. Mycologist 2000, 14, 153–159. [Google Scholar] [CrossRef]
- Wang, X.; Shi, F.; Wösten, H.A.B.; Hektor, H.; Poolman, B.; Robillard, G.T. The SC3 hydrophobin self-assembles into a membrane with distinct mass transfer properties. Biophys. J. 2005, 88, 3434–3443. [Google Scholar] [CrossRef]
- Bayry, J.; Aimanianda, V.; Guijarro, J.I.; Sunde, M.; Latgé, J.-P. Hydrophobins—Unique fungal proteins. PLoS Pathog. 2012, 8, e1002700. [Google Scholar] [CrossRef]
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.L.; Talbot, N.J. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 2001, 39, 385–417. [Google Scholar] [CrossRef]
- Whiteford, J.R.; Spanu, P.D. Hydrophobins and the interactions between fungi and plants: Hydrophobins and fungus-plant interactions. Mol. Plant Pathol. 2002, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ahn, I.-P.; Rho, H.-S.; Lee, Y.-H. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 2005, 57, 1224–1237. [Google Scholar] [CrossRef]
- Quarantin, A.; Hadeler, B.; Kröger, C.; Schäfer, W.; Favaron, F.; Sella, L.; Martínez-Rocha, A.L. Different hydrophobins of Fusarium graminearum are involved in hyphal growth, attachment, water-air interface penetration and plant infection. Front. Microbiol. 2019, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Maurus, I.; Leonard, M.; Nagel, A.; Starke, J.; Kronstad, J.W.; Harting, R.; Braus, G.H. Tomato xylem sap hydrophobins Vdh4 and Vdh5 are important for late stages of Verticillium dahliae plant infection. J. Fungi. 2022, 8, 1252. [Google Scholar] [CrossRef]
- Constantin, M.E.; Fokkens, L.; de Sain, M.; Takken, F.L.W.; Rep, M. Number of candidate effector genes in accessory genomes differentiates pathogenic from endophytic Fusarium oxysporum strains. Front. Plant Sci. 2021, 12, 761740. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yu, H.; Ma, L.-J. Accessory chromosomes in Fusarium oxysporum. Phytopathology 2020, 110, 1488–1496. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casado-del Castillo, V.; Benito, E.P.; Díaz-Mínguez, J.M. The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.). Pathogens 2023, 12, 380. https://doi.org/10.3390/pathogens12030380
Casado-del Castillo V, Benito EP, Díaz-Mínguez JM. The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.). Pathogens. 2023; 12(3):380. https://doi.org/10.3390/pathogens12030380
Chicago/Turabian StyleCasado-del Castillo, Virginia, Ernesto P. Benito, and José María Díaz-Mínguez. 2023. "The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.)" Pathogens 12, no. 3: 380. https://doi.org/10.3390/pathogens12030380
APA StyleCasado-del Castillo, V., Benito, E. P., & Díaz-Mínguez, J. M. (2023). The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.). Pathogens, 12(3), 380. https://doi.org/10.3390/pathogens12030380







