Non-Toxin-Based Clostridioides difficile Vaccination Approaches
Abstract
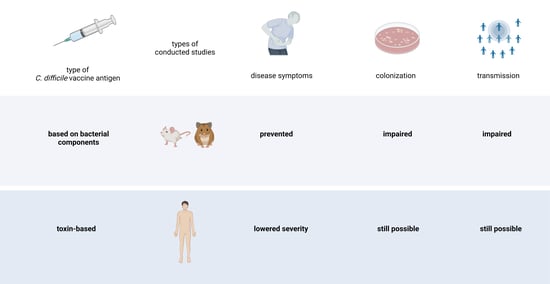
1. Introduction
2. Targeting CD Protein Surface Components
2.1. CD Spore Surface Proteins as Vaccine Candidates
2.2. Vegetative CD Surface Proteins as Vaccine Candidates
3. Targeting Intracellular Proteins
4. Targeting CD Surface Glycopolymers
5. Using Non-Toxigenic Strains
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbut, F.; Petit, J.C. Epidemiology of Clostridium difficile-Associated Infections. Clin. Microbiol. Infect. 2001, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased Diversity of the Fecal Microbiome in Recurrent Clostridium difficile-Associated Diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P. Can We Identify Patients at High Risk of Recurrent Clostridium difficile Infection? Predictors of C. Difficile Recurrence. Clin. Microbiol. Infect. 2012, 18, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Thelestam, M.; Chaves-Olarte, E. Cytotoxic Effects of the Clostridium difficile Toxins. In Clostridium difficile; Aktories, K., Wilkins, T.D., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2000; pp. 85–96. ISBN 978-3-662-06272-2. [Google Scholar]
- Rupnik, M.; Wilcox, M.H.; Gerding, D.N. Clostridium difficile Infection: New Developments in Epidemiology and Pathogenesis. Nat. Rev. Microbiol. 2009, 7, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Zaiß, N.H.; Witte, W.; Nübel, U. Fluoroquinolone Resistance and Clostridium difficile, Germany. Emerg. Infect. Dis. 2010, 16, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.S.; Jiang, Z.-D.; Garey, K.W.; Lasco, T.; DuPont, H.L. Use of Rifamycin Drugs and Development of Infection by Rifamycin-Resistant Strains of Clostridium difficile. Antimicrob. Agents Chemother. 2013, 57, 2690–2693. [Google Scholar] [CrossRef]
- Péchiné, S.; Collignon, A. Immune Responses Induced by Clostridium difficile. Anaerobe 2016, 41, 68–78. [Google Scholar] [CrossRef]
- Tait, D.R.; Hatherill, M.; Van Der Meeren, O.; Ginsberg, A.M.; Van Brakel, E.; Salaun, B.; Scriba, T.J.; Akite, E.J.; Ayles, H.M.; Bollaerts, A.; et al. Final Analysis of a Trial of M72/AS01E Vaccine to Prevent Tuberculosis. N. Engl. J. Med. 2019, 381, 2429–2439. [Google Scholar] [CrossRef]
- Péchiné, S.; Bruxelle, J.F.; Janoir, C.; Collignon, A. Targeting Clostridium difficile Surface Components to Develop Immunotherapeutic Strategies Against Clostridium difficile Infection. Front. Microbiol. 2018, 9, 1009. [Google Scholar] [CrossRef]
- Abhyankar, W.R.; Zheng, L.; Brul, S.; de Koster, C.G.; de Koning, L.J. Vegetative Cell and Spore Proteomes of Clostridioides difficile Show Finite Differences and Reveal Potential Protein Markers. J. Proteome Res. 2019, 18, 3967–3976. [Google Scholar] [CrossRef]
- Pizarro-Guajardo, M.; Ravanal, M.C.; Paez, M.D.; Callegari, E.; Paredes-Sabja, D. Identification of Clostridium difficile Immunoreactive Spore Proteins of the Epidemic Strain R20291. Proteom. Clin. Appl. 2018, 12, 1700182. [Google Scholar] [CrossRef]
- Mora-Uribe, P.; Miranda-Cárdenas, C.; Castro-Córdova, P.; Gil, F.; Calderón, I.; Fuentes, J.A.; Rodas, P.I.; Banawas, S.; Sarker, M.R.; Paredes-Sabja, D. Characterization of the Adherence of Clostridium difficile Spores: The Integrity of the Outermost Layer Affects Adherence Properties of Spores of the Epidemic Strain R20291 to Components of the Intestinal Mucosa. Front. Cell. Infect. Microbiol. 2016, 6, 99. [Google Scholar] [CrossRef]
- Maia, A.R.; Reyes-Ramírez, R.; Pizarro-Guajardo, M.; Saggese, A.; Castro-Córdova, P.; Isticato, R.; Ricca, E.; Paredes-Sabja, D.; Baccigalupi, L. Induction of a Specific Humoral Immune Response by Nasal Delivery of Bcla2ctd of Clostridioides difficile. Int. J. Mol. Sci. 2020, 21, 1277. [Google Scholar] [CrossRef]
- Aubry, A.; Zou, W.; Vinogradov, E.; Williams, D.; Chen, W.; Harris, G.; Zhou, H.; Schur, M.J.; Gilbert, M.; Douce, G.R.; et al. In Vitro Production and Immunogenicity of a Clostridium difficile Spore-Specific BclA3 Glycopeptide Conjugate Vaccine. Vaccines 2020, 8, 73. [Google Scholar] [CrossRef]
- Montes-Bravo, N.; Romero-Rodríguez, A.; García-Yunge, J.; Medina, C.; Pizarro-Guajardo, M.; Paredes-Sabja, D. Role of the Spore Coat Proteins CotA and CotB, and the Spore Surface Protein CDIF630_02480, on the Surface Distribution of Exosporium Proteins in Clostridioides difficile 630 Spores. Microorganisms 2022, 10, 1918. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Cervantes-Rincón, T.; Romero-Maldonado, A.; Monreal-Escalante, E.; Nieto-Gómez, R. Transgenic Plants Expressing a Clostridium difficile Spore Antigen as an Approach to Develop Low-Cost Oral Vaccines. Biotechnol. Prog. 2021, 37, e3141. [Google Scholar] [CrossRef]
- Ghose, C.; Eugenis, I.; Edwards, A.N.; Sun, X.; McBride, S.M.; Ho, D.D. Immunogenicity and Protective Efficacy of Clostridium difficile Spore Proteins. Anaerobe 2016, 37, 85–95. [Google Scholar] [CrossRef]
- Permpoonpattana, P.; Phetcharaburanin, J.; Mikelsone, A.; Dembek, M.; Tan, S.; Brisson, M.-C.; La Ragione, R.; Brisson, A.R.; Fairweather, N.; Hong, H.A.; et al. Functional Characterization of Clostridium difficile Spore Coat Proteins. J. Bacteriol. 2013, 195, 1492–1503. [Google Scholar] [CrossRef]
- Basak, S.; Deb, D.; Narsaria, U.; Kar, T.; Castiglione, F.; Sanyal, I.; Bade, P.D.; Srivastava, A.P. In Silico Designing of Vaccine Candidate against Clostridium difficile. Sci. Rep. 2021, 11, 14215. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Troncoso-Cotal, S.; Guerrero-Araya, E.; Paredes-Sabja, D. The Clostridioides difficile Cysteine-Rich Exosporium Morphogenetic Protein, CdeC, Exhibits Self-Assembly Properties That Lead to Organized Inclusion Bodies in Escherichia coli. mSphere 2020, 5, e01065-20. [Google Scholar] [CrossRef]
- Alves Feliciano, C.; Douché, T.; Giai Gianetto, Q.; Matondo, M.; Martin-Verstraete, I.; Dupuy, B. CotL, a New Morphogenetic Spore Coat Protein of Clostridium difficile. Environ. Microbiol. 2019, 21, 984–1003. [Google Scholar] [CrossRef] [PubMed]
- Díaz-González, F.; Milano, M.; Olguin-Araneda, V.; Pizarro-Cerda, J.; Castro-Córdova, P.; Tzeng, S.-C.; Maier, C.S.; Sarker, M.R.; Paredes-Sabja, D. Protein Composition of the Outermost Exosporium-like Layer of Clostridium difficile 630 Spores. J. Proteomics 2015, 123, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.E.; Shadid, T.M.; Lavey, N.P.; Kempher, M.L.; Ballard, J.D.; Duerfeldt, A.S. Identification of ClpP Dual Isoform Disruption as an Antisporulation Strategy for Clostridioides difficile. J. Bacteriol. 2022, 204, e00411-21. [Google Scholar] [CrossRef] [PubMed]
- DiCandia, M.A.; Edwards, A.N.; Jones, J.B.; Swaim, G.L.; Mills, B.D.; McBride, S.M. Identification of Functional Spo0A Residues Critical for Sporulation in Clostridioides difficile. J. Mol. Biol. 2022, 434, 167641. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, A.E.; Eckenroth, B.E.; Forster, E.R.; Kevorkian, Y.; Donnelly, M.L.; de la Puebla, H.B.; Doublié, S.; Shen, A. The CspC Pseudoprotease Regulates Germination of Clostridioides difficile Spores in Response to Multiple Environmental Signals. PLOS Genet. 2019, 15, e1008224. [Google Scholar] [CrossRef]
- Malyshev, D.; Baillie, L. Surface Morphology Differences in Clostridium difficile Spores, Based on Different Strains and Methods of Purification. Anaerobe 2020, 61, 102078. [Google Scholar] [CrossRef]
- Pechine, S. Immunological Properties of Surface Proteins of Clostridium difficile. J. Med. Microbiol. 2005, 54, 193–196. [Google Scholar] [CrossRef]
- Pechine, S.; Janoir, C.; Collignon, A. Variability of Clostridium difficile Surface Proteins and Specific Serum Antibody Response in Patients with Clostridium difficile-Associated Disease. J. Clin. Microbiol. 2005, 43, 5018–5025. [Google Scholar] [CrossRef]
- Kirk, J.A.; Banerji, O.; Fagan, R.P. Characteristics of the Clostridium difficile Cell Envelope and Its Importance in Therapeutics. Microb. Biotechnol. 2016, 10, 76–90. [Google Scholar] [CrossRef]
- Zhu, D.; Bullock, J.; He, Y.; Sun, X. Cwp22, a Novel Peptidoglycan Cross-Linking Enzyme, Plays Pleiotropic Roles in Clostridioides difficile. Environ. Microbiol. 2019, 21, 3076–3090. [Google Scholar] [CrossRef]
- Razim, A.; Pacyga, K.; Martirosian, G.; Szuba, A.; Gamian, A.; Myc, A.; Górska, S. Mapping Epitopes of a Novel Peptidoglycan Cross-Linking Enzyme Cwp22 Recognized by Human Sera Obtained from Patients with Clostridioides difficile Infection and Cord Blood. Microorganisms 2019, 7, 565. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rao, F.; Chen, Z.; Cheng, Y.; Zhang, Q.; Zhang, J.; Guan, Z.; He, Y.; Yu, W.; Cui, G.; et al. The Cwp66 Gene Affects Cell Adhesion, Stress Tolerance, and Antibiotic Resistance in Clostridioides difficile. Microbiol. Spectr. 2022, 10, e02704-21. [Google Scholar] [CrossRef] [PubMed]
- Kovacs-Simon, A.; Leuzzi, R.; Kasendra, M.; Minton, N.; Titball, R.W.; Michell, S.L. Lipoprotein CD0873 Is a Novel Adhesin of Clostridium difficile. J. Infect. Dis. 2014, 210, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.J.; Bruxelle, J.-F.; Kovacs-Simon, A.; Harmer, N.J.; Janoir, C.; Péchiné, S.; Acharya, K.R.; Michell, S.L. Molecular Features of Lipoprotein CD0873: A Potential Vaccine against the Human Pathogen Clostridioides difficile. J. Biol. Chem. 2019, 294, 15850–15861. [Google Scholar] [CrossRef] [PubMed]
- Karyal, C. Development of an Oral Vaccine against Clostridioides difficile. Available online: http://eprints.nottingham.ac.uk/65798/ (accessed on 6 July 2022).
- Bruxelle, J.-F.; Péchiné, S.; Collignon, A. Immunization Strategies Against Clostridium difficile. In Updates on Clostridium difficile in Europe: Advances in Microbiology, Infectious Diseases and Public Health Volume 8; Mastrantonio, P., Rupnik, M., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2018; pp. 197–225. ISBN 978-3-319-72799-8. [Google Scholar]
- Wang, S.; Ju, X.; Zhang, K.; Duan, Z.; Patabendige, H.M.L.W.; Zhao, S.; Sun, X. Recombinant Fusion Protein Vaccine Containing FliC and FliD Protects Mice Against Clostridioides difficile Infection. Res. Square 2022. [Google Scholar] [CrossRef]
- Razim, A.; Pacyga, K.; Naporowski, P.; Martynowski, D.; Szuba, A.; Gamian, A.; Górska, S. Identification of Linear Epitopes on the Flagellar Proteins of Clostridioides difficile. Sci. Rep. 2021, 11, 9940. [Google Scholar] [CrossRef]
- Cuenot, E.; Garcia-Garcia, T.; Douche, T.; Gorgette, O.; Courtin, P.; Denis-Quanquin, S.; Hoys, S.; Tremblay, Y.D.N.; Matondo, M.; Chapot-Chartier, M.-P.; et al. The Ser/Thr Kinase PrkC Participates in Cell Wall Homeostasis and Antimicrobial Resistance in Clostridium difficile. Infect. Immun. 2019, 87, e00005-19. [Google Scholar] [CrossRef]
- Deshpande, A.; Olaitan, A.O.; Mckelvey, A.M.; Rutherford, J.T.; Hurdle, J.G. The Ferrous Iron Transporter FeoB1 Is Essential for Clostridioides difficile Toxin Production and Pathogenesis in Mice. bioRxiv 2022. [Google Scholar] [CrossRef]
- Arato, V.; Gasperini, G.; Giusti, F.; Ferlenghi, I.; Scarselli, M.; Leuzzi, R. Dual Role of the Colonization Factor CD2831 in Clostridium difficile Pathogenesis. Sci. Rep. 2019, 9, 5554. [Google Scholar] [CrossRef]
- Aiman, S.; Ali, F.; Zia, A.; Aslam, M.; Han, Z.; Farooq, Q.u.A.; Shams, S.; Khan, A.; Li, C. Core Genome Mediated Potential Vaccine Targets Prioritization against Clostridium difficile via Reverse Vaccinology—An Immuno-Informatics Approach. J. Biol. Res. Thessalon. 2022, 29. [Google Scholar] [CrossRef]
- Zhu, D.; Patabendige, H.M.L.W.; Tomlinson, B.R.; Wang, S.; Hussain, S.; Flores, D.; He, Y.; Shaw, L.N.; Sun, X. Cwl0971, a Novel Peptidoglycan Hydrolase, Plays Pleiotropic Roles in Clostridioides difficile R20291. Environ. Microbiol. 2021, 23, 5222–5238. [Google Scholar] [CrossRef]
- Noori, M.; Ghalavand, Z.; Azimirad, M.; Yadegar, A.; Eslami, G.; Krutova, M.; Brajerova, M.; Goudarzi, M.; Zali, M.R. Genetic Diversity and Phylogenetic Analysis of the Surface Layer Protein A Gene (SlpA) among Clostridioides difficile Clinical Isolates from Tehran, Iran. Anaerobe 2021, 70, 102403. [Google Scholar] [CrossRef]
- Karpiński, P.; Wultańska, D.; Piotrowski, M.; Brajerova, M.; Mikucka, A.; Pituch, H.; Krutova, M. Motility and the Genotype Diversity of the Flagellin Genes FliC and FliD among Clostridioides difficile Ribotypes. Anaerobe 2022, 73, 102476. [Google Scholar] [CrossRef]
- Jain, S.; Smyth, D.; O’Hagan, B.M.G.; Heap, J.T.; McMullan, G.; Minton, N.P.; Ternan, N.G. Inactivation of the DnaK Gene in Clostridium difficile 630 Δerm Yields a Temperature-Sensitive Phenotype and Increases Biofilm-Forming Ability. Sci. Rep. 2017, 7, 17522. [Google Scholar] [CrossRef]
- Hennequin, C.; Porcheray, F.; Waligora-Dupriet, A.; Collignon, A.; Barc, M.; Bourlioux, P.; Karjalainen, T. GroEL (Hsp60) of Clostridium difficile Is Involved in Cell Adherence. Microbiol. Read. Engl. 2001, 147, 87–96. [Google Scholar] [CrossRef]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Franco-Serrano, L.; Cedano, J.; Perez-Pons, J.A.; Mozo-Villarias, A.; Piñol, J.; Amela, I.; Querol, E. A Hypothesis Explaining Why so Many Pathogen Virulence Proteins Are Moonlighting Proteins. Pathog. Dis. 2018, 76, fty046. [Google Scholar] [CrossRef]
- Péchiné, S.; Hennequin, C.; Boursier, C.; Hoys, S.; Collignon, A. Immunization Using GroEL Decreases Clostridium difficile Intestinal Colonization. PLoS ONE 2013, 8, e81112. [Google Scholar] [CrossRef]
- Razim, A.; Pacyga, K.; Aptekorz, M.; Martirosian, G.; Szuba, A.; Pawlak-Adamska, E.; Brzychczy-Włoch, M.; Myc, A.; Gamian, A.; Górska, S. Epitopes Identified in GAPDH from Clostridium difficile Recognized as Common Antigens with Potential Autoimmunizing Properties. Sci. Rep. 2018, 8, 13946. [Google Scholar] [CrossRef]
- Savijoki, K.; Nyman, T.A.; Kainulainen, V.; Miettinen, I.; Siljamäki, P.; Fallarero, A.; Sandholm, J.; Satokari, R.; Varmanen, P. Growth Mode and Carbon Source Impact the Surfaceome Dynamics of Lactobacillus Rhamnosus GG. Front. Microbiol. 2019, 10, 1272. [Google Scholar] [CrossRef]
- Robinson, M.W.; Buchtmann, K.A.; Jenkins, C.; Tacchi, J.L.; Raymond, B.B.A.; To, J.; Roy Chowdhury, P.; Woolley, L.K.; Labbate, M.; Turnbull, L.; et al. MHJ_0125 Is an M42 Glutamyl Aminopeptidase That Moonlights as a Multifunctional Adhesin on the Surface of Mycoplasma hyopneumoniae. Open Biol. 2013, 3, 130017. [Google Scholar] [CrossRef] [PubMed]
- Pacyga, K.; Razim, A.; Martirosian, G.; Aptekorz, M.; Szuba, A.; Gamian, A.; Myc, A.; Górska, S. The Bioinformatic and In Vitro Studies of Clostridioides difficile Aminopeptidase M24 Revealed the Immunoreactive KKGIK Peptide. Cells 2020, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Vedantam, G. Surface-Displayed Glycopolymers of Clostridioides difficile. Curr. Opin. Microbiol. 2022, 66, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.D.; St. Michael, F.; Aubry, A.; Strong, P.C.R.; Hayes, A.C.; Logan, S.M. Comparison of Polysaccharide Glycoconjugates as Candidate Vaccines to Combat Clostridiodes (Clostridium) Difficile. Glycoconj. J. 2021, 38, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Broecker, F.; Wegner, E.; Seco, B.M.S.; Kaplonek, P.; Bräutigam, M.; Ensser, A.; Pfister, F.; Daniel, C.; Martin, C.E.; Mattner, J.; et al. Synthetic Oligosaccharide-Based Vaccines Protect Mice from Clostridioides difficile Infections. ACS Chem. Biol. 2019, 14, 2720–2728. [Google Scholar] [CrossRef] [PubMed]
- Senoh, M.; Iwaki, M.; Yamamoto, A.; Kato, H.; Fukuda, T.; Shibayama, K. Development of Vaccine for Clostridium difficile Infection Using Membrane Fraction of Nontoxigenic Clostridium difficile. Microb. Pathog. 2018, 123, 42–46. [Google Scholar] [CrossRef]
- Hughes, J.; Aston, C.; Kelly, M.L.; Griffin, R. Towards Development of a Non-Toxigenic Clostridioides difficile Oral Spore Vaccine against Toxigenic C. Difficile. Pharmaceutics 2022, 14, 1086. [Google Scholar] [CrossRef]
- Oliveira Júnior, C.A.; Silva, R.O.S.; Cruz, D.S.G.; Pires, I.H.; Guedes, R.M.C.; Faria Lobato, F.C. The Non-Toxigenic Strain of Clostridioides difficile Z31 Can Prevent Infection by C. Difficile in Experimental Model Piglets. Anaerobe 2019, 55, 24–28. [Google Scholar] [CrossRef]
- Hahn, U.K.; Boehm, R.; Beyer, W. DNA vaccination against anthrax in mice—Combination of anti-spore and anti-toxin components. Vaccine 2006, 24, 4569–4571. [Google Scholar] [CrossRef]
- Steinberg, H.D.; Snitkin, E.S. Homologous recombination in Clostridioides difficile mediates diversification of cell surface features and transport systems. Msphere 2020, 5, e00799-20. [Google Scholar] [CrossRef]
- Harboe, M.; Quayle, A.J. Heat shock proteins: Friend and foe? Clin. Exp. Immunol. 1991, 86, 2. [Google Scholar] [CrossRef]
- Gerding, D.N.; Meyer, T.; Lee, C.; Cohen, S.H.; Murthy, U.K.; Poirier, A.; Van Schooneveld, T.C.; Pardi, D.S.; Ramos, A.; Barron, M.A.; et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: A randomized clinical trial. JAMA 2015, 313, 1719–1727. [Google Scholar] [CrossRef]
- Brouwer, M.S.; Roberts, A.P.; Hussain, H.; Williams, R.J.; Allan, E.; Mullany, P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat. Commun. 2013, 4, 2601. [Google Scholar] [CrossRef]

| Vaccine Antigen Type | Advantages | Disadvantages |
|---|---|---|
| Surface protein of the spore | Possible destruction of spores prior to germination [18]; no need to use whole inactivated spores. | Limited efficacy of vaccination when used alone [62]; spore coat proteins masking surface antigens [16]; lack of knowledge on the interaction between spores and immune system. |
| Surface protein of the vegetative cell | No need to use whole inactivated bacteria; proteins accessible for antibodies; potent immune stimulators. | Variability in surface proteins among strains (SLPs), also due to homologous recombination [63]. |
| Intracellular proteins | Major immunogens, highly expressed during infection. | High amino acid sequence homology to similar proteins in other organisms; possible autoimmune responses [52,64]; epitope mapping might be needed [55]. |
| Surface glycopolymers | Immunogenic, conserved structures, easily accessible on the surface [57]; possible chemical synthesis. | Typically, a carrier for proper presentation is needed. |
| Non-toxigenic strains | Administration well-tolerated; strains compete for the same niche; stimulates mucosal responses [65]. | Acquisition of toxin genes is possible by horizontal gene transfer [66]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razim, A.; Górska, S.; Gamian, A. Non-Toxin-Based Clostridioides difficile Vaccination Approaches. Pathogens 2023, 12, 235. https://doi.org/10.3390/pathogens12020235
Razim A, Górska S, Gamian A. Non-Toxin-Based Clostridioides difficile Vaccination Approaches. Pathogens. 2023; 12(2):235. https://doi.org/10.3390/pathogens12020235
Chicago/Turabian StyleRazim, Agnieszka, Sabina Górska, and Andrzej Gamian. 2023. "Non-Toxin-Based Clostridioides difficile Vaccination Approaches" Pathogens 12, no. 2: 235. https://doi.org/10.3390/pathogens12020235
APA StyleRazim, A., Górska, S., & Gamian, A. (2023). Non-Toxin-Based Clostridioides difficile Vaccination Approaches. Pathogens, 12(2), 235. https://doi.org/10.3390/pathogens12020235