Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense)
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of the RIL Population
2.2. Experimental Designs, Inoculation Methods, and Assessment of FOV 4 Resistance
2.3. Analysis of Variance
2.4. Linkage Mapping and QTL Analysis
3. Results and Discussion
3.1. Analysis of Variance of FOV4 Resistance Based on DSR
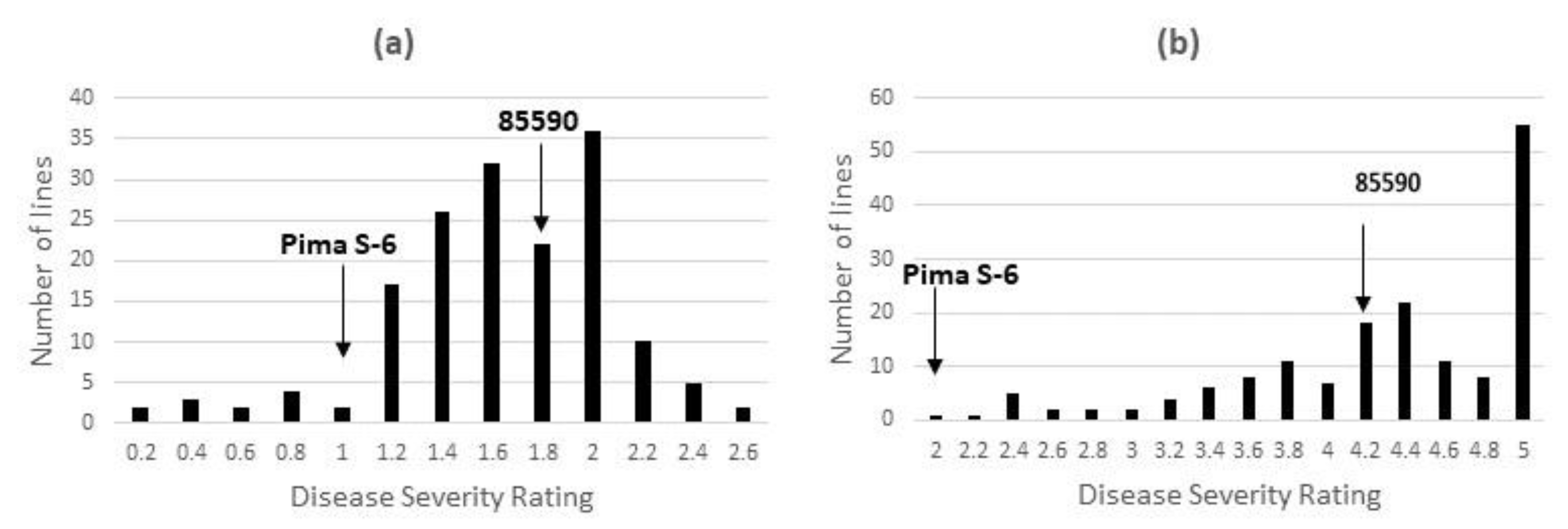
3.2. Performance of the RILs for FOV4 Resistance
3.3. QTL Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.F.; Percy, R.G.; McCarty, J.C. Introgression genetics and breeding between Upland and Pima cotton: A review. Euphytica 2014, 198, 1–12. [Google Scholar] [CrossRef]
- Hillocks, R.J. Fusarium wilt. In Cotton Diseases; CAB International: Wallingford, UK, 1992; pp. 127–160. [Google Scholar]
- Zhang, J.F.; Sanogo, S.; Ma, Z.Y.; Qu, Y.Y. Breeding, genetics, and quantitative trait locus mapping for Fusarium wilt resistance in cotton. Crop Sci. 2015, 55, 2435–2453. [Google Scholar] [CrossRef]
- Sanogo, S.; Zhang, J.F. Resistance sources, resistance screening techniques and disease management for Fusarium wilt in cotton. Euphytica 2016, 207, 255–271. [Google Scholar] [CrossRef]
- Kim, Y.; Hutmacher, R.B.; Davis, R.M. Characterization of California isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Dis. 2005, 89, 366–372. [Google Scholar] [CrossRef]
- Halpern, H.C.; Bell, A.A.; Wagner, T.A.; Liu, J.G.; Nichols, R.L.; Woodward, J.E.; Brewer, M.T. First report of Fusarium wilt of cotton caused by Fusarium oxysporum f. sp. vasinfectum race 4 in Texas, U.S.A. Plant Dis. 2018, 102, 446. [Google Scholar] [CrossRef]
- Zhu, Y.; Lujan, P.A.; Wedegaertner, T.; Nichols, R.; Abdelraheem, A.; Zhang, J.F.; Sanogo, S. First report of Fusarium oxysporum f. sp. vasinfectum race 4 causing Fusarium wilt of cotton in New Mexico, USA. Plant Dis. 2020, 104, 588. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Lujan, P.; Idowu, O.; Sullivan, P.; Nichols, R.; Wedegaertner, T.; Zhang, J.F. Detection and characterization of Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 in New Mexico, USA. Plant Dis. 2021, 105, 3353–3367. [Google Scholar] [CrossRef]
- Ulloa, M.; Percy, R.; Zhang, J.F.; Hutmacher, R.B.; Wright, S.D.; Davis, R.M. Registration of four pima cotton germplasm lines having good levels of Fusarium wilt race 4 resistance with moderate yields and good Fibers. J. Plant Reg. 2009, 3, 198–202. [Google Scholar] [CrossRef]
- Ulloa, M.; Hutmacher, R.B.; Roberts, P.A.; Wright, S.D.; Nichols, R.L.; Davis, R.M. Inheritance and QTL mapping of Fusarium wilt race 4 resistance in cotton. Theor. Appl. Genet. 2013, 126, 1405–1418. [Google Scholar] [CrossRef]
- Ulloa, M.; Hutmacher, R.B.; Percy, R.; Wright, S.D.; Burke, J. Registration of five Pima cotton germplasm lines (Pima SJ-FR05–Pima SJ-FR09) with improved resistance to Fusarium wilt race 4 and good lint yield and fiber quality. J. Plant Reg. 2016, 10, 154–158. [Google Scholar] [CrossRef]
- Zhang, J.; Abdelraheem, A.; Zhu, Y.; Wheeler, T.A.; Dever, J.K.; Elkins-Arce, H.; Nichols, R.; Wedegaertner, T. Pedigree selection under field conditions within Acala 1517-08 and its glandless derivatives for development of cotton resistant to Fusarium wilt caused by Fusarium oxysporum f. sp. vasinfectum race 4. Euphytica 2020, 216, 155. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Cooke, P.; Wheeler, T.; Dever, J.; Wedegaertner, T.; Hake, K.; Zhang, J. Comparative analysis of infection process in Pima cotton differing in resistance to Fusarium wilt caused by Fusarium oxysporum f. sp. vasinfectum race 4. Phytopathology 2022, 112, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Abdelraheem, A.; Liu, F.; Song, M.; Zhang, J.F. A meta-analysis of quantitative trait loci for abiotic and biotic stress resistance in tetraploid cotton. Mol. Genet. Genom. 2017, 292, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Abdelraheem, A.; Hanan, E.; Zhu, Y.; Kuraparthy, V.; Hinze, L.; Stelly, D.M.; Wedegaertner, T.; Zhang, J.F. Quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US Upland cotton. Theor. Appl. Genet. 2020, 133, 563–577. [Google Scholar] [CrossRef]
- Hulse-Kemp, A.M.; Lemm, J.; Lieske, J.; Ashrafi, H.; Buyyarapu, R.; Fang, D.D.; Frelichowski, J.; Giband, M.; Hague, S.; Hinze, L.L.; et al. Development of a 63K SNP array for cotton and high-density mapping of intra- and inter-specific populations of Gossypium spp. G3 Genes Genomes Genet. 2015, 5, 1187–1209. [Google Scholar]
- Wang, C.; Uolla, M.; Duong, T.; Robert, P.A. Quantitative trait loci mapping of multiple independent loci for resistance to Fusarium oxysporum f. sp. vasinfectum races 1 and 4 in an interspecific cotton population. Phytopathology 2018, 108, 759–767. [Google Scholar] [CrossRef]
- Hao, J.J.; Yang, M.E.; Davis, R.M. Effect of soil inoculum density of Fusarium oxysporum f. sp. vasinfectum race 4 on disease development in cotton. Plant Dis. 2009, 93, 1324–1328. [Google Scholar] [CrossRef]
- Zhang, J.; Abdelraheem, A.; Zhu, Y.; Wheeler, T.A.; Dever, J.K.; Nichols, R.; Wedegaertner, T. Importance of temperature in evaluating cotton for resistance to Fusarium wilt caused by Fusarium oxysporum f. sp. vasinfectum race 4. Crop Sci. 2021, 61, 1783–1796. [Google Scholar] [CrossRef]
- Zhang, J.; Abdelraheem, A.; Zhu, Y.; Wheeler, T.A.; Dever, J.K.; Frelichowski, J.; Love, J.; Ulloa, M.; Jenkins, J.N.; McCarty, J.C.; et al. Assessing genetic variation for Fusarium wilt race 4 resistance in tetraploid cotton by screening over three thousand germplasm lines under greenhouse or controlled conditions. Euphytica 2020, 216, 108. [Google Scholar] [CrossRef]
- Feaster, C.V.; Turcotte, E.L. Registration of Pima S-6 Cotton. Crop Sci. 1984, 24, 382. [Google Scholar] [CrossRef]
- Percy, R.G. Registration of extra-long staple cotton germplasm, 89590 and 8810. Crop Sci. 1998, 38, 1407. [Google Scholar] [CrossRef]
- Abdelraheem, A.; D Fang, D.D.; Dever, J.; Zhang, J.F. QTL analysis of agronomic, fiber quality, and abiotic stress tolerance traits in a recombinant inbred population of Pima cotton (Gossypium barbadense L.). Crop Sci. 2020, 40, 1823–1843. [Google Scholar] [CrossRef]
- Zhang, J.F.; Sanogo, S.; Flynn, R.; Baral, J.B.; Bajaj, S.; Hughs, S.E.; Percy, R.G. Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 2012, 187, 147–160. [Google Scholar] [CrossRef]
- Zhang, J.F.; Stewart, J.M. Economical and rapid method for extracting cotton genomic DNA. J. Cotton Sci. 2020, 4, 193–201. [Google Scholar]
- Van Ooijen, J.W. JoinMap 4: Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics. 2007, 175, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Qin, L.; Hu, B.; Guo, W.; Zhang, T.Z. Identification and molecular mapping of a Fusarium wilt resistant gene in upland cotton. Theor. Appl. Genet. 2009, 119, 733–739. [Google Scholar] [CrossRef]

| Sources of Variation | df | Mean Squares |
|---|---|---|
| Genotype | 162 | 0.85 ** |
| Test | 1 | 0.21 ** |
| Genotype × Test | 162 | 0.52 ** |
| Error | 324 | 0.38 |
| CV (%) | 31.2 | |
| h2 | 0.65 |
| Test | Mean DSR | Mean DI (%) | Mean Mortality (%) |
|---|---|---|---|
| 1 | 1.65 a | 82.18 a | 0.20 a |
| 2 | 4.67 b | 93.81 a | 83.65 b |
| Line | Test 1 | Test 2 | Line | Test 1 | Test 2 |
|---|---|---|---|---|---|
| 10 most susceptible lines | 10 most susceptible lines | ||||
| NMPRIL21 | 0.25 | 1.58 | NMPRIL6 | 2.63 | 3.95 |
| NMPRIL20 | 0.38 | 2.06 | NMRIL15 | 2.62 | 5.00 |
| NMPRIL24 | 0.44 | 2.22 | NMRIL140 | 2.50 | 4.67 |
| NMPRIL135 | 0.46 | 2.34 | NMRIL72 | 2.46 | 5.00 |
| NMPRIL12 | 0.48 | 2.41 | NMRIL67 | 2.41 | 5.00 |
| NMPRIL7 | 0.50 | 2.05 | NMRIL91 | 2.33 | 5.00 |
| NMPRIL55 | 0.60 | 2.04 | NMRIL82 | 2.29 | 4.86 |
| NMPRIL65 | 0.67 | 2.01 | NMRIL75 | 2.27 | 5.00 |
| NMPRIL71 | 0.72 | 1.98 | NMRIL101 | 2.25 | 4.83 |
| NMPRIL100 | 0.78 | 2.01 | NMRIL110 | 2.25 | 4.35 |
| LSD (0.05) | 1.12 | 1.18 | |||
| QTL Name | Chr. | Left Marker | Right Marker | Physical Location (Mb) | LOD | PVE (%) | Direction |
|---|---|---|---|---|---|---|---|
| qFOV.c14-1 | c14 | BNL3932-300 | BNL1897B-300 | 6.25–69.61 | 22.54 | 16.54 | Pima S-6 |
| qFOV.c17-1 | c17 | BNL3408a-200 | CIR347-280 | 48.63–48.81 | 21.54 | 29.01 | Pima S-6 |
| qFOV.c17-2 | c17 | BNL3408a-200 | CIR347-280 | 48.63–48.81 | 21.54 | 29.01 | Pima S-6 |
| qFOV.c19-1 | c19 | BNL1611-430 | DPL0071 | 10.56–21.19 | 11.58 | 18.57 | Pima S-6 |
| qFOV.c21-1 | c21 | BNL1053a-230 | BNL3997-430 | 17.52–26.24 | 9.98 | 15.00 | Pima S-6 |
| qFOV.c24-1 | c24 | MUSS260a-300 | BNL2961b-400 | 56.77–56.97 | 10.25 | 19.87 | Pima S-6 |
| qFOV.c24-2 | c24 | BNL2961b-400 | BNL1521-400 | 56.77–56.97 | 11.89 | 19.87 | Pima S-6 |
| qFOV.c25-1 | c25 | MUSS122 | JESPR056 | 52.85 | 13.54 | 21.84 | Pima S-6 |
| qFOV.c25-2 | c25 | MUSS122 | JESPR056 | 52.85 | 13.54 | 21.84 | Pima S-6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelraheem, A.; Zhu, Y.; Zhang, J. Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense). Pathogens 2022, 11, 1143. https://doi.org/10.3390/pathogens11101143
Abdelraheem A, Zhu Y, Zhang J. Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense). Pathogens. 2022; 11(10):1143. https://doi.org/10.3390/pathogens11101143
Chicago/Turabian StyleAbdelraheem, Abdelraheem, Yi Zhu, and Jinfa Zhang. 2022. "Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense)" Pathogens 11, no. 10: 1143. https://doi.org/10.3390/pathogens11101143
APA StyleAbdelraheem, A., Zhu, Y., & Zhang, J. (2022). Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense). Pathogens, 11(10), 1143. https://doi.org/10.3390/pathogens11101143






