Abstract
Fusarium graminearum causing head scab (HS) or head blight (HB) disease in wheat is one of the nasty fungi reported to cause significant grain quality and yield loss. Biological control using endophytic bacteria has emerged as a prospective option for containing fungal diseases in an environmentally benevolent, durable, and sustainable manner. In this regard, 112 endophytic bacilli were isolated from the anthesis stage (Zadok’s growth stage 65) from five different wheat genotypes with an aim to identify prospective antagonistic strains against F. graminearum. The molecular identity of the strains was confirmed by matching 16S rRNA sequences of bacterial strains with the gene sequences of type strains available in the National Center for Biotechnology Information database and reported 38 different species of Bacillus in all the five wheat cultivars. Further, it has been observed that only fourteen strains (B. clarus NOK09, B. mojavensis NOK16, B. subtilis NOK33, B. rugosus NOK47, B. mojavensis NOK52, B. clarus NOK59, B. coahuilensis NOK72, B. cabrialesii NOK78, B. cabrialesii NOK82, B. rugosus NOK85, B. amyloliquefaciens NOK89, B. australimaris NOK95, B. pumilus NOK103, and B. amyloliquefaciens NOK109) displayed in-vitro antagonistic effect against Fusarium graminearum fungus. Furthermore, the three endophytic Bacillus strains showing the strongest antagonistic effect (>70% of growth inhibition of fungal mycelium) under in-vitro antagonistic assay were selected for field experiments. In a two-year consecutive field study, a combination of three strains (B. clarus NOK09 + B. subtilis NOK33 + B. amyloliquefaciens NOK109) displayed a remarkable reduction in HS disease index by 81.47% and 77.85%, respectively. Polymerase chain reaction assay detected three genes (ituD, bmyC, and srfA) involved in antibiotic biosynthesis pathways. Additional attributes such as potassium solubilization, siderophore release, and hydrolytic enzyme (protease, lipase, amylase, chitinase, and pectinase) synthesis have been observed in these strains. Overall, the present study was successful in profiling endophytic bacilli and selecting the combination of effective antagonistic endophytic Bacillus strains that could be the best alternative for the sustainable and ecological sound management of HS disease in wheat under field conditions.
1. Introduction
Fusariumgraminearum causing head scab (HS) or head blight (HB) disease in wheat is one of the nasty fungi reported to cause significant grain quality and yield loss. The major outbreaks of HS disease in wheat from major wheat-growing countries of the world, including Australia, Asia, Canada, Europe, and South America, have been well documented [1]. In India, noteworthy wheat yield loss has been noticed due to HS disease, specifically when rain coincides with the anthesis stage of the wheat [2]. In wheat fields, HS symptoms can be seen on the glumes and rachis as water-soaked spots. Later, the disease dispersed within the wheat ear heads, and infected ear heads showed partial to complete bleaching. Under warm, humid conditions, orange sporulation can also be seen on infected wheat spikelets. Published literature revealed that crop rotation, agronomic interventions, and avoidance of the cultivation of susceptible varieties are not useful in providing an adequate level of protection against diseases. Moreover, none of these approaches alone or in an integrated manner can control the disease in an effective manner [3,4,5]. Growing HS tolerant cultivars is one of the most effective options. However, in India, a majority of the varieties under cultivation have a low level of resistance against F. graminearum due to a lack of complete HS-resistant wheat cultivars. Although adequate levels of wheat protection from HS can be achieved with agrochemicals and resistance inducers, their efficacy varies with each fungal strain [6,7,8,9,10]. Moreover, recommended fungicides had adverse effects on soil microbiota and the environment, warranting new technological innovations with minimal reliance on agrochemicals [11]. Recently published reports indicated that agrochemicals are regularly losing their ground in the management of diseases under field conditions due to the rapid evolution of pathogen races and mutations resulting in resistance phenotypes [12,13] and thus warrants alternative molecules with novel modes of action. Recently, de Chaves et al. [14] noticed Tebuconazole and Prochloraz resistant isolates of F. graminearum in wheat fields, where the continuous application of fungicides was utilized for a long time. At present, a limited variety of novel molecules for tackling the rising problem of fungicide resistance development in F. graminearum is available. Moreover, the successful and timely management of HS by fungicides is difficult to attain as the effect of fungicide is highly influenced by the optimal timing of the fungicide application, which is too short in the case of HS disease [3]. Therefore, HS management by employing eco-compatible technologies such as antagonistic endophytic bacteria could be of great benefit.
Several studies highlighted the escalating research interest in exploring the prospective role of endophytic bacteria in mitigating the adverse effects on agricultural crops as a consequence of biotic and abiotic stressors [15,16]. Generally, endophytic bacteria reside inside the plant host without displaying any prominent disease symptoms. Here, it is important to mention that the prime benefit of harnessing endophytes as potential antagonists is their strong adaptation character to stay within the plants, which makes them suitable candidates as crop stress defenders from biotic anomalies [17]. The majority of the benefits of endophytic bacteria are analogous to rhizospheric bacteria. However, emerging research evidence ranked endophytic bacteria over traditional antagonists due to their ability to transfer to the next generation in sustainable manner [18,19]. Among various endophytic bacteria, the exploration of endophytic antagonistic Bacillus has emerged as one of the highly prospective and eco-friendly substitutes due to its unique inherent endospore formation capacity. This capacity helps to get high resistance against stressors, omnipresence, and stability in extreme environments [20,21]. Besides this, numerous studies illustrate the remarkable contribution of antimicrobial peptides (AMPs) released by Bacillus strains in the microbial management of fungal plant pathogens [22,23,24,25,26]. Unfortunately, inadequate information exists with respect to the occurrence of genes associated with the synthesis of AMPs in Bacillus species allied with wheat hosts as endophytes.
There have been a number of reports of functional characterization and evaluation of endophytic Bacillus strains for the management of fungal pathogens attacking different crops. For instance, endophytic Bacillus mojavensis has been reported to enhance maize growth when attacked with Fusarium verticillioides [27]. Similarly, Pan et al. [17] noticed that endophytic B. megaterium and B. subtilis derived from wheat grain markedly suppressed the fungal growth of F. graminearum. Besides these, several other Bacillus strains of endopphytic origin, such as B. amyloliquefaciens YN201732 [28], B. velezensis strain OEE1 [29], B. thuringiensis [30], and B. safensis B21 [31] have been recognized as ideal candidates for the bio-control of myriads of fungal pathogens attacking diverse types of agricultural crops. In a recent study, Munakata et al. [32] made a functional comparison of endophytic microbiota of vetiver root from different ecological niches across Africa and Europe and noticed a strong antagonistic character of Bacillus origin endophytes towards F. graminearum. All these research efforts indicate the wide spectrum bio-control potential of endophytic Bacillus strains. However, despite the huge economic significance of wheat, very limited research efforts have been made to explore the diversity of endophytic Bacillus strains as potential antagonists for the management of wheat diseases in an ecologically sound manner. In wheat, the anthesis period is identified as the most susceptible stage for F. graminearum infection. Further, it has been observed that anthers serve as a frequent path of entrance into the wheat host and markedly important growth stage for the application of bio-control agents or fungicides to restrict the fungal infection in a spatio-temporal manner, even under favorable environmental conditions [33]. Thus, the present study was planned with following prime objectives: (i) to profile the endophytic bacilli associated with ears of different wheat genotypes and evaluate their antagonistic features towards F. graminearum, and (ii) to assess the potential of identified antagonistic endophytic Bacillus species individually or in combination for the bio-control of HS under field conditions.
2. Materials and Methods
2.1. Field Sampling for the Isolation and Identification of Endophytic Bacillus
Healthy ear head samples were collected from wheat genotypes (viz., DBW187, HD2967, PBW343, HD3086, and Agra local). Every sample comprised of three plants of 65 days old wheat crop (anthesis stage) was gathered in sterilized plastic sampling bags. A complete surface sterilization procedure reported by Kushwaha et al. [15] was employed for the isolation of endophytic bacteria from wheat ear heads. In brief, preliminary treatment of sampled wheat ear heads was performed with tap water, followedwashing the sample with double distilled sterilized water. After preliminary washing, treated samples were placed in 70% ethanol for 1 min and consequently soaked in 3% sodium hypochlorite for 3 min and 70% ethanol again for 30 s in a sequential manner. The final treatment of the sample was done with autoclaved double distilled water. This step was repeated three times before the final drying of the treated samples on sterile Whatman filter paper. Approximately 1 g of each tissue was finely crushed and suspended in phosphate-buffered saline (PBS) solution (Hi-Media Laboratories Pvt Ltd. India). Heat treatment of the macerated tissue suspension was performed for the isolation of Bacillus species, according to Sharma et al. [34]. An aliquot (100 mL) from each heat-treated serial dilution (10−2 to 10−6) sample was streaked on Petri plates containing different media [e.g., nutrient agar (NA), Luria Bertani agar, and tryptic soy agar] and incubated at 37 ± 2 °C temperature for two days. An aliquot (100 µL) of remaining water after the last wash was inoculated on NA Petri plates to inspect the occurrence of different bacterial colonies for seven days. Different microscopy and biochemical tests (Gram staining, motility, microscopic appearance, oxidase test, catalase test, reduction of nitrate to nitrite) were also conducted in three replicates for preliminary confirmation of the Bacillus spp. [35]. Bacterial colonies with different morphotypes were selected and maintained in glycerol (50%) at −80 °C [36].
For the accurate identification of bacterial endophytes and detection of genes linked with antimicrobial peptides (AMPs) produced by Bacillus, isolation of the total genomic DNA of each bacterium was carried out as per the protocol of Pospiech and Neumann [37]. Amplification of 16S rRNA region and AMP genes was carried out in a thermocycler machine (Q cycler 96, Hain Lifescience UK Ltd., Surrey, UK), and conditions of PCR reaction are described in Table 1. For the identification of bacterial endophytes, generated amplicons of ~1500 bp were sent to Eurofins genomics sequencing services, India, for DNA sequence analysis. The matching of 16S rRNA gene sequences of bacterial endophytes was made against the sequences available in the EZ Biocloud e-server (https://www.ezbiocloud.net/, accessed on 10 December 2021) to identify their nearest match. Molecular Evolutionary Genetics Analysis (MEGA) version 11 [38] tool was used for the construction of phylogenetic tree with nearest type strain sequences of Bacillus sp. available in National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/, accessed on 2 February 2022) database. The gene sequences were deposited in the NCBI databank to get the gene accession numbers (Figure 1, Figure 2, Figure 3, Figure 4 and Figure 5). The alignments of the 16S rRNA gene sequences of bacterial endophytes, along with matching type strains, were conducted with the help of Clustal W [39]. The multiple sequence alignment profiles were used to build the best fit phylogenetic tree using the neighbor-joining method [40] with Kimura’s two-parameter model [41] executed in MEGA version 11 software [38]. Bootstrap analysis was performed to assess confidence levels for the branches with 1000 replicates [42].

Table 1.
List of primers and thermal profile employed for PCR based amplification of target genomic region of endophytic Bacillus strains.
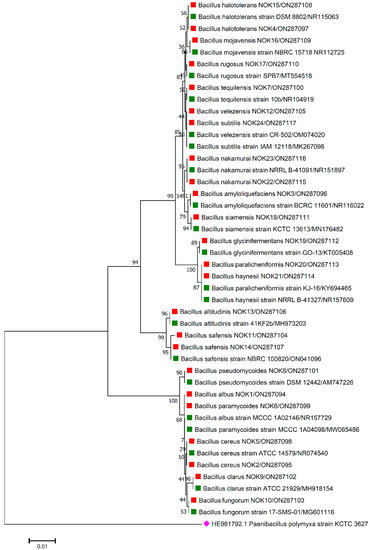
Figure 1.
Phylogenetic tree based on partial 16S rRNA sequences of endophytic Bacillus strains (indicated by red colour square) allied with anthesis stage of wheat (cv. DBW187) along with those of maximum similar entries of type strains (indicated by green colour square) from database. The tree is constructed by neighbour-joining method with 1000 bootstrap replications. The scale bar represents the number of changes per base position. Paenibacillus polymyxa strain KCTC 3627 (HE981792.1) was used as an out-group strain (indicated by pink colour rhombus).
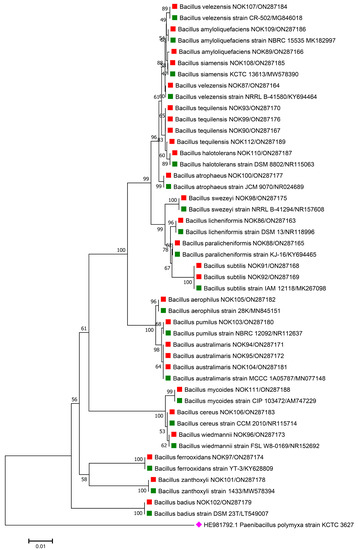
Figure 2.
Phylogenetic tree based on partial 16S rRNA sequences of endophytic Bacillus strains (indicated by red colour squares) allied with anthesis stage of wheat (cv. HD2967) along with those of maximum similar entries of type strains (indicated by green colour squares) from database. The tree is constructed by neighbour-joining method with 1000 bootstrap replications. Scale bar represents the number of changes per base position. Paenibacillus polymyxa strain KCTC 3627 (HE981792.1) was used as an out-group strain (indicated by a pink colour rhombus).

Figure 3.
Phylogenetic tree based on partial 16S rRNA sequences of endophytic Bacillus strains (indicated by red colour squares) allied with anthesis stage of wheat (cv. PBW343) along with those of maximum similar entries of type strains (indicated by green colour squares) from database. The tree is constructed by neighbour-joining method with 1000 bootstrap replications. Scale bar represents the number of changes per base position. Paenibacillus polymyxa strain KCTC 3627 (HE981792.1) was used as an out-group strain (indicated by a pink colour rhombus).
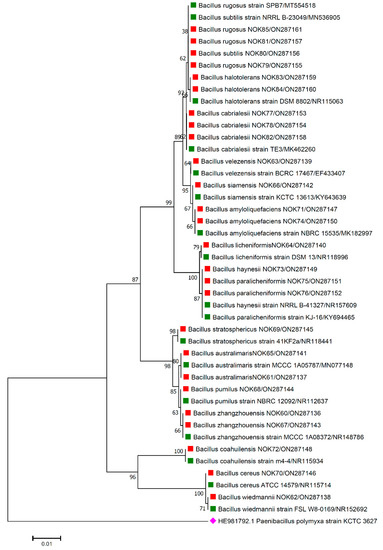
Figure 4.
Phylogenetic tree based on partial 16S rRNA sequences of endophytic Bacillus strains (indicated by red colour squares) allied with anthesis stage of wheat (cv. HD3086) along with those of maximum similar entries of type strains (indicated by green colour squares) from database. The tree is constructed by neighbour-joining method with 1000 bootstrap replications. Scale bar represents the number of changes per base position. Paenibacillus polymyxa strain KCTC 3627 (HE981792.1) was used as an out-group strain (indicated by a pink colour rhombus).
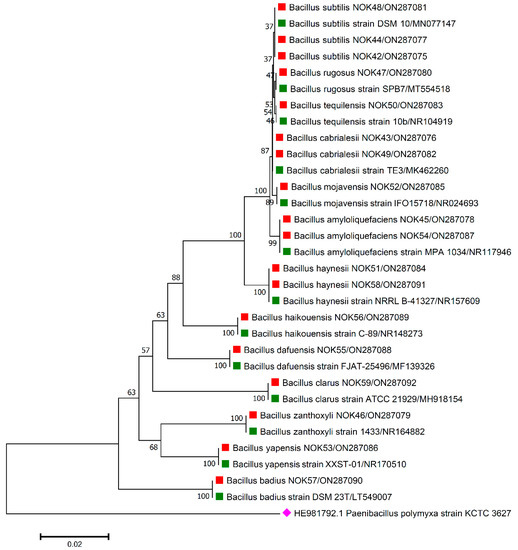
Figure 5.
Phylogenetic tree based on partial 16S rRNA sequences of endophytic Bacillus strains (indicated by red square box symbols) allied with anthesis stage of wheat (cv. Agra Local) along with those of maximum similar entries of type strains (indicated by green square box symbols) from database. The tree is constructed by neighbour-joining method with 1000 bootstrap replications. Scale bar represents the number of changes per base position. Paenibacillus polymyxa strain KCTC 3627 (HE981792.1) was used as an out-group strain (indicated by a pink colour rhombus).
2.2. In-Vitro Determination of Antagonistic Activity
Dual-culture plate assay was used to confirm the antagonistic capabilities of endophytic bacterial strains against F. graminearum. Briefly, a 5 mm diameter segment of fungal mass of highly virulent F. graminearum NFG1 isolate was positioned in the mid-point of Petri plates amended with PDA: NA (1:1). Each strain (~5 × 108 cfu ml−1) was streaked in a straight line closer to the border of Petri plate. The inoculated plates were incubated at 30 ± 2 °C. F. graminearum inoculated Petri plate without endophytic strain, and neutral bacterial strain (Bacillus pumilus NOK68) served as a control. Data on the growth of F. graminearum was recorded in mm till the control plate (without endophytic strain) was completely filled. F. graminearum growth inhibition (%) by each endophytic strain was computed by the formula quoted by Sharma et al. [34]. The assay was performed with three independent repetitions.
2.3. Field Trial of Antagonists against FHB
Three Bacillus strains that reflected maximum antagonistic action towards F. graminearum under in-vitro conditions were chosen for testing their bio-control potentialities against HS disease in susceptible wheat genotype (PBW 343) at Crop Protection Experiment Area, ICAR-Indian Institute of Wheat and Barley Research, Karnal, Haryana, India. Under each field treatment, wheat seeds were sown in six lines (each line of 3 m long) with 22.5 cm line spacing. All the experiments were planned in a randomized complete block design with five replicates per treatment. Different treatments comprised of antagonistic endophytic bacteria (~107 cfu ml−1) and without endophytic bacteria were used to inoculate 150 wheat ear heads (50 heads per replicate, three replicates per treatment) at anthesis time (Zadoks growth stage 65). The bacterial suspension (2 mL per wheat ear head) in each plot was applied by a compressed air sprayer in the late afternoon (at 4:00PM). The control treatment (T8) was only sprayed with highly virulent F. graminearum isolate (NFG1). An additional control treatment (T9) comprised of fungicide (Propiconazole @ 0.1%, Syngenta, Pune, India) sprayed alone. Field assessment of HS incidence and HS severity was made at the late milk stage. Disease data was recorded on 40 ear heads per replicate (200 ear heads treatment −1) as per the disease rating described by Stack and McMullen [46]. Disease index (DI) computation was made by using the formula: (incidence × severity)/100).
2.4. Strain Characterization for Siderophore Production and Hydrolytic Enzyme Activities
Chrome azurol (CAS) agar assay [47] was carried out to know the potential of 14 endophytic antagonistic bacterial strains for siderophore synthesis. For assessing the potential of endophytic bacterial strains for potassium (K) solubilization, the spot inoculation procedure quoted by Hu et al. [48] was used, where potassium feldspar powder was used as an insoluble phosphate source. The potential of endophytic bacterial strains for siderophore production and potassium solubilization was checked for halo development around the colony, and halo size was recorded in mm. Similarly, the potential of the extracellular hydrolytic enzymes (chitinase, amylase, cellulase, protease, and lipase) production by each strain was determined according to Kushwaha et al. [21] and Sharma et al. [34], where the formation of clear halo zone by each strains was observed on NA Petri plates amended with a substrate such as chitin, soluble starch, carboxy methyl cellulose, casein, and tributerin for the detection of chitinase, amylase, cellulase, protease, and lipase activities, respectively. The halo zone around the bacterial colony was indicative of positive enzymatic activity and measured in mm. Each assay was repeated three times.
2.5. Detection of Antimicrobial Peptide Gene (s)
Three endophytic strains (NOK9, NOK33, and NOK109) displaying strong antifungal activities (>70%) were tested for the presence of different AMP genes in Bacillus strains. The polymerase chain reaction temperature profiles and primers information given in Table 1 was used for the amplification of the surfactin, bacillomycin, and iturin genes, respectively.
2.6. Statistical Analysis
The field experiments performed to check the antagonistic effects of selected bacteria were arranged in complete randomized block design (CRD) with three replicates. A one-way analysis of variance (ANOVA) was performed to test the significance of each treatment. A post hoc comparison of mean values was carried out by performing Duncan’s multiple range test (DMRT).
3. Results
3.1. Species Diversity of Endophytic Bacillus in Tissues of Wheat of Different Genotypes
The 16S rRNA amplicon sequencing results illustrated that all the 112 strains isolated from wheat ear heads of five different cultivars (viz., DBW187, HD2967, PBW343, HD3086, and Agra local) represent the Bacillus genus. The genotypes DBW187, HD2967, PBW343, HD3086, and Agra local contain 24, 27, 17, 26, and 18 Bacillus strains of endophytic origin, respectively. The bacterial strains belonged to 28 different species (B. aerophilus, B. albus, B. altitudinis, B. amyloliquefaciens, B. atrophaeus, B. australimaris, B. badius, B. cabrialesii, B. cereus, B. clarus, B. coahuilensis, B. dafuensis, B. ferrooxidans, B. fungorum, B. glycinifermentans, B. haikouensis, B. halotolerans, B. haynesii, B. licheniformis, B. mojavensis, B. mycoides, B. nakamurai, B. paralicheniformis, B. paramycoides, B. pseudomycoides, B. pumilus, B. rugosus, B. safensis, B. siamensis, B. stratosphericus, B. subtilis, B. subtilis subsp. spizizenii, B. swezeyi, B. tequilensis, B. velezensis, B. wiedmannii, B. yapensis, B. zanthoxyli, and B. zhangzhouensis) and reveal a high degree (100%) of sequence resemblance with the type strain sequences in available in EZ Biocloud e-server (https://www.ezbiocloud.net/, accessed on 10 December 2021). It has been observed that the wheat genotype DWB 187 contained the maximum number of distinct Bacillus species (20 species; Figure 1) followed by HD 2967 (19 species; Figure 2), DBW 343 (12 species; Figure 3), HD 3086 (17 species; Figure 4), and Agra local (13 species; Figure 5). Additionally, the gene sequences of 16S rRNA were deposited at NCBI GenBank with accession number mentioned in Figure 1, Figure 2, Figure 3, Figure 4 and Figure 5.
3.2. Identification and Selection of Antagonistic Endophytes
A total of 112 endophytic Bacillus strains were recovered from five different wheat cultivars, but only fourteen strains (B. clarus NOK09, B. mojavensis NOK16, B. subtilis NOK33, B. rugosus NOK47, B. mojavensis NOK52, B. clarus NOK59, B. coahuilensis NOK72, Bacillus cabrialesii NOK78, B. cabrialesii NOK82, B. rugosus NOK85, B. amyloliquefaciens NOK89, B. australimaris NOK95, B. pumilus NOK103, and B. amyloliquefaciens NOK109) displayed antagonist action towards highly virulent F. graminearum NFG1 isolate (Table 2). The strain NOK68 did not show any antagonistic effects against F. graminearum NFG1 (Figure 6). All the antagonists presented a significant reduction in the F. graminearum growth (p < 0. 05), among which B. clarus NOK09 (77.3% F. graminearum growth inhibition), B. subtilis NOK33 (71.9% F. graminearum growth inhibition), and B. amyloliquefaciens NOK109 (79.4% F. graminearum growth inhibition) displayed more than a 70 % inhibitory effect on fungal growth (Table 2).

Table 2.
In-vitro evaluation of the antagonism against Fusarium graminearum and different activities of production of siderophore and hydrolytic enzymes and potassium solubilisation displayed by endophytic Bacillus strains isolated at anthesis stage from different wheat genotypes.
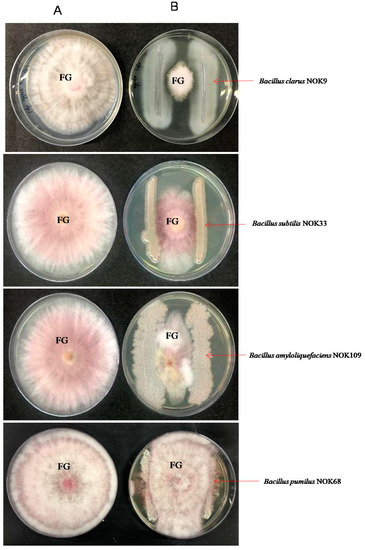
Figure 6.
In-vitro interaction between endophytic Bacillus strains and F. graminearum NFG1 in a dual culture on NA: PDA plate at 5th day after incubation at 28 ± 2 °C. (A) A 5-mm agar plug of F. graminearum NFG1 on center of PDA plate and (B) Endophytic Bacillus strains inoculated on two corners of PDA plate with equal distance from the colony of F. graminearum placed at the centre. FG = F. graminearum NFG1. Bacillus pumilus NOK68 serve as a neutral bacterial strain.
3.3. Strain Characterization for Potassium Solubilization, Siderophores Release and Hydrolytic Enzyme Activity
Among fourteen shortlisted strains on the criterion of in vitro antifungal activity, NOK09, NOK16, NOK33, NOK47, NOK52, NOK59, NOK72, NOK78, NOK82, NOK85, NOK89, NOK95, NOK103, and NOK109 were found siderophore producing in nature and displayed clear halo zone (10.11–21.11 mm) formation (Table 2). It has been noticed that strain NOK109, followed by NOK09 and NOK33, released an amazingly high amount of siderophore and displayed more than 20.62 mm clear halo zone formation. Likewise, NOK09, NOK16, NOK33, NOK47, NOK52, NOK59, NOK78, NOK82, NOK89, NOK103, and NOK109 strains were observed as efficient phosphate solubilizers, showing a prominent, clear halo zone in the range of 9.42 mm to 17.86 mm. Similarly, NOK09, NOK33, NOK47, NOK52, NOK59, NOK78, NOK85, NOK89, NOK95, NOK103, and NOK109 were recorded as lipase producers, displayinga halo zone in the range of 10.71 mm to 22.07 mm. Besides this, all the fourteen strains had the potential to synthesize extracellular enzymes (protease, amylase, chitinase, and pectinase) at variable levels (Table 2). Three strains, i.e., NOK9, NOK33, and NOK109, showed the highest activities for protease, lipase, amylase, chitinase, and pectinase (Table 2).
3.4. Detection of Antimicrobial Peptides (AMPs) Biosynthesis Associated Genes
PCR amplification of AMP genes in three endophytic Bacillus strains (B. clarus NOK09, B. subtilis NOK33, and B. amyloliquefaciens NOK109) were displayed in Figure 7. Amplification of the iturin (ituD), bacillomycin (bmyC), and surfactin (srfA) gene produced a single specific amplicon of 647, 395, and 201 bp, respectively, in NOK9, NOK33, and NOK109 strains.
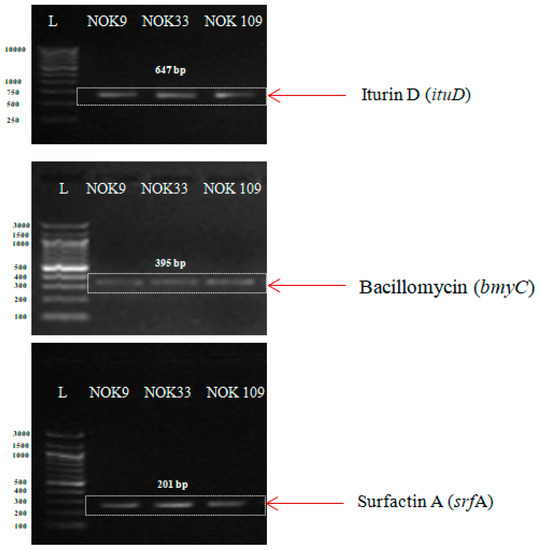
Figure 7.
Agarose gel-electrophoresis of PCR products for revealing the presence of antimicrobial peptides (AMPs) i.e., iturin, bacillomycin, and surfactin A genes in promising antagonistic endophytic Bacillus strains. L is the DNA ladder. Numbers on left size indicates base pairs of the steps in the ladder.
3.5. Field Evaluation of Antagonistic Strains against F. graminearum
Three endophytic Bacillus strains (B. clarus NOK09, B. subtilis NOK33, and B. amyloliquefaciens NOK109) which showed the greatest bio-control activity (>70%) towards F. graminearum NFG1 under in-vitro conditions were chosen to appraise their bio-control potential to contain HS disease in wheat under field conditions and observations were presented in Figure 8, Figure 9 and Figure 10 and Table 3. The perusal of data indicated that in 2020–2021, the disease severity varied between 15.29 to 87.92 % of infected spikelets (Figure 9), while in 2021–2022, it ranged from 15.05 to 87.80% (Figure 9). The wheat plants sprayed at the anthesis stage with different treatments of endophytic antagonists individually (NOK09, NOK33, and NOK109) or in combination (NOK09 + NOK33, NOK33+. NOK109, NOK09 + NOK109, and NOK09 + NOK33+ NOK109) significantly reduced the impact of HS incidence, severity, and disease index compared to the control check i.e., T8 (sprayed with only highly virulent F. graminearum NFG1 isolate) in both the years (Figure 8 and Figure 9). A noteworthy decline in DS was recorded in plots sprayed with a combination of three strains composed of NOK 9 + NOK 33 + NOK 109 (61.18 and 59.14%) followed by NOK 9 + NOK109 (50.96 and 50.12%), NOK 33 and NOK 109 (45.57 and 45.80%), NON 9 + NOK33 (42.72 and 41.63%), NOK 109 (40.02 and 35.64%), NOK 9 (27.32 and 24.42%) and NOK33 (33.20 and 30.05%), in both the years, respectively. Similar trends were observed in the case of disease incidence in both years (Figure 8). It has been noticed that among the treatment of individual strains, a significant reduction in DI (Figure 8), DS (Figure 9), and FHB index (Table 3) was observed in the case of NOK109 followed by NOK33 and NOK9 relative to untreated control. Based on field data from both years, all the treatments of antagonists (T1 to T7), either singly or in combination, show a significant gain in grain yield (p < 0.05) relative to the untreated endophyte control (T8) (Figure 10). Maximum disease reduction effects were observed for T9 (Propiconazole @ 0.1%), which reduced disease severity by 82.62 and 82.90% in both years, respectively (Table 3). In both the years, treatment with NOK09 + NOK33 + NOK109 was the most effective among all antagonist treatments.
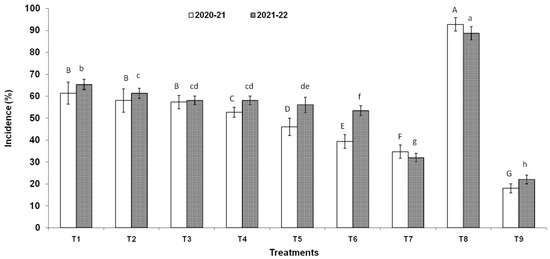
Figure 8.
Disease incidence (%) of Fusarium head blight disease observed during 2020-21 and 2021-22 field trials conducted on susceptible wheat cultivar (cv. PBW343). On x axis, treatments are: T1 (NOK9 + NFG1), T2 (NOK33 + NFG1), T3 (NOK109 + NFG1), T4 (NOK9 + NBOK33 + NFG1), T5 (NOK 9 + NOK109 + NFG1), T6 (NOK33 + NOK109 + NFG1), T7 (NOK9 + NOK33 + NOK109 + NFG1), T8 (F. graminearum NFG1), and T9 (Propiconazole @ 0.1%+ + NFG1). Data were analyzed for significance with an analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference.
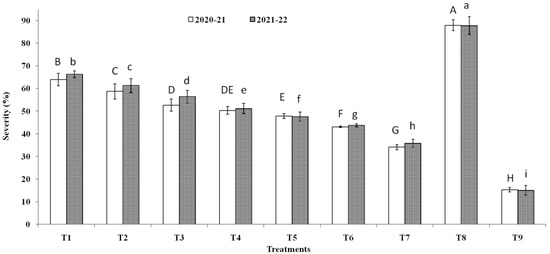
Figure 9.
Disease severity (%) of Fusarium head blight disease observed during 2020-21 and 2021-22 field trials conducted on susceptible wheat cultivar (cv. PBW343). On X-axis, treatments are: T1 (NOK9 + NFG1), T2 (NOK33 + NFG1), T3 (NOK109 + NFG1), T4 (NOK9 + NBOK33 + NFG1), T5 (NOK 9 + NOK109 + NFG1), T6 (NOK33 + NOK109 + NFG1), T7 (NOK9 + NOK33 + NOK109 + NFG1), T8 (F. graminearum NFG1), and T9 (Propiconazole @0.1% + NFG1). Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference.
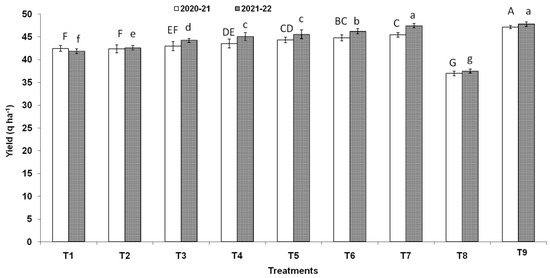
Figure 10.
Influence of antagonists on the wheat (cv. PBW343) yield under field trials conducted during 2021–2021 and 2021–2022. On X-axis, treatments are: T1 (NOK9 + NFG1), T2 (NOK33 + NFG1), T3 (NOK109 + NFG1), T4 (NOK9 + NBOK33 + NFG1), T5 (NOK 9 + NOK109 + NFG1), T6 (NOK33 + NOK109 + NFG1), T7 (NOK9 + NOK33 + NOK109 + NFG1), T8 (F. graminearum NFG1), and T9 (Propiconazole @ 0.1% + NFG1). Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference Y-axis represent grain yield in q ha−1.

Table 3.
Effect of endophytic Bacillus strains on Fusarium head scab incited of wheat (cv. PBW343).
4. Discussion
Head scab (HS) is acknowledged as one of the prime diseases of wheat because of its global presence in all wheat-growing countries and is ranked as the fourth biggest threat to successful quality wheat production [49]. Due to the unavailability of completely resistant cultivars, the management of HS is heavily relying on fungicides. Nevertheless, with injudicious and excessive use of recommended fungicides, the cases of resistance development in F. graminearum against fungicides are mounting and ultimately resulted in the loss of their field efficacy [6,50]. Furthermore, the farmers many times miss the optimal fungicide application time due to their inability to predict the right infection time of the wheat spike. As a result, it becomes necessary to devise new strategies for the effective and timely management of HS disease in wheat. Therefore, the major goal of the present research is to decipher the diversity and antagonistic potential of endophytic Bacillus spp. allied to the anthesis stage of wheat for the eco-friendly and sustainable management of HS under natural field conditions. In the current research investigation, the diversity of cultivable endophytic Bacillus strains allied with healthy wheat ear heads of five different wheat cultivars (DBW187, HD2967, PBW343, HD3086, and Agra local) has been explored. It is important to mention here that healthy wheat ears were selected for the exploration of Bacillus endophytes because of the fact that the healthy plant parts harbor a diverse and distinct type of microbial population of endophytic bacteria than unhealthy plants [51,52]. Earlier published literature reported that the composition of bacterial endophytes was greatly influenced by plant type, growth stage, and soil nutrient availability [53]. Marag et al. [54] reported the flowering stage as the prime site for maximal abundance and population of endophytic bacteria than other crop growth stages. Most importantly, endophytic Bacillus strains displaying excellent antagonistic activity in different crops have been reported by various workers [15,55,56,57]. It is important to mention that the anthesis or wheat ear emergence stage is the most susceptible window for F. graminearum infection [33]. Moreover, because phyllosphere endophytes reside in a similar ecological niche as foliar pathogens [58], there is a great probability that the bacterial endophytes associated with wheat ear head might serve as excellent bio-control agents against HS fungus. Thus, keeping these facts in mind, the present study was undertaken to profile the diversity of endophytic strains of Bacillus species from wheat genotypes and explore their antifungal activity for the field management of HS in wheat.
A series of published literature indicated that investigation of the diversity of the plant microbiome is one of the potential approaches to identify novel and effective microorganisms as a fungal antagonist and plant growth promoter [56,57,58]. By following a cultivation-dependent approach, a total of 112 Bacillus strains were recovered from wheat ear heads of five different cultivars. These results are in harmony with earlier published literature, where a culture-dependent approach was employed to isolate the bacterial endophytes from different regions of the plant [59,60,61]. Interestingly, this study showed the endophytic association of B. aerophilus, B. albus, B. atrophaeus, B. australimaris, B. badius, B. cabrialesii, B. cereus, B. clarus, B. coahuilensis, B. dafuensis, B. ferrooxidans, B. fungorum, B. glycinifermentans, B. haikouensis, B. halotolerans, B. haynesii, B. mojavensis, B. mycoides, B. nakamurai, B. paralicheniformis, B. paramycoides, B. pseudomycoides, B. rugosus, B. safensis, B. siamensis, B. stratosphericus, B. swezeyi, B. tequilensis, B. velezensis, B. wiedmannii, B. yapensis, B. zanthoxyli, and B. zhangzhouensis with wheat ear head for the first time.
A large body of published reports on bacterial endophytes has focused on the plausible applications of isolated strains in managing agriculturally important diseases and established dual-culture assays as one of the golden standards for assessing the antagonistic effectiveness of isolated Bacillus strains prior to field validations [62,63]. Similarly, in the current study, a dual-culture growth inhibition test was performed, and the obtained results revealed 12.5% of the isolated Bacillus strains of antagonistic nature against F. graminearum. Based on the 16S rRNA sequence analysis performed in the present study, the antagonistic bacteria recovered from wheat ear heads were identified as B. clarus NOK09, B. mojavensis NOK16, B. subtilis NOK33, B. rugosus NOK47, B. mojavensis NOK52, B. clarus NOK59, B. coahuilensis NOK72, Bacillus cabrialesii NOK78, B. cabrialesii NOK82, B. rugosus NOK85, B. amyloliquefaciens NOK89, B. australimaris NOK95, B. pumilus NOK103, and B. amyloliquefaciens NOK109. These research findings agree with earlier reports, where B. amyloliquefaciens, B. pumilus and B. subtilis strains have been described as potential antagonists [64,65,66].
In the current study, three endophytic Bacillus strains (B. clarus NOK09, B. subtilis NOK33, and B. amyloliquefaciens NOK109) showing maximum bio-control activity against F. graminearum reflected strong character for potassium solubilization and siderophore production. These results are in harmony with earlier reports, where the presence of endophytic bacterial strains showing strong attributes of potassium solubilization and siderophore production were observed [67,68,69]. Kushwaha et al. [15] also recorded potassium solubilizing and siderophore-producing traits in endophytic strains of B. cereus, B. amyloliquefaciens, and B. subtilis subsp. subtilis from pearl millet host and further suggesting their function as a growth promoter, nutrient mobilizer, and disease defender. Here, it is important to mention that siderophore is a small, low molecular weight (500-1000 Daltons) iron-chelating agent which binds to the available iron making it unavailable for the phytopathogens. It also supports the plant by making iron easily available for various biological processes operating inside the plant system. Numerous recent studies reported siderophore production as the most common trait in bacterial endophytes associated with plants [69,70,71,72]. Apart from plant growth promoting traits, another mechanism that bacterial endophytes employ to combat fungal plant pathogens is the synthesis of extracellular hydrolytic enzymes [73]. In the present study, the three above-mentioned endophytic antagonistic Bacillus strains were also found to be positive for the production of chitinase, protease, lipase, amylase, and pectinase enzymes. Additionally, it appears that these hydrolytic enzymes could be responsible for the remarkable inhibition of F. graminearum. Here, it is worth mentioning that with the help of these extracellular hydrolytic enzymes, endophytes can penetrate the plant tissue and play a vital role in endophytic colonization and establishment inside the host plant [74,75].
PCR detection of AMP genes indicated that three antagonistic strains showing maximum bio-control activity against F. graminearum had three diverse antibiotic biosynthesis genes (ituD, bmyC, and srfA) that are linked with the production of the antibiotics iturin, bacillomycin, and surfactin. The obtained results are in conformity with other workers that revealed the prevalence and linkages of numerous antifungal peptide genes in diverse types of Bacillus strains [15,24,76,77,78,79,80]. Based on the published reports, it can be inferred that Bacillus species contain diverse types of AMP genes that have a role in the biosynthesis of antibiotics with specific modes of action [78,81]. For instance, the itu gene is essential in enhancing fungal cell membrane permeability [23]. On the other hand, the function of the srf gene is linked with biofilm synthesis [82] and bmy associated with the degradation and alterations of the cell wall and cell membrane of fungal hypha [76,83].
A large number of bacterial endophytes from diverse types of plants have been identified and evaluated for bio-control activities for several decades to control fungal diseases in India [15,84,85]. However, the research investigation with respect to the application of bio-control agents has been restricted to the in vitro identification and characterization of microbes against wheat pathogens. No field experimentation-based research report is documented for the suppression of wheat head scabs in India. Keeping these facts in mind, attempts were made to validate the hypothesis that combinations of endophytic bacteria are highly effective and versatile than individual endophytic strains in the bio-control of HS disease in wheat. Field testing of the biocontrol potential of endophytic bacterial strains (B. clarus NOK09, B. subtilis NOK33, and B. amyloliquefaciens NOK109) reflecting >70% mycelial growth inhibition against F. graminearum under in-vitro conditions in single or in combination was performed to test the hypothesis. Here, it is pertinent to mention that microbial consortia function in a strong and structured network and offers additional protection for the host to survive better than individual microorganisms under adverse conditions [86,87]. For instance, a combination of Paenibacillus sp. strain B2 and Arhtrobacter spp. strain AA was found more effective than a single-strain inoculum for promoting resistance against Septoria tritici blotch disease under drought stress [88]. Similarly, three strain mix (B. subtilis B2, B. thuringiensis B10, and Enterobacter cloacae B16) also showed noteworthy bio-control effects against Sclerotinia stem rot disease in tomatoes [89]. On parallel lines, the application of two formulated biological control agents (Bacillus subtilis RC 218 and Brevibacillus sp. RC 263) under semi-controlled field conditions resulted in a significant reduction in the HS severity level in wheat [90]. These studies strengthen the notion of evaluating the effect of the combination of antagonistic Bacillus strains of endophytic origin along with individual antagonists. The results emanating from the present investigation clearly evidenced the effectiveness of the combination of three distinct strains (B. clarus NOK09 + B. subtilis NOK33 and B. amyloliquefaciens NOK109) followed by the consortia of two strains in suppressing HS infection in wheat. However, positive and noteworthy antagonistic effects were achieved with all the test strains when used singly or in combination under in-vivo bio-control experiments. The reason that a single strain may be less effective can be associated with poor competitiveness against native microflora and fluctuating environmental conditions [91]. Here, it becomes important to underline that the repetitive success of the consortia in both the years under field conditions also supports the phenomenon of strain compatibility. In fact, the field results of triple endophyte consortia in bio-control were significantly different and better than the individual strain. These observations were also harmonized with the data obtained from the analysis of both years that yield gain was more pronounced in triple entophyte consortia relative to two strain consortia or individual strain application. These results were corroborated with the findings of Muhae-Ud-Din et al. [92], who noticed wheat grain yield gain by 30.8% over control check when a combination of endophytic microbes (Bacillus sp. MN54 + Trichoderma sp. MN6) was applied. Besides this, the results obtained in the current study with respect to the bio-control efficacy agree with earlier published literature, where a significant level of disease protection has been documented by spraying microbial consortia [87,88]. In addition, another imperative observation noticed in the current study was the additive effect of the microbial consortia composed of B. clarus NOK09 + B. subtilis NOK33 and B. amyloliquefaciens NOK109 when compared with the antagonistic effect of individual strain treatments on HS disease containment. Similar types of observation regarding the effectiveness of endophytic bacterial consortia in controlling the purple blotch disease and enhancing the growth and yield of shallots have been demonstrated by Resti et al. [93]. In a parallel fashion, Sundaramoorthy et al. [94] also documented the significant effect of the combination of P. fluorescens Pf1 and B. subtilis (EPCO16 and EPC5) in controlling the Fusarium wilt incidence in chilli by 17–30%. However, in the current study, better percent disease suppression in terms of HS incidence (62.52 to 63.86%) and severity (59.14–61.18%) was obtained when the combination of B. clarus NOK09 + B. subtilis NOK33 and B. amyloliquefaciens NOK109 spray were inoculated on the wheat plants.
In conclusion, it can be inferred that wheat genotypes at the anthesis stage harbor 28 distinct species of Bacillus of entophytic nature, where only ten species displayed antagonism towards F. graminearum. Based on field results, it can be proposed that the combination of three endophytic Bacillus mixes (B. clarus NOK09 + B. subtilis NOK33 + B. amyloliquefaciens NOK109) in place of a single strain could be an important approach for the fieldmanagement of HS disease of wheat in an eco-friendly and sustainable manner.
Author Contributions
Conceptualization, N.K., P.L.K. and D.S.; Data curation, N.K., S.K. and D.S.; Formal analysis, N.K. and P.L.K.; Funding acquisition, P.L.K. and G.P.S.; Investigation, N.K., P.L.K., S.K. and G.P.S.; Methodology, N.K.; Project administration, P.L.K.; Resources, N.K. and G.P.S.; Software, N.K. and P.L.K.; Supervision, P.L.K., S.K. and G.P.S.; Validation, N.K., P.L.K. and D.S.; Visualization, N.K.; Writing—original draft, N.K.; Writing—review & editing, P.L.K., S.K., D.S. and G.P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Indian Council of Agricultural Research.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Acknowledgments
The authors thanks to ICAR-IIWBR, Karnal (Haryana) for providing required support for conducting the research work and acknowledge Amity Institute of Microbial technologies, Amity University, Rajasthan, India. This work is the part of PhD programme of first author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaul, N.; Kashyap, P.L.; Kumar, S.; Singh, D.; Singh, G.P. Genetic Diversity and Population Structure of Head Blight Disease Causing Fungus Fusarium graminearum in Northern Wheat Belt of India. J. Fungi 2022, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Saharan, M.S.; Kumar, H.M.A.; Gurjar, M.S.; Aggarwal, R. Fusarium head blight of wheat in India-variability in pathogens associated and sources of resistance: An overview. Indian Phytopathol. 2021, 74, 345–353. [Google Scholar] [CrossRef]
- Wegulo, S.N.; Baenziger, P.S.; Nopsa, J.H.; Bockus, W.W.; Hallen-Adams, H. Management of Fusarium head blight of wheat and barley. Crop Prot. 2015, 73, 100–107. [Google Scholar] [CrossRef]
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium Toxins in Cereals: Occurrence, Legislation, Factors Promoting the Appearance and Their Management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef] [PubMed]
- Degrassi, V.C.-P.G. Biological Control of Fusarium Head Blight by Bacterial Endophytes and Reduction of Deoxynivalenol in Wheat. Adv. Biochem. Biotechnol. 2020, 5, 10103. [Google Scholar] [CrossRef]
- Tini, F.; Beccari, G.; Onofri, A.; Ciavatta, E.; Gardiner, D.M.; Covarelli, L. Fungicides may have differential efficacies towards the main causal agents of Fusarium head blight of wheat. Pest Manag. Sci. 2020, 76, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Mesterházy, Á.; Bartók, T.; Lamper, C. Influence of Wheat Cultivar, Species of Fusarium, and Isolate Aggressiveness on the Efficacy of Fungicides for Control of Fusarium Head Blight. Plant Dis. 2003, 87, 1107–1115. [Google Scholar] [CrossRef]
- Ramírez, M.L.; Chulze, S.; Magan, N. Impact of environmental factors and fungicides on growth and deoxynivalenol production by Fusarium graminearum isolates from Argentinian wheat. Crop Prot. 2004, 23, 117–125. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, Identification, and Complete Genome Assembly of an Endophytic Bacillus velezensis YB-130, Potential Biocontrol Agent against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef]
- Kashyap, P.L.; Kumar, S.; Aggarwal, S.K.; Kaul, N.; Jasrotia, P.; Gupta, A.; Singh, G.P. Resistance inducers and their role in reinforcing wheat defense system against fungal pathogens. J. Cereal Res. 2021, 13, 229–254. [Google Scholar] [CrossRef]
- Fatima, S.; Anjum, T. Identification of a Potential ISR Determinant from Pseudomonas aeruginosa PM12 against Fusarium Wilt in Tomato. Front. Plant Sci. 2017, 8, 848. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cui, J.; Tian, B.; Cao, S.; Zhang, X.; Chen, H. Resistance risk assessment for Fusarium graminearum to pydiflumetofen, a new succinate dehydrogenase inhibitor. Pest Manag. Sci. 2020, 76, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Hou, Y.; Cai, Y.; Zhang, Y.; Li, Y.; Zhou, M. Whole-genome sequencing reveals that mutations in myosin-5 confer resistance to the fungicide phenamacril in Fusarium graminearum. Sci. Rep. 2015, 5, 8248. [Google Scholar] [CrossRef] [PubMed]
- de Chaves, M.A.; Reginatto, P.; da Costa, B.S.; de Paschoal, R.I.; Teixeira, M.L.; Fuentefria, A.M. Fungicide Resistance in Fusarium graminearum Species Complex. Curr. Microbiol. 2022, 79, 62. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.; Kashyap, P.L.; Srivastava, A.K.; Tiwari, R.K. Plant growth promoting and antifungal activity in endophytic Bacillus strains from pearl millet (Pennisetumglaucum). Braz. J. Microbiol. 2020, 51, 229–241. [Google Scholar] [CrossRef]
- Ahlawat, O.P.; Yadav, D.; Kashyap, P.L.; Khippal, A.; Singh, G. Wheat endophytes and their potential role in managing abiotic stress under changing climate. J. Appl. Microbiol. 2022, 132, 2501–2520. [Google Scholar] [CrossRef]
- Pan, D.; Mionetto, A.; Tiscornia, S.; Bettucci, L. Endophytic bacteria from wheat grain as biocontrol agents of Fusarium graminearum and deoxynivalenol production in wheat. Mycotoxin Res. 2015, 31, 137–143. [Google Scholar] [CrossRef]
- Hong, C.E.; Park, J.M. Endophytic bacteria as biocontrol agents against plant pathogens: Current state-of-the-art. Plant Biotechnol. Rep. 2016, 10, 353–357. [Google Scholar] [CrossRef]
- Frank, A.C.; Guzmán, J.P.S.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Bhardwaj, A.K.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K. Bacterial endophyte mediated plant tolerance to salinity: Growth responses and mechanisms of action. World J. Microbiol. Biotechnol. 2020, 36, 26. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K. Functional characterization of endophytic bacilli from pearl millet (Pennisetumglaucum) and their possible role in multiple stress tolerance. Plant Biosyst.-Int. J. Deal. Asp. Plant Biol. 2020, 154, 503–514. [Google Scholar] [CrossRef]
- Shahid, I.; Han, J.; Hanooq, S.; Malik, K.A.; Borchers, C.H.; Mehnaz, S. Profiling of Metabolites of Bacillus spp. And Their Application in Sustainable Plant Growth Promotion and Biocontrol. Front. Sustain. Food Syst. 2021, 5, 605195. [Google Scholar] [CrossRef]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.-W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The Iturin and Fengycin Families of Lipopeptides Are Key Factors in Antagonism of Bacillus subtilis toward Podosphaerafusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Kong, H.; Buyer, J.S.; Lakshman, D.K.; Lydon, J.; Kim, S.D.; Roberts, D.P. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soil borne pathogens of cucumber and pepper. Appl. Microbiol. Biotechnol. 2008, 80, 115–123. [Google Scholar] [CrossRef]
- Mora, I.; Cabrefiga, J.; Montesinos, E. Cyclic Lipopeptide Biosynthetic Genes and Products, and Inhibitory Activity of Plant-Associated Bacillus against Phytopathogenic Bacteria. PLoS ONE 2015, 10, e0127738. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthegrisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef]
- Kalai-Grami, L.; Ben Slimane, I.; Mnari-Hattab, M.; Rezgui, S.; Aouani, M.A.; Hajlaoui, M.R.; Limam, F. Protective effect of Bacillus amyloliquefaciens against infections of Citrus aurantium seedlings by Phomatracheiphila. World J. Microbiol. Biotechnol. 2014, 30, 529–538. [Google Scholar] [CrossRef]
- Jiao, R.; Cai, Y.; He, P.; Munir, S.; Li, X.; Wu, Y.; Wang, J.; Xia, M.; He, P.; Wang, G.; et al. Bacillus amyloliquefaciens YN201732 Produces Lipopeptides with Promising Biocontrol Activity Against Fungal Pathogen Erysiphe cichoracearum. Front. Cell. Infect. Microbiol. 2021, 11, 598999. [Google Scholar] [CrossRef]
- Cheffi, M.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef]
- Tao, A.; Pang, F.; Huang, S.; Yu, G.; Li, B.; Wang, T. Characterisation of endophytic Bacillus thuringiensis strains isolated from wheat plants as biocontrol agents against wheat flag smut. Biocontrol Sci. Technol. 2014, 24, 901–924. [Google Scholar] [CrossRef]
- Rong, S.; Xu, H.; Li, L.; Chen, R.; Gao, X.; Xu, Z. Antifungal activity of endophytic Bacillus safensis B21 and its potential application as a biopesticide to control rice blast. Pestic. Biochem. Physiol. 2020, 162, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Munakata, Y.; Gavira, C.; Genestier, J.; Bourgaud, F.; Hehn, A.; Slezack-Deschaumes, S. Composition and functional comparison of vetiver root endophytic microbiota originating from different geographic locations that show antagonistic activity towards Fusarium graminearum. Microbiol. Res. 2021, 243, 126650. [Google Scholar] [CrossRef]
- da Luz, W.C.; Stockwell, C.A.; Bergstrom, G.C. Biological control of Fusarium graminearum. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; APS Press: St. Paul, MN, USA, 2003; pp. 381–394. [Google Scholar]
- Sharma, A.; Kashyap, P.L.; Srivastava, A.K.; Bansal, Y.K.; Kaushik, R. Isolation and characterization of halotolerant bacilli from chickpea (Cicer arietinum L.) rhizosphere for plant growth promotion and biocontrol traits. Eur. J. Plant Pathol. 2018, 153, 787–800. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual, 10th ed.; Benjamin/Cunnings Science Publishing: San Francisco, CA, USA, 2013; p. 560. [Google Scholar]
- Kumar, S.; Kashyap, P.L.; Singh, R.; Srivastava, A.K. Preservation and Maintenance of Microbial Cultures. In Analyzing Microbes. Springer Protocols Handbooks; Arora, D., Das, S., Sukumar, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 135–152. [Google Scholar] [CrossRef]
- Pospiech, A. A Versatile Quick-Prep of Genomic DNA from Gram-Positive Bacteria. Trends Genet. 1995, 11, 217–218. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-F.; Xu, X.-M.; Zhu, Q.; Deng, M.-C.; Feng, L.; Peng, J.; Yuan, J.-P.; Wang, J.-H. An effective method for the detoxification of cyanide-rich wastewater by Bacillus sp. CN-22. Appl. Microbiol. Biotechnol. 2014, 98, 3801–3807. [Google Scholar] [CrossRef]
- Mora, I.; Cabrefiga, J. Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol. 2011, 14, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Ramarathnam, R.; Bo, S.; Chen, Y.; Fernando, W.G.; Xuewen, G.; De Kievit, T. Molecular and biochemical detection of fengycin and bacillomycin D-producing Bacillus spp., antagonistic to fungal pathogens of canola and wheat. Can. J. Microbiol. 2007, 53, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Stack, R.W.; McMullen, M.P. A Visual Scale to Estimate Severity of Fusarium Head Blight in Wheat; North Dakota State University Extension Service: Fargo, ND, USA, 1995. [Google Scholar]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Hu, X.F.; Chen, J.; Guo, J.F. Two phosphate and potassium solubilizing bacterial isolated from Tiannu Mountain, Zhijiang, China. World J. Microbiol. Biotechnol. 2006, 22, 983–990. [Google Scholar] [CrossRef]
- Dutilloy, E.; Oni, F.E.; Esmaeel, Q.; Clément, C.; Barka, E.A. Plant Beneficial Bacteria as Bioprotectants against Wheat and Barley Diseases. J. Fungi 2022, 8, 632. [Google Scholar] [CrossRef]
- Becher, R.; Hettwer, U.; Karlovsky, P.; Deising, H.B.; Wirsel, S.G.R. Adaptation of Fusarium graminearum to Tebuconazole Yielded Descendants Diverging for Levels of Fitness, Fungicide Resistance, Virulence, and Mycotoxin Production. Phytopathology 2010, 100, 444–453. [Google Scholar] [CrossRef]
- Sessitsch, A.; Reiter, B.; Pfeifer, U.; Wilhelm, E. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 2002, 39, 23–32. [Google Scholar] [CrossRef]
- Pang, F.; Tao, A.; Ayra-Pardo, C.; Wang, T.; Yu, Z.; Huang, S. Plant organ- and growth stage-diversity of endophytic bacteria with potential as biofertilisers isolated from wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 276. [Google Scholar] [CrossRef]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 2016, 405, 381–396. [Google Scholar] [CrossRef]
- Marag, P.S.; Suman, A.; Gond, S. Prospecting endophytic bacterial colonisation and their potential plant growth promoting attributes in hybrid maize (Zea mays L.). Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1292–1304. [Google Scholar] [CrossRef]
- Wang, G.; Meng, J.; Tian, T.; Xiao, X.; Zhang, B.; Xiao, Y. Endophytic Bacillus velezensis strain B-36 is a potential biocontrol agent against lotus rot caused by Fusarium oxysporum. J. Appl. Microbiol. 2020, 128, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, D.J.; Goswami, G.; Gautom, T.; Parveen, A.; Das, P.; Barooah, M.; Boro, R.C. Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 2019, 19, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guan, Y.; Dong, Y.; Zhao, L.; Rong, S.; Chen, W.; Lv, M.; Xu, H.; Gao, X.; Chen, R.; et al. Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaportheoryzae. PLoS ONE 2018, 13, e0203505. [Google Scholar] [CrossRef]
- Larran, S.; Simon, M.R.; Moreno, M.V.; Siurana, M.S.; Perelló, A. Endophytes from wheat as biocontrol agents against tan spot disease. Biol. Control 2016, 92, 17–23. [Google Scholar] [CrossRef]
- Kashyap, P.L.; Srivastava, A.K.; Tiwari, S.P.; Kumar, S. Microbes for Climate Resilient Agriculture; John Wiley & Sons: Hoboken, NJ, USA, 2018; p. 358. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Kashyap, P.L.; Srivastava, M. The Plant Microbiome in Sustainable Agriculture; John Wiley & Sons: Hoboken, NJ, USA, 2021; p. 320. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Kaur, T.; Sheikh, I.; Yadav, A.N.; Kumar, V.; Suman, A.; Dhaliwal, H.S. Endophytic Microbes from Diverse Wheat Genotypes and Their Potential Biotechnological Applications in Plant Growth Promotion and Nutrient Uptake. Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2020, 90, 969–979. [Google Scholar] [CrossRef]
- Solanki, M.K.; Kumar, S.; Pandey, A.K.; Srivastava, S.; Singh, R.K.; Kashyap, P.L.; Srivastava, A.K.; Arora, D.K. Diversity and antagonistic potential of Bacillus spp. associated to the rhizosphere of tomato for the management of Rhizoctoniasolani. Biocontrol Sci. Technol. 2012, 22, 203–217. [Google Scholar] [CrossRef]
- Besset-Manzoni, Y.; Joly, P.; Brutel, A.; Gerin, F.; Soudière, O.; Langin, T.; Prigent-Combaret, C. Does in vitro selection of biocontrol agents guarantee success in planta? A study case of wheat protection against Fusarium seedling blight by soil bacteria. PLoS ONE 2019, 14, e0225655. [Google Scholar] [CrossRef]
- Gao, X.; Gong, Y.; Huo, Y.; Han, Q.; Kang, Z.; Huang, L. Endophytic Bacillus subtilis Strain E1R-J Is a Promising Biocontrol Agent for Wheat Powdery Mildew. Biomed Res. Int. 2015, 2015, 462645. [Google Scholar] [CrossRef]
- Comby, M.; Gacoin, M.; Robineau, M.; Rabenoelina, F.; Ptas, S.; Dupont, J.; Profizi, C.; Baillieul, F. Screening of wheat endophytes as biological control agents against Fusarium head blight using two different in vitro tests. Microbiol. Res. 2017, 202, 11–20. [Google Scholar] [CrossRef]
- Jamali, H.; Sharma, A.; Kashyap, P.L.; Hi, R.; Srivastava, A.K. Exploitation of Multifarious Abiotic Stresses, Antagonistic Activity and Plant Growth Promoting Attributes of Bacillus amyloliquefaciens AH53 for Sustainable Agriculture Production. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 751–763. [Google Scholar] [CrossRef]
- Shukla, N.; Singh, D.; Tripathi, A.; Kumari, P.; Gupta, R.K.; Singh, S.; Shanker, K.; Singh, A. Synergism of endophytic Bacillus subtilis and Klebsiella aerogenes modulates plant growth and bacoside biosynthesis in Bacopa monnieri. Front. Plant Sci. 2022, 13, 896856. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Verma, A.; Pal, G.; Anubha; White, J.F.; Verma, S.K. Seed Endophytic Bacteria of Pearl Millet (Pennisetumglaucum L.) Promote Seedling Development and Defend Against a Fungal Phytopathogen. Front. Microbiol. 2021, 12, 774293. [Google Scholar] [CrossRef] [PubMed]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Kumar, K.; Pal, G.; Verma, A.; Verma, S.K. Seed inhabiting bacterial endophytes of finger millet (Eleusinecoracana L.) promote seedling growth and development, and protect from fungal disease. S. Afr. J. Bot. 2020, 134, 91–98. [Google Scholar] [CrossRef]
- Khanna, A.; Raj, K.; Kumar, P.; Wati, L. Antagonistic and growth-promoting potential of multifarious bacterial endophytes against Fusarium wilt of chickpea. Egypt. J. Biol. Pest Control 2022, 32, 17. [Google Scholar] [CrossRef]
- Agarwal, H.; Dowarah, B.; Baruah, P.M.; Bordoloi, K.S.; Krishnatreya, D.B.; Agarwala, N. Endophytes from Gnetumgnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol. Res. 2020, 238, 126503. [Google Scholar] [CrossRef]
- Ahmed, A.; Munir, S.; He, P.; Li, Y.; He, P.; Yixin, W.; He, Y. Biocontrol arsenals of bacterial endophyte: An imminent triumph against clubroot disease. Microbiol. Res. 2020, 241, 126565. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Dogan, G.; Taskin, B. Hydrolytic Enzymes Producing Bacterial Endophytes of Some Poaceae Plants. Pol. J. Microbiol. 2021, 70, 297–304. [Google Scholar] [CrossRef]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef]
- Singh, R.K.; Kumar, D.P.; Singh, P.; Solanki, M.K.; Srivastava, S.; Kashyap, P.L.; Kumar, S.; Srivastava, A.K.; Singhal, P.K.; Arora, D.K. Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. Plant Growth Regul. 2014, 73, 91–101. [Google Scholar] [CrossRef]
- Vinodkumar, S.; Nakkeeran, S.; Renukadevi, P.; Malathi, V.G. Biocontrol Potentials of Antimicrobial Peptide Producing Bacillus Species: Multifaceted Antagonists for the Management of Stem Rot of Carnation Caused by Sclerotiniasclerotiorum. Front. Microbiol. 2017, 8, 446. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Singh, R.K.; Srivastava, S.; Kumar, S.; Kashyap, P.L.; Srivastava, A.K. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctoniasolani in tomato. J. Basic Microbiol. 2015, 55, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Kim, P.I.; Ryu, J.; Kim, Y.H.; Chi, Y.-T. Production of Biosurfactant Lipopeptides Iturin A, Fengycin and Surfactin A from Bacillus subtilis CMB32 for Control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 2010, 20, 138–145. [Google Scholar] [CrossRef]
- Toral, L.; Rodríguez, M.; Béjar, V.; Sampedro, I. Antifungal Activity of Lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea. Front. Microbiol. 2018, 9, 1315. [Google Scholar] [CrossRef]
- Nandhini, S.; Sendhilvel, V.; Babu, S. Endophytic bacteria from tomato and their efficacy against Fusarium oxysporum f.sp. lycopersici, the wilt pathogen. J. Biopest. 2012, 5, 178–185. [Google Scholar]
- Sharma, C.K.; Vishnoi, V.K.; Dubey, R.C.; Maheshwari, D.K. A twin rhizospheric bacterial consortium induces systemic resistance to a phytopathogen Macrophominaphaseolina in mug bean. Rhizosphere 2018, 5, 71–75. [Google Scholar] [CrossRef]
- Hays, S.G.; Patrick, W.G.; Ziesack, M.; Oxman, N.; A Silver, P. Better together: Engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 2015, 36, 40–49. [Google Scholar] [CrossRef]
- Minchev, Z.; Kostenko, O.; Soler, R.; Pozo, M.J. Microbial Consortia for Effective Biocontrol of Root and Foliar Diseases in Tomato. Front. Plant Sci. 2021, 12, 756368. [Google Scholar] [CrossRef] [PubMed]
- Samain, E.; Ernenwein, C.; Aussenac, T.; Selim, S. Effective and durable systemic wheat-induced resistance by a plant-growth-promoting rhizobacteria consortium of Paenibacillus sp. strain B2 and Arthrobacter spp. strain AA against Zymoseptoriatritici and drought stress. Physiol. Mol. Plant Pathol. 2022, 119, 101830. [Google Scholar] [CrossRef]
- Abdeljalil, N.O.-B.; Renault, D.; Gerbore, J.; Vallance, J.; Daami-Remadi, P.R.A.M. Comparative Efficacy of Three Tomato-Associated Rhizobacteria used Singly or in Combination in Suppressing Rhizoctonia Root Rot and Enhancing Tomato Growth. J. Microb. Biochem. Technol. 2016, 8, 110–119. [Google Scholar] [CrossRef]
- Palazzini, J.M.; Alberione, E.; Torres, A.; Donat, C.; Köhl, J.; Chulze, S. Biological control of Fusarium graminearum sensu stricto, causal agent of Fusarium head blight of wheat, using formulated antagonists under field conditions in Argentina. Biol. Control. 2016, 94, 56–61. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Muhae-Ud-Din, G.; Ali, M.A.; Naveed, M.; Naveed, K.; Abbas, A.; Anwar, J.; Tanveer, M.H. Consortium application of endophytic bacteria and fungi improves grain yield and physiological attributes in advanced lines of bread wheat. Turk. J. Agric.-Food Sci. Technol. 2018, 6, 136–144. [Google Scholar] [CrossRef]
- Resti, Z.; Warnita; Liswarni, Y. Endophytic bacterial consortia as biocontrol of purple blotch and plant growth promoters of shallots. IOP Conf. Ser. Earth Environ. Sci. 2021, 741, 012009. [Google Scholar] [CrossRef]
- Sundaramoorthy, S.; Raguchander, T.; Ragupathi, N.; Samiyappan, R. Combinatorial effect of endophytic and plant growth promoting rhizobacteria against wilt disease of Capsicum annum L. caused by Fusarium solani. Biol. Control 2012, 60, 59–67. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).