Epidemic Characteristics of HIV Drug Resistance in Hefei, Anhui Province
Abstract
:1. Introduction
2. Results
2.1. Characteristics of the Study Population
2.2. Drug Resistance Analysis
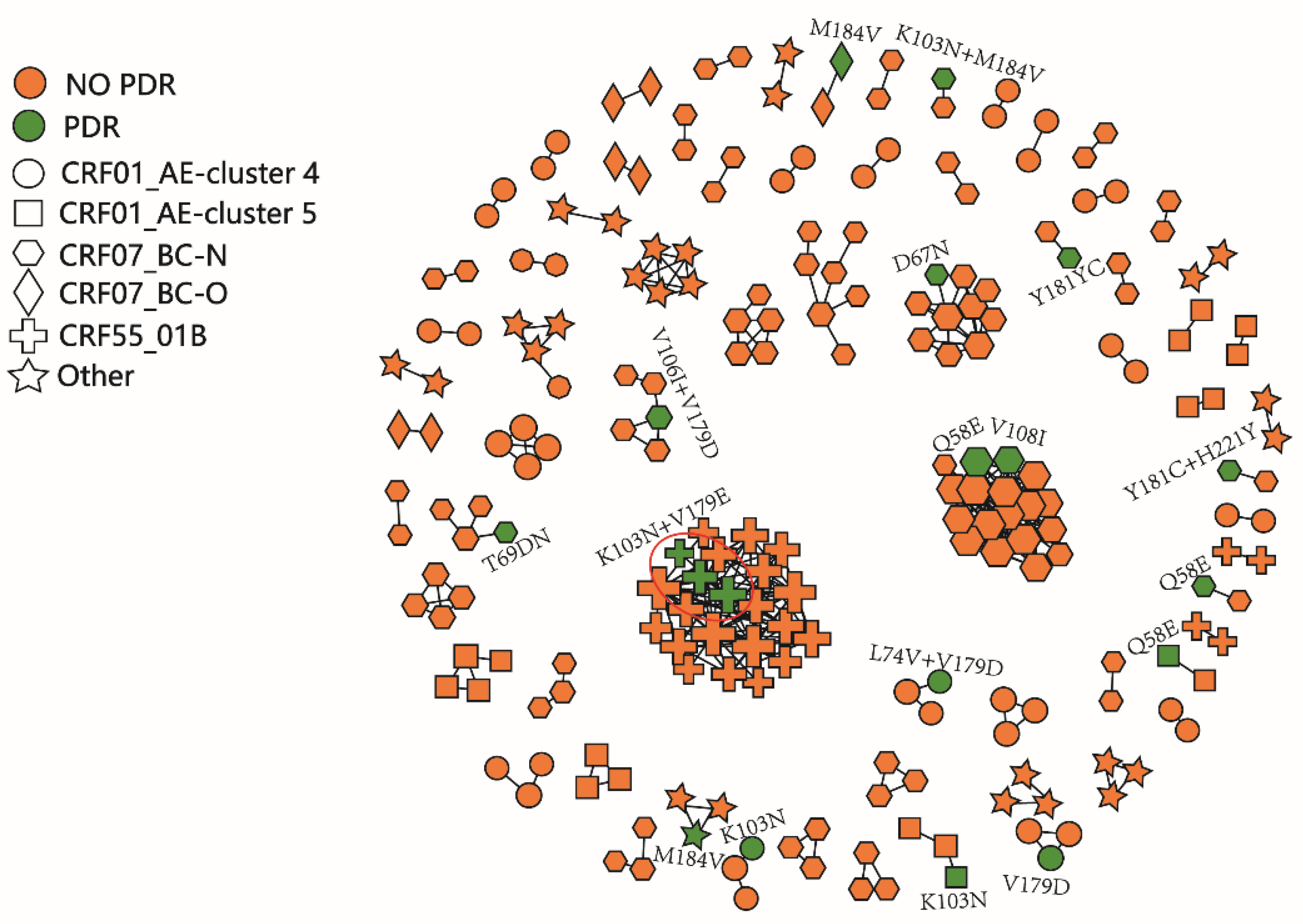
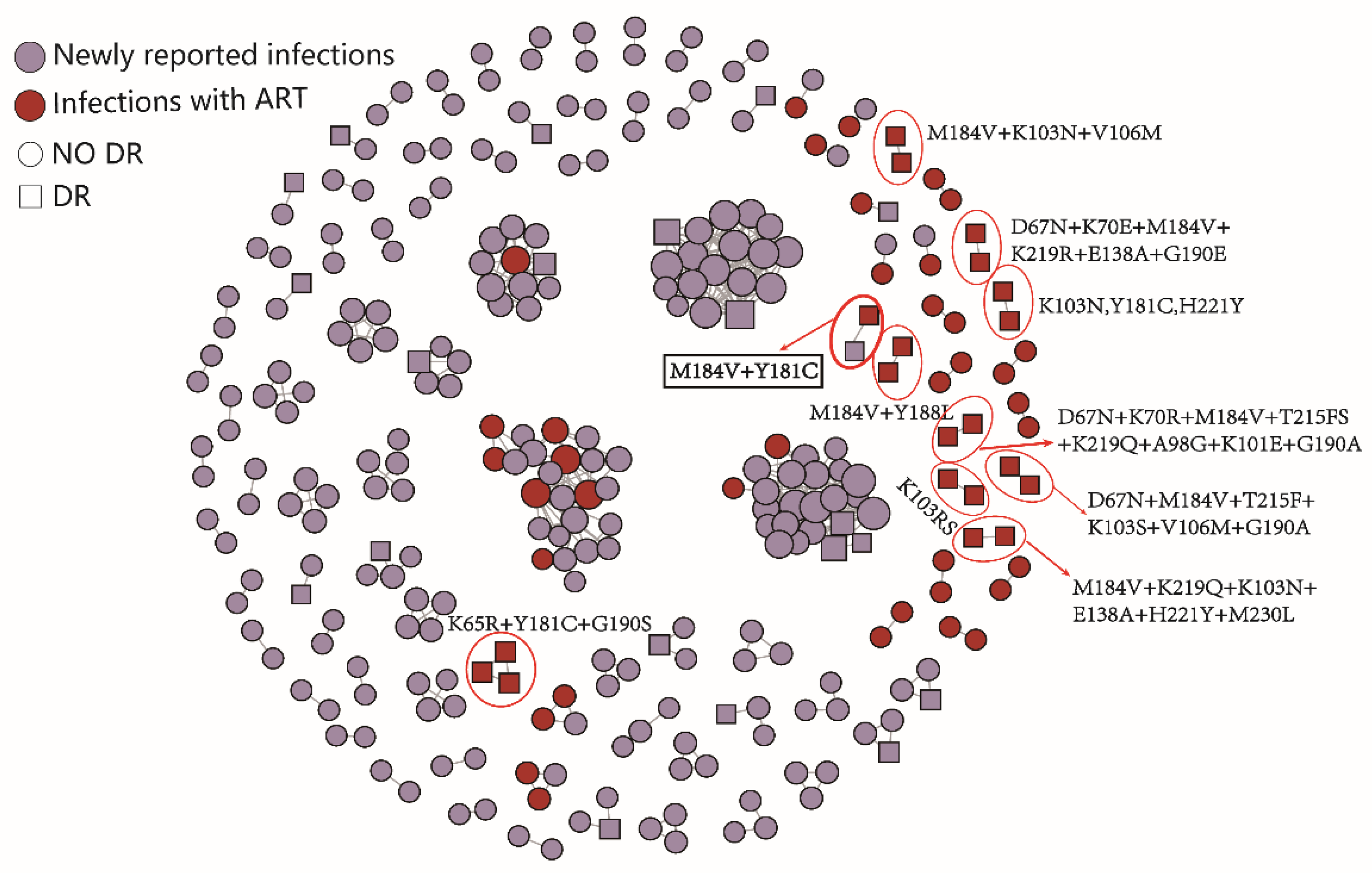
2.3. HIV Molecular Network Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population and Design
4.2. Laboratory Tests
4.3. Subtype and Drug Resistance Analysis
4.4. HIV Molecular Network Construction
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- UNAIDS. UNAIDS Data 2021. Available online: https://www.unaids.org/en/resources/documents/2021/2021_unaids_data (accessed on 29 November 2021).
- Wertheim, J.O.; Oster, A.M.; Johnson, J.A.; Switzer, W.M.; Saduvala, N.; Hernandez, A.L.; Hall, H.I.; Heneine, W. Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol. 2017, 3, vex008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Luo, L.; Song, C.; Li, J.; Chen, H.; Zhu, Q.; Lan, G.; Liang, S.; Shen, Z.; Cao, Z.; et al. Survey of pretreatment HIV drug resistance and the genetic transmission networks among HIV-positive individuals in southwestern China, 2014–2020. BMC Infect. Dis. 2021, 21, 1153. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Li, J.; Chen, H.; Tang, Z.; Pan, S.W.; Luo, L.; Zhu, Q.; Lan, G.; Liang, S.; Lu, H.; et al. Using longitudinal genetic-network study to understand HIV treatment-as-prevention. AIDS 2021, 35, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, J.; Zhang, Y.; Shen, Y.; Dai, S.; Wang, X.; Xing, H.; Lin, J.; Han, J.; Li, J.; et al. Genetic characterization of HIV-1 epidemic in Anhui Province, China. Virol. J. 2020, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Tao, H.-D.; Liu, J.; Fan, Y.-G.; Zhang, C.; Li, P.; Li, L.-J.; Huang, Q.; Zhao, W.; Ye, D.-Q. Prevalence and associated factors of HIV infection among men who have sex with men in Hefei, China, 2013–2014: A cross-sectional study. Int. J. STD AIDS 2015, 27, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Nelson, L.E.; Nyblade, L.; Torpey, K.; Logie, C.H.; Qian, H.-Z.; Manu, A.; Gyamerah, E.; Boakye, F.; Appiah, P.; Turner, D.; et al. Multi-level intersectional stigma reduction intervention to increase HIV testing among men who have sex with men in Ghana: Protocol for a cluster randomized controlled trial. PLoS ONE 2021, 16, e0259324. [Google Scholar] [CrossRef] [PubMed]
- Babel, R.A.; Wang, P.; Alessi, E.J.; Raymond, H.F.; Wei, C. Stigma, HIV Risk, and Access to HIV Prevention and Treatment Services Among Men Who have Sex with Men (MSM) in the United States: A Scoping Review. AIDS Behav. 2021, 25, 3574–3604. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Zheng, S.; Hao, J.; Ruan, Y.; Liao, L.; Shao, Y.; Feng, Y.; Xing, H. The prevalence of CRF55_01B among HIV-1 strain and its connection with traffic development in China. Emerg. Microbes Infect. 2021, 10, 256–265. [Google Scholar] [CrossRef]
- Qian, Y.; Lei, Y.; Shen, Y.; Wang, X.; Li, Y.; Yao, H. Study of HIV-1 subtype diversity and drug resistance among antiretroviral treatment-naive patients in Hefei district. Anhui J. Prev. Med. 2013, 19, 333–335. [Google Scholar]
- Hao, J.; Zheng, S.; Gan, M.; Dong, A.; Kang, R.; Li, M.; Zhao, S.; Hu, J.; Song, C.; Liao, L.; et al. Changing Proportions of HIV-1 Subtypes and Transmitted Drug Resistance Among Newly Diagnosed HIV/AIDS Individuals—China, 2015 and 2018. China CDC Wkly. 2021, 3, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhu, Q.; Xing, H.; Chen, H.; Jin, X.; Dong, L.; Dai, J.; Yang, M.; Yang, C.; Jia, M.; et al. The characteristics of pretreatment HIV-1 drug resistance in western Yunnan, China. Epidemiol. Infect. 2020, 148, e102. [Google Scholar] [CrossRef]
- Zeng, R.; Ren, M.D.; Gong, X.; Wei, M.; Gao, L.; Yu, A.; Zhang, D.; Mi, Y.; Ma, P. HIV-1 Genetic Diversity and High Prevalence of Pretreatment Drug Resistance in Tianjin, China. AIDS Res. Hum. Retroviruses 2020, 36, 852–861. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Wang, X.; Han, J.; Jia, L.; Li, T.; Li, J.; Li, L. Natural presence of V179E and rising prevalence of E138G in HIV-1 reverse transcriptase in CRF55_01B viruses. Infect. Genet. Evol. 2019, 77, 104098. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Liu, K.; Liu, H.; Hu, Y.; Zhang, Z.; Qin, J.; Xu, Q.; Peng, K.; Jin, X.; Wang, J.-H.; et al. Trend of HIV-1 drug resistance in China: A systematic review and meta-analysis of data accumulated over 17 years (2001–2017). eClinicalMedicine 2020, 18, 100238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winand, R.; Theys, K.; Eusébio, M.; Aerts, J.; Camacho, R.J.; Gomes, P.; Suchard, M.A.; Vandamme, A.-M.; Abecasis, A.B. Assessing transmissibility of HIV-1 drug resistance mutations from treated and from drug-naive individuals. AIDS 2015, 29, 2045–2052. [Google Scholar] [CrossRef] [Green Version]
- LIAO, L.; Xing, H.; Shao, Y.; Shang, H.; Li, J.; Zhong, P.; Kang, L.; Cheng, H.; Si, X.; Jiang, S.; et al. The Prevalence of Transmitted Antiretroviral Drug Resistance in Treatment-Naive HIV-Infected Individuals in China: China Meets New AIDS Challenges. J. Acquir. Immune Defic. Syndr. 1999, 2010, 53. [Google Scholar] [CrossRef]
- World Health Organization. Surveillance of HIV Drug Resistance in Populations Initiating Antiretroviral Therapy (Pre-Treatment HIV Drug Resistance): Concept Note. Available online: https://apps.who.int/iris/handle/10665/112802 (accessed on 16 May 2014).
- Kosakovsky Pond, S.L.K.; Weaver, S.; Leigh Brown, A.J.; Wertheim, J.O. HIV-TRACE (TRAnsmission Cluster Engine): A Tool for Large Scale Molecular Epidemiology of HIV-1 and Other Rapidly Evolving Pathogens. Mol. Biol. Evol. 2018, 35, 1812–1819. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- PS18-1802 Attachments | Announcements | Funding | HIV/AIDS | CDC. 2020. Available online: https://www.cdc.gov/hiv/funding/announcements/ps18-1802/attachments.html (accessed on 1 January 2020).
| Variable | Number | % |
|---|---|---|
| Total | 816 | 100.0 |
| Age (years) | ||
| 18–29 | 318 | 39.0 |
| 30–39 | 147 | 18.0 |
| 40–49 | 100 | 12.3 |
| ≥50 | 121 | 14.8 |
| Unknown | 130 | 15.9 |
| Sex | ||
| Male | 627 | 76.8 |
| Female | 59 | 7.2 |
| Unknown | 130 | 15.9 |
| Ethnicity | ||
| Han | 680 | 83.3 |
| Other | 6 | 0.7 |
| Unknown | 130 | 15.9 |
| Education | ||
| Illiterate | 81 | 9.9 |
| Primary or junior high school | 143 | 17.5 |
| Senior high school or higher | 462 | 56.6 |
| Unknown | 130 | 15.9 |
| Marital status | ||
| Single | 379 | 46.4 |
| Married | 210 | 25.7 |
| Divorced/widowed | 97 | 11.9 |
| Unknown | 130 | 15.9 |
| Route of infection * | ||
| MSM | 505 | 61.9 |
| HET | 178 | 21.8 |
| IDU | 3 | 0.4 |
| Unknown | 130 | 15.9 |
| Subtype | ||
| CRF55_01B | 51 | 6.3 |
| CRF01_AE-cluster 4 | 199 | 24.4 |
| CRF01_AE-cluster 5 | 66 | 8.1 |
| CRF07_BC-N | 285 | 34.9 |
| Other | 215 | 26.3 |
| CD4 count before ART (cells/µL) | ||
| <200 | 214 | 26.2 |
| 200–350 | 196 | 24.0 |
| 350–500 | 140 | 17.2 |
| >500 | 136 | 16.7 |
| Unknown | 130 | 15.9 |
| Sampling time | ||
| 2017 | 66 | 8.1 |
| 2018 | 162 | 19.9 |
| 2019 | 215 | 26.3 |
| 2020 | 373 | 45.7 |
| Variable | Number | % |
|---|---|---|
| Total | 127 | 100.0 |
| Age (years) | ||
| 18–29 | 24 | 18.9 |
| 30–39 | 53 | 41.7 |
| 40–49 | 15 | 11.8 |
| ≥50 | 33 | 26.0 |
| Unknown | 2 | 1.6 |
| Sex | ||
| Male | 109 | 85.8 |
| Female | 16 | 12.6 |
| Unknown | 2 | 1.6 |
| Ethnicity | ||
| Han | 126 | 99.2 |
| Other | 1 | 0.8 |
| Marital status | ||
| Single | 75 | 59.1 |
| Married | 35 | 27.6 |
| Divorced/widowed | 15 | 11.7 |
| Unknown | 2 | 1.6 |
| Route of infection * | ||
| MSM | 72 | 56.7 |
| HET | 49 | 38.6 |
| IDU | 1 | 0.8 |
| Unknown | 5 | 3.9 |
| VL (copies/mL) | ||
| 1000–10,000 | 58 | 45.7 |
| 10,000–50,000 | 41 | 32.3 |
| >50,000 | 28 | 22.0 |
| ART regimen * | ||
| EFV+3TC+TDF | 47 | 37.0 |
| EFV+3TC+AZT | 16 | 12.6 |
| LPV/r+3TC+TDF | 24 | 18.9 |
| LPV/r+3TC+AZT | 19 | 15.0 |
| NVP+3TC+AZT | 12 | 9.4 |
| Other | 9 | 7.1 |
| Sampling time | ||
| 2018 | 59 | 46.5 |
| 2019 | 68 | 53.5 |
| Antiretroviral Drug | Number | % | HIV Drug Resistance Mutations (n, %) |
|---|---|---|---|
| Total | 52 | 6.4 | |
| PIs | 16 | 2.0 | |
| ATV | 1 | 0.1 | D30N (1, 0.1), K20T (2, 0.2), L10F (1, 0.1), L90M (1, 0.1), M46I (7, 0.9), Q58E (7, 0.9) |
| FPV | 2 | 0.2 | |
| IDV | 1 | 0.1 | |
| LPV/r | 1 | 0.1 | |
| NFV | 9 | 1.1 | |
| SQV | 1 | 0.1 | |
| TPV | 7 | 0.9 | |
| NRTIs | 10 | 1.2 | |
| ABC | 8 | 1.0 | A62V (2, 0.2), D67N (2, 0.2), K219E/R (3, 0.4), K65R (1, 0.1), K70T (1, 0.1), L74L/V (2, 0.2), M184V (4, 0.5), T215I (1, 0.1), T69DN (1, 0.1) |
| AZT | 2 | 0.2 | |
| D4T | 4 | 0.5 | |
| DDI | 6 | 0.7 | |
| FTC | 5 | 0.6 | |
| 3TC | 5 | 0.6 | |
| TDF | 2 | 0.2 | |
| NNRTIs | 31 | 3.8 | |
| DOR | 8 | 1.0 | V179E/D/T (101, 12.4), K103N (11, 1.3), V106I/M (11, 1.3), V108I (4, 0.5), E138G/A/Q (5, 0.6), Y181H/C (3, 0.3), G190S (1, 0.1), K101E (1, 0.1), P225H (1, 0.1), F227L (1, 0.1), H221Y (1, 0.1) |
| EFV | 20 | 2.5 | |
| ETR | 8 | 1.0 | |
| NVP | 25 | 3.1 | |
| RPV | 13 | 1.6 |
| HIV Drug Resistance Mutations | Number (%) | Level of Drug Resistance * | NNRTIs |
|---|---|---|---|
| Total | 101 | ||
| V179D/E/T | 90 (89.1) | - | - |
| V179D | 1 (1.0) | I | EFV, NVP |
| 1 (1.0) | L | RPV | |
| E138A, V179E | 1 (1.0) | L | ETR, RPV |
| E138G, V179E | 3 (3.0) | L | EFV, ETR, NVP, RPV |
| V106I, V179D | 1 (1.0) | L | ETR, NVP, RPV |
| K103N, V179E | 4 (4.0) | H | EFV, NVP |
| Variable | Number | Drug Resistance, n (%) | OR * (95% CI) | p | AOR * (95% CI) | p |
|---|---|---|---|---|---|---|
| Total | 816 | 52 (6.4) | ||||
| Age (years) | ||||||
| 18–29 | 318 | 17 (5.3) | 1.00 | |||
| 30–39 | 147 | 9 (6.1) | 1.16 (0.50–0.66) | 0.735 | ||
| 40–49 | 100 | 6 (6.0) | 1.13 (0.43–2.95) | 0.803 | ||
| ≥50 | 121 | 12 (9.9) | 1.95 (0.90–4.21) | 0.090 | ||
| Unknown | 130 | 8 (6.2) | 1.16 (0.49–2.76) | 0.736 | ||
| Sex | ||||||
| Male | 627 | 39 (6.2) | 1.00 | |||
| Female | 59 | 5 (8.5) | 1.40 (0.53–3.69) | 0.501 | ||
| Unknown | 130 | 8 (6.2) | 0.99 (0.45–2.17) | 0.977 | ||
| Ethnicity | ||||||
| Han | 680 | 44 (6.5) | 1.00 | |||
| Other | 6 | 0 (0.0) | - | - | ||
| Unknown | 130 | 8 (6.2) | 0.95 (0.44–2.06) | 0.893 | ||
| Education | ||||||
| Illiterate | 81 | 4 (4.9) | 1.00 | |||
| Primary or junior high school | 143 | 15 (10.5) | 2.26 (0.72–7.04) | 0.161 | ||
| Senior high school or higher | 462 | 25 (5.4) | 1.10 (0.37–3.25) | 0.861 | ||
| Unknown | 130 | 8 (6.2) | 1.26 (0.37–4.33) | 0.711 | ||
| Marital status | ||||||
| Single | 379 | 20 (5.3) | 1.00 | |||
| Married | 210 | 14 (6.7) | 1.28 (0.63–2.59) | 0.490 | ||
| Divorced/widowed | 97 | 10 (10.3) | 2.06 (0.93–4.57) | 0.074 | ||
| Unknown | 130 | 8 (6.2) | 1.18 (0.51–2.74) | 0.705 | ||
| Route of infection * | ||||||
| MSM | 505 | 29 (5.7) | 1.00 | |||
| HET | 178 | 15 (8.4) | 1.51 (0.79–2.89) | 0.212 | ||
| IDU | 3 | 0 (0.0) | - | - | ||
| Unknown | 130 | 8 (6.1) | 1.08 (0.48–2.41) | 0.858 | ||
| Subtype | ||||||
| CRF55_01B | 51 | 6 (11.8) | 1.00 | 1.00 | ||
| CRF01_AE-cluster 4 | 199 | 8 (4.0) | 0.31 (0.10–0.95) | 0.040 | 0.26 (0.08–0.81) | 0.021 |
| CRF01_AE-cluster 5 | 66 | 3 (4.5) | 0.36 (0.09–1.50) | 0.160 | 0.33 (0.07–1.42) | 0.036 |
| CRF07_BC-N | 285 | 15 (5.2) | 0.42 (0.15–1.13) | 0.086 | 0.35 (0.12–0.98) | 0.045 |
| Other | 215 | 20 (9.3) | 0.77 (0.29–2.03) | 0.595 | 0.60 (0.21–1.70) | 0.334 |
| CD4 count before ART (cells/µL) | ||||||
| <200 | 214 | 15 (7.0) | 1.00 | |||
| 200–350 | 196 | 13 (6.6) | 0.94 (0.44–2.03) | 0.880 | ||
| 350–500 | 140 | 7 (5.0) | 0.70 (0.28–1.76) | 0.446 | ||
| >500 | 136 | 9 (6.6) | 0.94 (0.40–2.21) | 0.888 | ||
| Unknown | 130 | 8 (6.2) | 0.87 (0.36–2.11) | 0.758 | ||
| Sampling time | ||||||
| 2017 | 66 | 4 (6.1) | 1.00 | |||
| 2018 | 162 | 8 (4.9) | 0.81 (0.23–2.77) | 0.731 | ||
| 2019 | 215 | 10 (4.7) | 0.76 (0.23–2.50) | 0.646 | ||
| 2020 | 373 | 30 (8.0) | 1.36 (0.46–3.98) | 0.580 |
| Antiretroviral Drug | Number | % | HIV Drug Resistance Mutations (n, %) |
|---|---|---|---|
| Total | 49 | 38.6 | |
| PIs | 3 | 2.4 | |
| ATV | 2 | 1.6 | M46I (2, 1.6), I54V (2, 1.6), L76V (1, 0.8), V82A (2, 1.6), L10F (2, 1.6) |
| DRV | 1 | 0.8 | |
| FPV | 2 | 1.6 | |
| IDV | 2 | 1.6 | |
| LPV/r | 2 | 1.6 | |
| NFV | 2 | 1.6 | |
| SQV | 2 | 1.6 | |
| TPV | 3 | 2.4 | |
| NRTIs | 37 | 29.1 | |
| ABC | 36 | 28.3 | D67N/T (9, 7.1), K70R/E (8, 6.3), M184V (31, 24.4), T215F/Y (10, 7.9), K219Q/E/R (7, 5.5), K65R (9, 7.1), K70Q (2, 1.6), Y115F (7, 5.5), A62V (3, 2.4), L74I (3, 2.4), M41L (5, 3.9), V75M/I (3, 2.4), E44A (1, 0.8), L210L/W (1, 0.8) |
| AZT | 11 | 8.7 | |
| D4T | 25 | 19.7 | |
| DDI | 27 | 21.3 | |
| FTC | 36 | 28.3 | |
| 3TC | 36 | 28.3 | |
| TDF | 22 | 17.3 | |
| NNRTIs | 46 | 36.2 | |
| DOR | 36 | 28.3 | V179D/E (9, 7.1), Y181C (14, 11.0), K101E (8, 6.3), G190S/A/E (13, 10.2), K103N/S (20, 15.7), E138A (7, 5.5), H221Y (6, 4.7), M230L (3, 2.4), V106M/I (13, 10.2), F227L (2, 1.6), A98G (3, 2.4), P225H (3, 2.4), V108I (2, 1.6), Y188L (2, 1.6), L234I (2, 1.6) |
| EFV | 45 | 35.4 | |
| ETR | 24 | 18.9 | |
| NVP | 45 | 35.4 | |
| RPV | 31 | 24.4 |
| VL * (Copies/mL) | Total | Link to Newly Reported Infections | Z | p |
|---|---|---|---|---|
| 1000–10,000 | 58 | 13 (21.7) | −2.98 | 0.001 |
| 10,000–50,000 | 41 | 21 (35.0) | ||
| >50,000 | 28 | 26 (43.3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Wu, J.; Hao, J.; Wang, D.; Hu, Z.; Liu, L.; Song, C.; Hu, J.; Lei, Y.; Wang, H.; et al. Epidemic Characteristics of HIV Drug Resistance in Hefei, Anhui Province. Pathogens 2022, 11, 866. https://doi.org/10.3390/pathogens11080866
Zheng S, Wu J, Hao J, Wang D, Hu Z, Liu L, Song C, Hu J, Lei Y, Wang H, et al. Epidemic Characteristics of HIV Drug Resistance in Hefei, Anhui Province. Pathogens. 2022; 11(8):866. https://doi.org/10.3390/pathogens11080866
Chicago/Turabian StyleZheng, Shan, Jianjun Wu, Jingjing Hao, Dong Wang, Zhongwang Hu, Lei Liu, Chang Song, Jing Hu, Yanhua Lei, Hai Wang, and et al. 2022. "Epidemic Characteristics of HIV Drug Resistance in Hefei, Anhui Province" Pathogens 11, no. 8: 866. https://doi.org/10.3390/pathogens11080866
APA StyleZheng, S., Wu, J., Hao, J., Wang, D., Hu, Z., Liu, L., Song, C., Hu, J., Lei, Y., Wang, H., Liao, L., Feng, Y., Shao, Y., Ruan, Y., & Xing, H. (2022). Epidemic Characteristics of HIV Drug Resistance in Hefei, Anhui Province. Pathogens, 11(8), 866. https://doi.org/10.3390/pathogens11080866





