New Insights into the Biology of the Emerging Tembusu Virus
Abstract
1. Introduction
2. Tembusu Virus
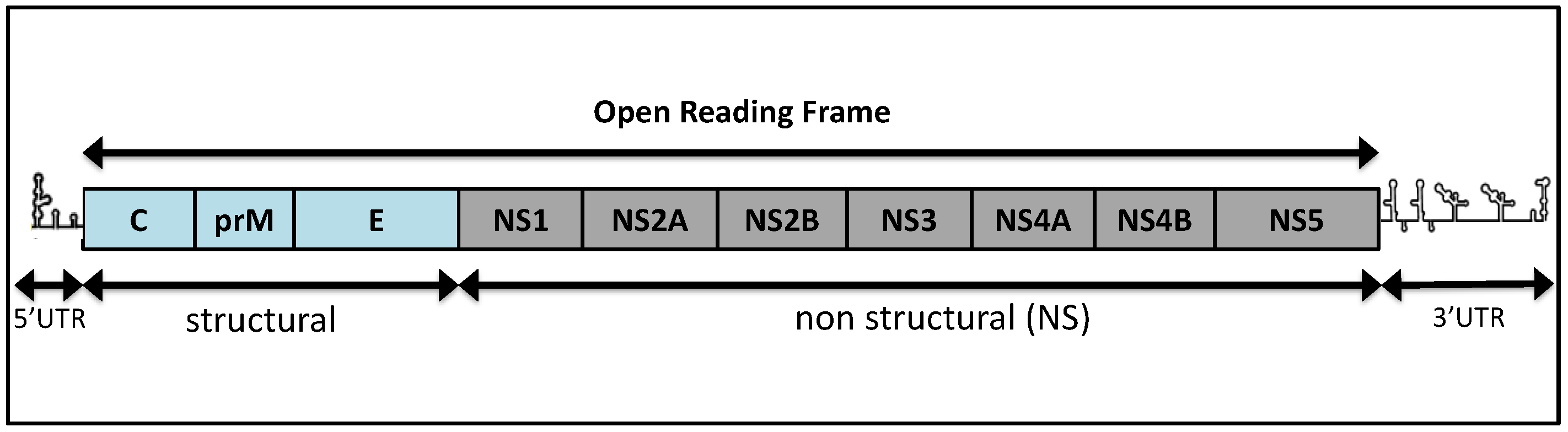
2.1. Genomic Organization and Replication
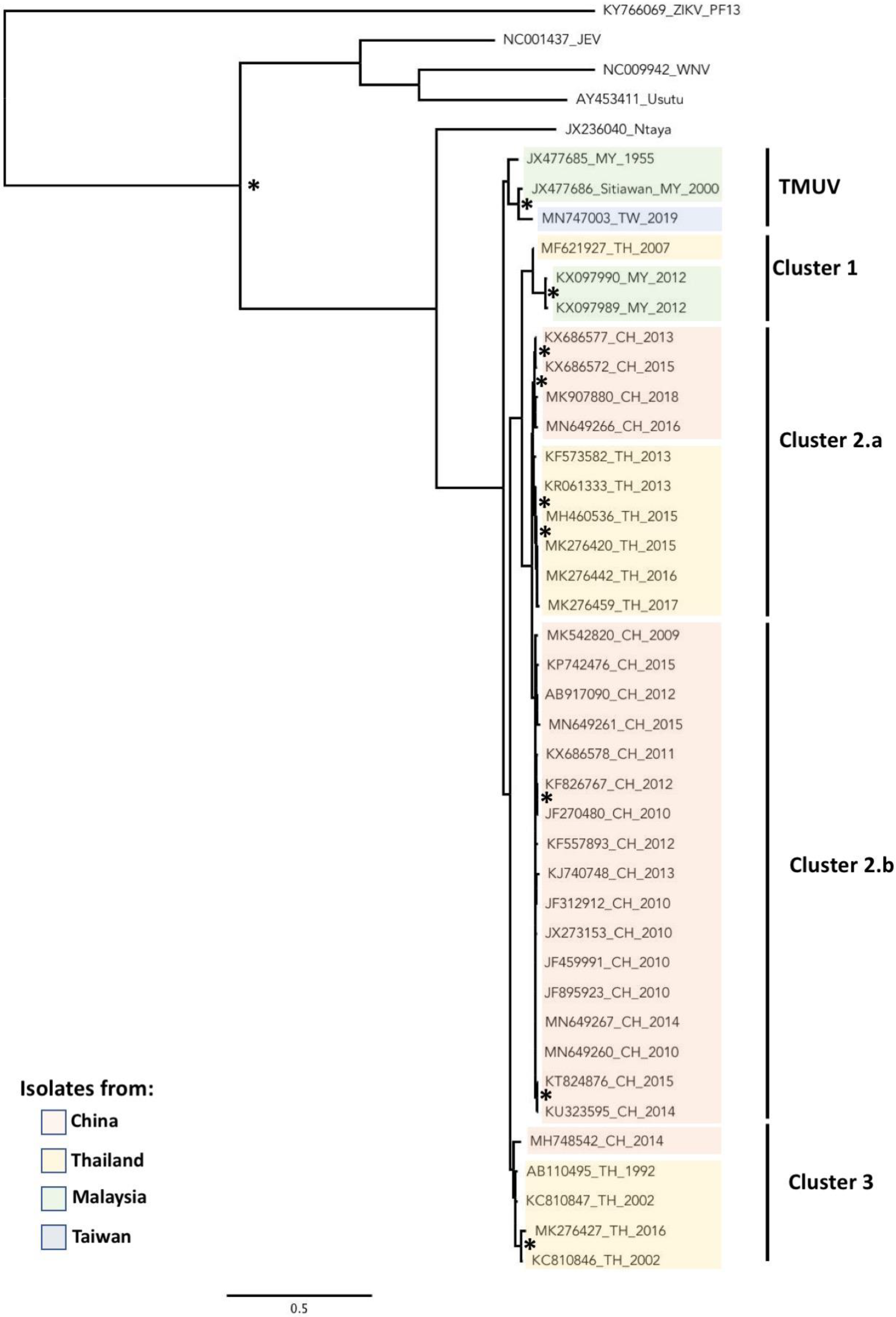
2.2. Phylogeny
3. Epidemiology
3.1. Vector
3.2. Vertebrate Host
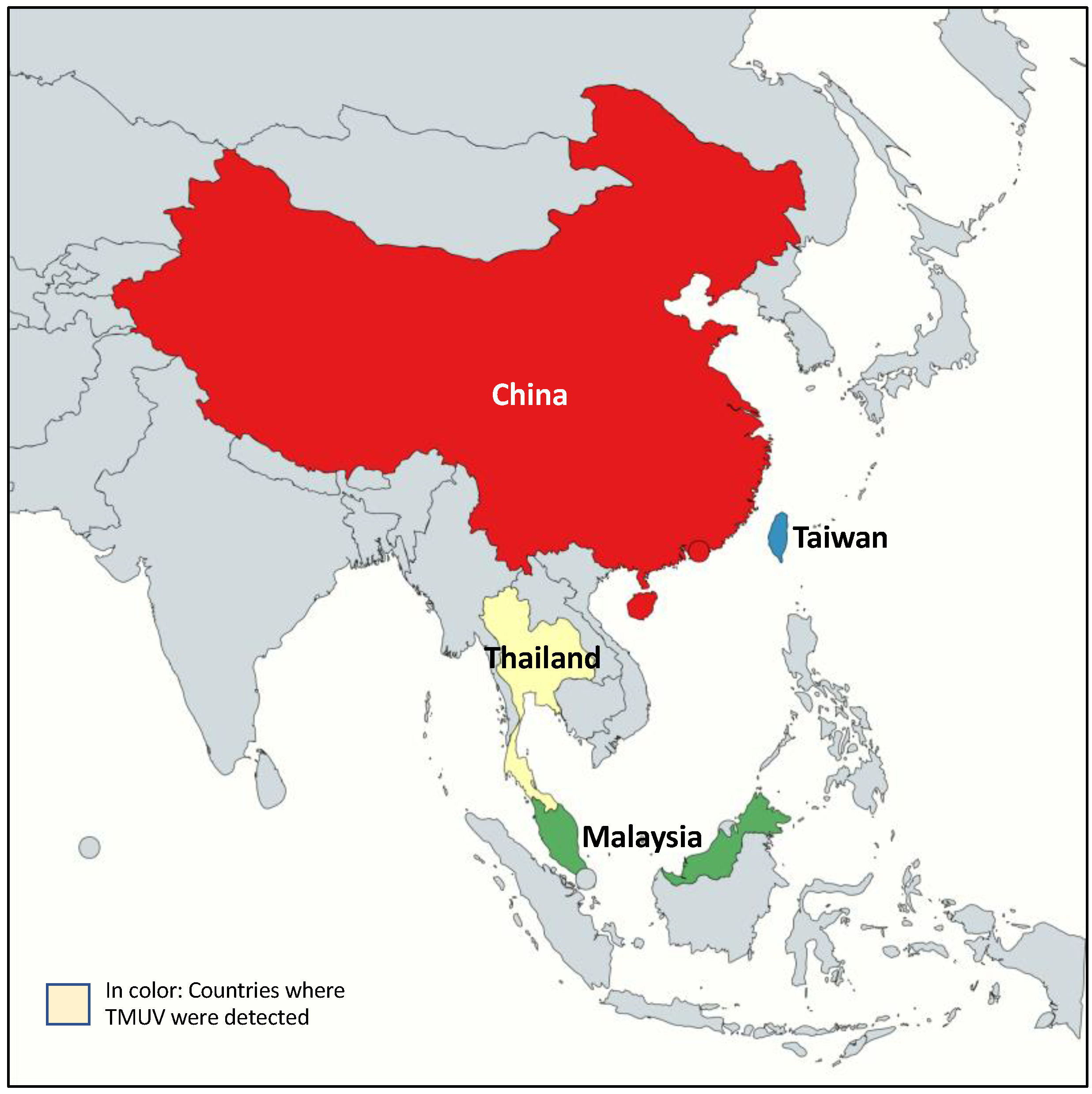
3.3. Geographic Distribution
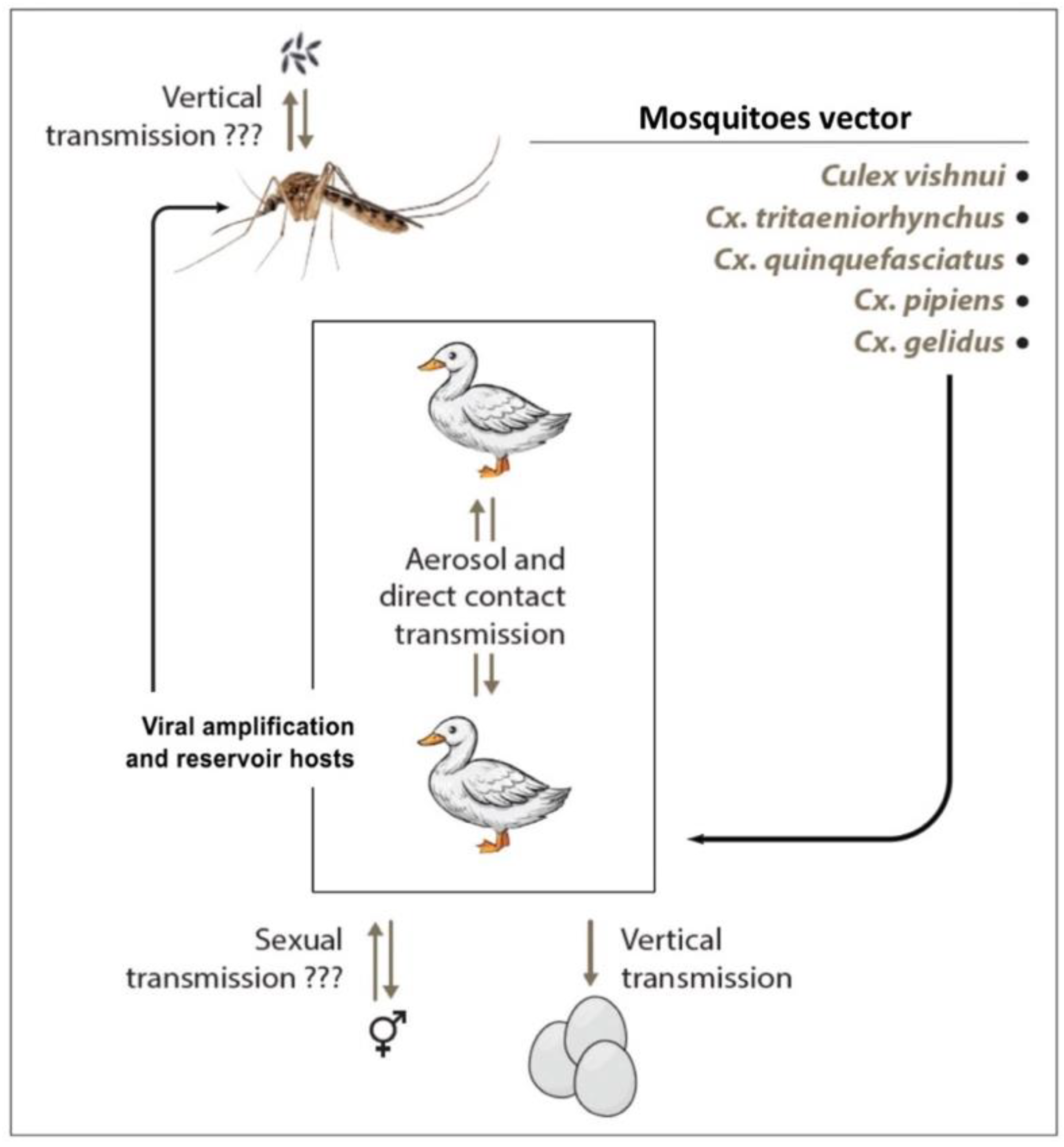
3.4. Transmission
4. Clinical Features and Pathogenicity
4.1. Symptomatology/Disease Presentations
4.2. Detection and Diagnostics
4.3. Cell Biology Tools
4.4. Innate Immune Response and Viral Evasion
4.5. Treatment and Vaccine Development
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Pandey, B.D.; Karabatsos, N.; Cropp, B.; Tagaki, M.; Tsuda, Y.; Ichinose, A.; Igarashi, A. Identification of a flavivirus isolated from mosquitos in Chiang Mai Thailand. Southeast Asian J. Trop. Med. Public Health 1999, 30, 161–165. [Google Scholar]
- Platt, G.S.; Way, H.J.; Bowen, E.T.; Simpson, D.I.; Hill, M.N.; Kamath, S.; Bendell, P.J.; Heathcote, O.H. Arbovirus infections in Sarawak, October 1968–February 1970 Tembusu and Sindbis virus isolations from mosquitoes. Ann. Trop. Med. Parasitol. 1975, 69, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Homonnay, Z.G.; Kovács, E.W.; Bányai, K.; Albert, M.; Fehér, E.; Mató, T.; Tatár-Kis, T.; Palya, V. Tembusu-like flavivirus (Perak virus) as the cause of neurological disease outbreaks in young Pekin ducks. Avian Pathol. 2014, 43, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Li, S.; Hu, X.; Yu, X.; Wang, Y.; Liu, P.; Lu, X.; Zhang, G.; Liu, D.; Li, X.; et al. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS ONE 2011, 6, e18106. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Tsukamoto, K.; Abd Hamid, M.; Darus, A.; Lian, T.C.; Sam, L.S.; Yok, C.N.; Di, K.B.; Lim, K.T.; Yamaguchi, S.; et al. Encephalitis and retarded growth of chicks caused by Sitiawan virus, a new isolate belonging to the genus Flavivirus. Am. J. Trop. Med. Hyg. 2000, 63, 94–101. [Google Scholar] [CrossRef]
- Benzarti, E.; Linden, A.; Desmecht, D.; Garigliany, M. Mosquito-borne epornitic flaviviruses: An update and review. J. Gen. Virol. 2019, 100, 119–132. [Google Scholar] [CrossRef]
- Yun, T.; Ye, W.; Ni, Z.; Zhang, D.; Zhang, C. Identification and molecular characterization of a novel flavivirus isolated from Pekin ducklings in China. Vet. Microbiol. 2012, 157, 311–319. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus entry receptors: An update. Viruses 2014, 6, 69–88. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar]
- Wu, S.; Wu, Z.; Wu, Y.; Wang, T.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; et al. Heparin sulfate is the attachment factor of duck Tembus virus on both BHK21 and DEF cells. Virol. J. 2019, 16, 134. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Helenius, A. Virus entry at a glance. J. Cell Sci. 2013, 126, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, D.; Han, K.; Huang, X.; Liu, Y.; Liu, Q.; Yang, J.; Li, S.; Li, Y. Tembusu virus enters BHK-21 cells through a cholesterol-dependent and clathrin-mediated endocytosis pathway. Microb. Pathog. 2020, 147, 104242. [Google Scholar] [CrossRef] [PubMed]
- Baloch, A.S.; Liu, C.; Liang, X.; Liu, Y.; Chen, J.; Cao, R.; Zhou, B. Avian Flavivirus Enters BHK-21 Cells by a Low pH-Dependent Endosomal Pathway. Viruses 2019, 11, 1112. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, W.; Wu, Y.; Wang, T.; Wu, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; et al. Binding of the Duck Tembusu Virus Protease to STING Is Mediated by NS2B and Is Crucial for STING Cleavage and for Impaired Induction of IFN-β. J. Immunol. 2019, 203, 3374–3385. [Google Scholar] [CrossRef]
- Finol, E.; Ooi, E.E. Evolution of Subgenomic RNA Shapes Dengue Virus Adaptation and Epidemiological Fitness. iScience 2019, 16, 94–105. [Google Scholar] [CrossRef]
- Slonchak, A.; Khromykh, A.A. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antivir. Res. 2018, 159, 13–25. [Google Scholar] [CrossRef]
- Pompon, J.; Manuel, M.; Ng, G.K.; Wong, B.; Shan, C.; Manokaran, G.; Soto-Acosta, R.; Bradrick, S.S.; Ooi, E.E.; Missé, D.; et al. Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog. 2017, 13, e1006535. [Google Scholar] [CrossRef]
- Papageorgiou, L.; Loukatou, S.; Koumandou, V.L.; Makałowski, W.; Megalooikonomou, V.; Vlachakis, D.; Kossida, S. Structural models for the design of novel antiviral agents against Greek Goat Encephalitis. PeerJ 2014, 2, e664. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Jimenez-Clavero, M.A.; Leblond, A.; Durand, B.; Nowotny, N.; Leparc-Goffart, I.; Zientara, S.; Jourdain, E.; Lecollinet, S. Flaviviruses in Europe: Complex circulation patterns and their consequences for the diagnosis and control of West Nile disease. Int. J. Environ. Res. Public Health 2013, 10, 6049–6083. [Google Scholar] [CrossRef]
- Liu, P.; Lu, H.; Li, S.; Moureau, G.; Deng, Y.Q.; Wang, Y.; Zhang, L.; Jiang, T.; de Lamballerie, X.; Qin, C.F.; et al. Genomic and antigenic characterization of the newly emerging Chinese duck egg-drop syndrome flavivirus: Genomic comparison with Tembusu and Sitiawan viruses. J. Gen. Virol. 2012, 93, 2158–2170. [Google Scholar] [CrossRef]
- Cao, Z.; Zhang, C.; Liu, Y.; Ye, W.; Han, J.; Ma, G.; Zhang, D.; Xu, F.; Gao, X.; Tang, Y.; et al. Tembusu virus in ducks, china. Emerg. Infect. Dis. 2011, 17, 1873–1875. [Google Scholar] [CrossRef]
- Wallace, H.; Rudinick, A.; Rajagopal, V. Activity of Tembusu and Umbre viruses in a Malaysian community: Mosquito studies. Mosquito News 1977, 37, 35–42. [Google Scholar]
- Leake, C.J.; Ussery, M.A.; Nisalak, A.; Hoke, C.H.; Andre, R.G.; Burke, D.S. Virus isolations from mosquitoes collected during the 1982 Japanese encephalitis epidemic in northern Thailand. Trans. R. Soc. Trop Med. Hyg. 1986, 80, 831–837. [Google Scholar] [CrossRef]
- Peng, S.H.; Su, C.L.; Chang, M.C.; Hu, H.C.; Yang, S.L.; Shu, P.Y. Genome Analysis of a Novel Tembusu Virus in Taiwan. Viruses 2020, 12, 567. [Google Scholar] [CrossRef]
- Cheng, M.C.; Lee, M.S.; Ho, Y.H.; Chyi, W.L.; Wang, C.H. Avian influenza monitoring in migrating birds in Taiwan during 1998–2007. Avian Dis. 2010, 54, 109–114. [Google Scholar] [CrossRef]
- Gao, X.; Liu, H.; Wang, H.; Fu, S.; Guo, Z.; Liang, G. Southernmost Asia is the source of Japanese encephalitis virus (genotype 1) diversity from which the viruses disperse and evolve throughout Asia. PLoS Negl. Trop. Dis. 2013, 7, e2459. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Ninvilai, P.; Nonthabenjawan, N.; Limcharoen, B.; Tunterak, W.; Oraveerakul, K.; Banlunara, W.; Amonsin, A.; Thontiravong, A. The presence of duck Tembusu virus in Thailand since 2007: A retrospective study. Transbound. Emerg. Dis. 2018, 65, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Cui, Y.; Li, Y.; Wang, Y. The spread of Tembusu virus in China from 2010 to 2019. Virus Res. 2021, 198374. [Google Scholar] [CrossRef]
- Ninvilai, P.; Tunterak, W.; Oraveerakul, K.; Amonsin, A.; Thontiravong, A. Genetic characterization of duck Tembusu virus in Thailand, 2015–2017: Identification of a novel cluster. Transbound. Emerg. Dis. 2019, 66, 1982–1992. [Google Scholar] [CrossRef]
- Guo, X.; Jiang, T.; Jiang, Y.; Zhao, T.; Li, C.; Dong, Y.; Xing, D.; Qin, C. Potential Vector Competence of Mosquitoes to Transmit Baiyangdian Virus, a New Tembusu-Related Virus in China. Vector Borne Zoonotic Dis. 2020, 20, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Nitatpattana, N.; Apiwatanason, C.; Nakgoi, K.; Sungvornyothin, S.; Pumchompol, J.; Wanlayaporn, D.; Chaiyo, K.; Siripholvat, V.; Yoksan, S.; Gonzalez, J.-P. Isolation of Tembusu virus from Culex quinquefasciatus in Kanchanaburi Province, Thailand. Southeast Asian J. Trop. Med. Public Health 2017, 48, 546–551. [Google Scholar]
- Sanisuriwong, J.; Yurayart, N.; Thontiravong, A.; Tiawsirisup, S. Vector competence of Culex tritaeniorhynchus and Culex quinquefasciatus (Diptera: Culicidae) for duck Tembusu virus transmission. Acta Trop. 2021, 214, 105785. [Google Scholar] [CrossRef]
- Sanisuriwong, J.; Yurayart, N.; Thontiravong, A.; Tiawsirisup, S. Duck Tembusu virus detection and characterization from mosquitoes in duck farms, Thailand. Transbound. Emerg. Dis. 2020, 67, 1082–1088. [Google Scholar] [CrossRef]
- O’Guinn, M.L.; Turell, M.J.; Kengluecha, A.; Jaichapor, B.; Kankaew, P.; Miller, R.S.; Endy, T.P.; Jones, J.W.; Coleman, R.E.; Lee, J.S. Field detection of Tembusu virus in western Thailand by rt-PCR and vector competence determination of select culex mosquitoes for transmission of the virus. Am. J. Trop. Med. Hyg. 2013, 89, 1023–1028. [Google Scholar] [CrossRef]
- Tang, Y.; Diao, Y.; Chen, H.; Ou, Q.; Liu, X.; Gao, X.; Yu, C.; Wang, L. Isolation and genetic characterization of a tembusu virus strain isolated from mosquitoes in Shandong, China. Transbound. Emerg. Dis. 2015, 62, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Guo, X.; Fu, S.; Feng, Y.; Tao, X.; Gao, X.; Song, J.; Yang, Z.; Zhou, H.; Liang, G. The genetic characteristics and evolution of Tembusu virus. Vet. Microbiol. 2017, 201, 32–41. [Google Scholar] [CrossRef]
- Yu, G.; Lin, Y.; Tang, Y.; Diao, Y. Evolution of Tembusu Virus in Ducks, Chickens, Geese, Sparrows, and Mosquitoes in Northern China. Viruses 2018, 10, 485. [Google Scholar] [CrossRef]
- Tanner, W.D.; Toth, D.J.; Gundlapalli, A.V. The pandemic potential of avian influenza A(H7N9) virus: A review. Epidemiol. Infect. 2015, 143, 3359–3374. [Google Scholar] [CrossRef] [PubMed]
- Tunterak, W.; Prakairungnamthip, D.; Ninvilai, P.; Bunyapisitsopa, S.; Oraveerakul, K.; Sasipreeyajan, J.; Amonsin, A.; Thontiravong, A. Serological evidence of duck Tembusu virus infection in free-grazing ducks, Thailand. Transbound. Emerg. Dis. 2018, 65, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Mogasale, V.; Lim, J.K.; Carabali, M.; Lee, K.S.; Sirivichayakul, C.; Dang, D.A.; Palencia-Florez, D.C.; Nguyen, T.H.A.; Riewpaiboon, A.; et al. A multi-country study of the economic burden of dengue fever: Vietnam, Thailand, and Colombia. PLoS Negl. Trop. Dis. 2017, 11, e0006037. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Vasilakis, N.; Musso, D. History and Emergence of Zika Virus. J. Infect. Dis. 2017, 216, S860–S867. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, N.D.; Kilbourn, A.M.; Karesh, W.B.; Rahman, H.A.; Bosi, E.J.; Cropp, B.C.; Andau, M.; Spielman, A.; Gubler, D.J. Sylvatic transmission of arboviruses among Bornean orangutans. Am. J. Trop. Med. Hyg. 2001, 64, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gao, X.; Diao, Y.; Feng, Q.; Chen, H.; Liu, X.; Ge, P.; Yu, C. Tembusu virus in human, China. Transbound. Emerg. Dis. 2013, 60, 193–196. [Google Scholar] [CrossRef]
- Yan, P.; Zhao, Y.; Zhang, X.; Xu, D.; Dai, X.; Teng, Q.; Yan, L.; Zhou, J.; Ji, X.; Zhang, S.; et al. An infectious disease of ducks caused by a newly emerged Tembusu virus strain in mainland China. Virology 2011, 417, 1–8. [Google Scholar] [CrossRef]
- Yu, K.; Sheng, Z.Z.; Huang, B.; Ma, X.; Li, Y.; Yuan, X.; Qin, Z.; Wang, D.; Chakravarty, S.; Li, F.; et al. Structural, antigenic, and evolutionary characterizations of the envelope protein of newly emerging Duck Tembusu Virus. PLoS ONE 2013, 8, e71319. [Google Scholar] [CrossRef]
- Yun, T.; Zhang, D.; Ma, X.; Cao, Z.; Chen, L.; Ni, Z.; Ye, W.; Yu, B.; Hua, J.; Zhang, Y.; et al. Complete genome sequence of a novel flavivirus, duck tembusu virus, isolated from ducks and geese in china. J. Virol. 2012, 86, 3406–3407. [Google Scholar] [CrossRef]
- Huang, X.; Qiu, H.; Peng, X.; Zhao, W.; Lu, X.; Mo, K.; Yan, Y.; Liao, M.; Zhou, J. Molecular analysis and serological survey of Tembusu virus infection in Zhejiang, China, 2010–2016. Arch. Virol. 2018, 163, 3225–3234. [Google Scholar] [CrossRef]
- Liu, M.; Chen, S.; Chen, Y.; Liu, C.; Yin, X.; Li, G.; Zhang, Y. Adapted Tembusu-like virus in chickens and geese in China. J. Clin. Microbiol. 2012, 50, 2807–2809. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, K.; Zhao, D.; Liu, Y.; Zhang, J.; Niu, H.; Zhang, K.; Zhu, J.; Wu, D.; Gao, L.; et al. Identification and molecular characterization of a novel flavivirus isolated from geese in China. Res. Vet. Sci. 2013, 94, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Huang, X.; Li, Y.; Zhao, D.; Liu, Y.; Zhou, X.; You, Y.; Xie, X. Complete genome sequence of goose tembusu virus, isolated from jiangnan white geese in Jiangsu, China. Genome Announc. 2013, 1, e0023612. [Google Scholar] [CrossRef]
- Tang, Y.; Diao, Y.; Yu, C.; Gao, X.; Ju, X.; Xue, C.; Liu, X.; Ge, P.; Qu, J.; Zhang, D. Characterization of a Tembusu virus isolated from naturally infected house sparrows (Passer domesticus) in Northern China. Transbound. Emerg. Dis. 2013, 60, 152–158. [Google Scholar] [CrossRef]
- Li, L.; An, H.; Sun, M.; Dong, J.; Yuan, J.; Hu, Q. Identification and genomic analysis of two duck-origin Tembusu virus strains in southern China. Virus Genes 2012, 45, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, X.; Tang, Y.; Zhang, Y.; Ti, J.; Gao, X.; Diao, Y. Complete genome sequences of two waterfowl-origin tembusu virus strains isolated in Shandong province, China. Genome Announc. 2013, 1, e00789-13. [Google Scholar] [CrossRef]
- Wang, Q.; Wen, Y.; Huang, Y.; Wu, Y.; Cai, Y.; Xu, L.; Wang, C.; Li, A.; Wu, B.; Chen, J. Isolation and identification of Duck tembusu virus strain lH and development of latex-agglutination diagnostic method for rapid detection of antibodies. Avian Dis. 2014, 58, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Chakritbudsabong, W.; Taowan, J.; Lertwatcharasarakul, P.; Phattanakunanan, S.; Munkhong, A.; Songserm, T.; Chaichoun, K. Genomic characterization of a new Tembusu flavivirus from domestic ducks in Thailand. Thai J. Vet. Med. 2015, 45, 419. [Google Scholar]
- Zhu, K.; Huang, J.; Jia, R.; Zhang, B.; Wang, M.; Zhu, D.; Chen, S.; Liu, M.; Yin, Z.; Cheng, A. Identification and molecular characterization of a novel duck Tembusu virus isolate from Southwest China. Arch. Virol. 2015, 160, 2781–2790. [Google Scholar] [CrossRef]
- Xie, Z.; Zeng, T.; Xie, L.; Deng, X.; Liu, J.; Fan, Q.; Pang, Y.; Luo, S. Genome Analysis of a Tembusu Virus, GX2013H, Isolated from a Cheery Valley Duck in Guangxi, China. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Zeng, T.; Xie, Z.; Xie, L.; Deng, X.; Huang, L.; Luo, S.; Huang, J. Identification and Whole-Genome Sequence Analysis of Tembusu Virus GX2013G, Isolated from a Cherry Valley Duckling in Southern China. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, C.; Wang, H.; Yan, Y.; Ding, C.; Sun, J. Complete genome sequence of duck tembusu virus isolated from pekin ducks in Shanghai, China. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Thontiravong, A.; Ninvilai, P.; Tunterak, W.; Nonthabenjawan, N.; Chaiyavong, S.; Angkabkingkaew, K.; Mungkundar, C.; Phuengpho, W.; Oraveerakul, K.; Amonsin, A. Tembusu-Related Flavivirus in Ducks, Thailand. Emerg. Infect. Dis. 2015, 21, 2164–2167. [Google Scholar] [CrossRef] [PubMed]
- Tunterak, W.; Prakairungnamthip, D.; Ninvilai, P.; Bunyapisitsopa, S.; Oraveerakul, K.; Sasipreeyajan, J.; Amonsin, A.; Thontiravong, A. Response to “A comment on ‘Serological evidence of duck Tembusu virus infection in free-grazing ducks, Thailand’”. Transbound. Emerg. Dis. 2019, 66, 1098–1099. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Wang, H.; Wei, L.; Zhang, M.; Yang, J.; Chen, H.; Tang, Y.; Diao, Y. Epidemiological investigation of H9 avian influenza virus, Newcastle disease virus, Tembusu virus, goose parvovirus and goose circovirus infection of geese in China. Transbound. Emerg. Dis. 2018, 65, e304–e316. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liu, Y.; Zhao, D.; Huang, X.; Yang, J.; Liu, Q.; An, F.; Xu, T.; Li, Y. Tembusu Virus Strain TMUV-JS06, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KR869106 (accessed on 24 March 2021).
- Zhou, X.; Zhang, T.; Song, D.; Huang, T.; Peng, Q.; Chen, Y.; Li, A.; Zhang, F.; Wu, Q.; Ye, Y.; et al. Whole-Genome Sequence of Duck Tembusu Virus Strain DTMUV/CH/2014, Isolated in China. Genome Announc. 2016, 4, e01657-15. [Google Scholar] [CrossRef]
- Li, Y.; Hu, F.; Liu, C.; Yu, K.; Ma, X.; Huang, B.; Song, M.; Wu, J. Tembusu Virus Isolate SD14, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MH748542 (accessed on 24 March 2021).
- Li, L.; Sun, M.; Dong, J.; Kuang, R.; Zhang, J.; Liu, Z. Tembusu Virus Isolate HZ4-2015, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX686571 (accessed on 24 March 2021).
- Li, L.; Sun, M.; Dong, J.; Kuang, R.; Zhang, J.; Liu, Z. Tembusu Virus Isolate HZ1-2015, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX686570 (accessed on 24 March 2021).
- Yan, Z.; Shen, H.; Wang, Z.; Lin, W.; Xie, Q.; Bi, Y.; Chen, F. Isolation and Characterization of a Novel Tembusu Virus Circulating in Muscovy Ducks in South China. Transbound. Emerg. Dis. 2017, 64, e15–e17. [Google Scholar] [CrossRef]
- Tunterak, W.; Prakairungnamthip, D.; Ninvilai, P.; Tiawsirisup, S.; Oraveerakul, K.; Sasipreeyajan, J.; Amonsin, A.; Thontiravong, A. Patterns of duck Tembusu virus infection in ducks, Thailand: A serological study. Poult. Sci. 2021, 100, 537–542. [Google Scholar] [CrossRef]
- Pulmanausahakul, R.; Ketsuwan, K.; Jaimipuk, T.; Smith, D.R.; Auewarakul, P.; Songserm, T. Detection of antibodies to duck tembusu virus in human population with or without the history of contact with ducks. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Martins, M.M.; Alves da Cunha, A.J.L.; Robaina, J.R.; Raymundo, C.E.; Barbosa, A.P.; Medronho, R.A. Fetal, neonatal, and infant outcomes associated with maternal Zika virus infection during pregnancy: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0246643. [Google Scholar] [CrossRef]
- Li, X.; Shi, Y.; Liu, Q.; Wang, Y.; Li, G.; Teng, Q.; Zhang, Y.; Liu, S.; Li, Z. Airborne Transmission of a Novel Tembusu Virus in Ducks. J. Clin. Microbiol. 2015, 53, 2734–2736. [Google Scholar] [CrossRef]
- Ninvilai, P.; Limcharoen, B.; Tunterak, W.; Prakairungnamthip, D.; Oraveerakul, K.; Banlunara, W.; Thontiravong, A. Pathogenesis of Thai duck Tembusu virus in Cherry Valley ducks: The effect of age on susceptibility to infection. Vet. Microbiol. 2020, 243, 108636. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Li, R.; Liu, X.; Li, N.; Liu, S. Pathogenicity comparison of duck Tembusu virus in different aged Cherry Valley breeding ducks. BMC Vet. Res. 2019, 15, 282. [Google Scholar] [CrossRef] [PubMed]
- Ti, J.; Zhang, L.; Li, Z.; Zhao, D.; Zhang, Y.; Li, F.; Diao, Y. Effect of age and inoculation route on the infection of duck Tembusu virus in Goslings. Vet. Microbiol. 2015, 181, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Huang, Y.; Shi, Y.; Bai, X.; Yang, P.; Chen, Q. Tembusu Virus entering the central nervous system caused nonsuppurative encephalitis without disrupting the blood-brain barrier. J. Virol. 2021, 95, e02191-20. [Google Scholar] [CrossRef] [PubMed]
- Yurayart, N.; Ninvilai, P.; Chareonviriyaphap, T.; Kaewamatawong, T.; Thontiravong, A.; Tiawsirisup, S. Pathogenesis of Thai duck Tembusu virus in BALB/c mice: Descending infection and neuroinvasive virulence. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Li, X.; Li, G.; Teng, Q.; Yu, L.; Wu, X.; Li, Z. Development of a blocking ELISA for detection of serum neutralizing antibodies against newly emerged duck Tembusu virus. PLoS ONE 2012, 7, e53026. [Google Scholar] [CrossRef]
- Zhou, Q.; Bi, Z.; Yin, D.; Gu, X.; Xu, Z.; Huang, R.; Xing, X.; Qi, K.; Wang, G. Development and application of an indirect ELISA for the serological detection of duck Tembusu virus infection based on the NS1 protein antigen. Arch. Virol. 2020, 165, 709–714. [Google Scholar] [CrossRef]
- Kanai, R.; Kar, K.; Anthony, K.; Gould, L.H.; Ledizet, M.; Fikrig, E.; Marasco, W.A.; Koski, R.A.; Modis, Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 2006, 80, 11000–11008. [Google Scholar] [CrossRef]
- Luca, V.C.; AbiMansour, J.; Nelson, C.A.; Fremont, D.H. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 2012, 86, 2337–2346. [Google Scholar] [CrossRef]
- Nybakken, G.E.; Nelson, C.A.; Chen, B.R.; Diamond, M.S.; Fremont, D.H. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 2006, 80, 11467–11474. [Google Scholar] [CrossRef]
- Rothman, A.L. Immunity to dengue virus: A tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011, 11, 532–543. [Google Scholar] [CrossRef]
- Wahala, W.M.; Silva, A.M. The human antibody response to dengue virus infection. Viruses 2011, 3, 2374–2395. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Lin, W.; Li, T.; Li, X.; Bai, X.; Wulin, S.; Zhang, Q.; Li, S.; Liu, M.; et al. A Novel Neutralizing Antibody Targeting a Unique Cross-Reactive Epitope on the hi Loop of Domain II of the Envelope Protein Protects Mice against Duck Tembusu Virus. J. Immunol. 2020, 204, 1836–1848. [Google Scholar] [CrossRef]
- Elizalde, M.; Cano-Gómez, C.; Llorente, F.; Pérez-Ramírez, E.; Casades-Martí, L.; Aguilera-Sepúlveda, P.; Ruiz-Fons, F.; Jiménez-Clavero, M.; Fernández-Pinero, J. A Duplex Quantitative Real-Time Reverse Transcription-PCR for Simultaneous Detection and Differentiation of Flaviviruses of the Japanese Encephalitis and Ntaya Serocomplexes in Birds. Front. Vet. Sci. 2020, 7, 203. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, X.; Gao, Y.; Song, S.; Xu, D.; Yan, L. Development and application of multiplex PCR method for simultaneous detection of seven viruses in ducks. BMC Vet. Res. 2019, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yao, M.; Tang, Z.; Xu, D.; Luo, Y.; Gao, Y.; Yan, L. Development and application of a triplex real-time PCR assay for simultaneous detection of avian influenza virus, Newcastle disease virus, and duck Tembusu virus. BMC Vet. Res. 2020, 16, 203. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, L.; Cai, G.; Zhang, D.; Lin, J. Establishment of a simultaneous detection method for ten duck viruses using MALDI-TOF mass spectrometry. J. Virol. Methods 2019, 273, 113723. [Google Scholar] [CrossRef]
- Yan, D.; Shi, Y.; Wang, H.; Li, G.; Li, X.; Wang, B.; Su, X.; Wang, J.; Teng, Q.; Yang, J.; et al. A Single Mutation at Position 156 in the Envelope Protein of Tembusu Virus Is Responsible for Virus Tissue Tropism and Transmissibility in Ducks. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Pan, Y.; Cheng, A.; Zhang, X.; Wang, M.; Chen, S.; Zhu, D.; Liu, M.; Yang, Q.; Wu, Y.; et al. Autophagy Promotes Duck Tembusu Virus Replication by Suppressing p62/SQSTM1-Mediated Innate Immune Responses In Vitro. Vaccines 2020, 8, 22. [Google Scholar] [CrossRef]
- Wang, H.J.; Li, X.F.; Liu, L.; Xu, Y.P.; Ye, Q.; Deng, Y.Q.; Huang, X.Y.; Zhao, H.; Qin, E.D.; Shi, P.Y.; et al. The Emerging Duck Flavivirus Is Not Pathogenic for Primates and Is Highly Sensitive to Mammalian Interferon Antiviral Signaling. J. Virol. 2016, 90, 6538–6548. [Google Scholar] [CrossRef]
- Ruangrung, K.; Chakritbudsabong, W.; Rungarunlert, S.; Smith, D.R.; Hongeng, S.; Sirinonthanawech, N.; Boonarkart, C.; Pulmanausahakul, R.; Suptawiwat, O.; Auewarakul, P. Analysis of Tembusu virus infection of human cell lines and human induced pluripotent stem cell derived hepatocytes. Virus Res. 2021, 292, 198252. [Google Scholar] [CrossRef]
- Ma, Y.; Liang, Y.; Wang, N.; Cui, L.; Chen, Z.; Wu, H.; Zhu, C.; Wang, Z.; Liu, S.; Li, H. Avian Flavivirus Infection of Monocytes/Macrophages by Extensive Subversion of Host Antiviral Innate Immune Responses. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Wang, J.; Lei, C.Q.; Ji, Y.; Zhou, H.; Ren, Y.; Peng, Q.; Zeng, Y.; Jia, Y.; Ge, J.; Zhong, B.; et al. Duck Tembusu Virus Nonstructural Protein 1 Antagonizes IFN-β Signaling Pathways by Targeting VISA. J. Immunol. 2016, 197, 4707–4713. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Liu, F.; Yang, Q.; Wu, Y.; Sun, K.; et al. Antigen distribution of TMUV and GPV are coincident with the expression profiles of CD8α-positive cells and goose IFNγ. Sci. Rep. 2016, 6, 25545. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Diamond, M.S. New insights into innate immune restriction of West Nile virus infection. Curr. Opin. Virol. 2015, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Aguirre, S.; Fernandez-Sesma, A. Innate immunity evasion by Dengue virus. Viruses 2012, 4, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, A.; Wang, M. Innate sensing of viruses by pattern recognition receptors in birds. Vet. Res. 2013, 44, 82. [Google Scholar] [CrossRef]
- Nazmi, A.; Dutta, K.; Hazra, B.; Basu, A. Role of pattern recognition receptors in flavivirus infections. Virus Res. 2014, 185, 32–40. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Li, R.; Liu, J.; Zhang, J.; Cai, Y.; Liu, S.; Chai, T.; Wei, L. Immune responses of ducks infected with duck Tembusu virus. Front. Microbiol. 2015, 6, 425. [Google Scholar] [CrossRef]
- Zhang, J.; An, D.; Fan, Y.; Tang, Y.; Diao, Y. Effect of TMUV on immune organs of TMUV infected ducklings. Vet. Microbiol. 2021, 255, 109033. [Google Scholar] [CrossRef]
- Fu, G.; Chen, C.; Huang, Y.; Cheng, L.; Fu, Q.; Wan, C.; Shi, S.; Chen, H.; Liu, W. Comparative analysis of transcriptional profiles of retinoic-acid-induced gene I-like receptors and interferons in seven tissues from ducks infected with avian Tembusu virus. Arch. Virol. 2016, 161, 11–18. [Google Scholar] [CrossRef]
- Chen, S.; Luo, G.; Yang, Z.; Lin, S.; Wang, S.; Goraya, M.U.; Chi, X.; Zeng, X.; Chen, J.L. Avian Tembusu virus infection effectively triggers host innate immune response through MDA5 and TLR3-dependent signaling pathways. Vet. Res. 2016, 47, 74. [Google Scholar] [CrossRef]
- Hua, K.; Li, Y.; Chen, H.; Ni, J.; Bi, D.; Luo, R.; Jin, H. Functional characterization of duck TBK1 in IFN-β induction. Cytokine 2018, 111, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Baran, M.; Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef]
- Li, N.; Jiang, S.; Zhao, J.; Yang, Y.; Deng, K.; Wei, L.; Cai, Y.; Li, B.; Liu, S. Molecular identification of duck DDX3X and its potential role in response to Tembusu virus. Dev. Comp. Immunol. 2020, 106, 103599. [Google Scholar] [CrossRef]
- Han, K.; Zhao, D.; Liu, Y.; Liu, Q.; Huang, X.; Yang, J.; An, F.; Li, Y. Quantitative Proteomic Analysis of Duck Ovarian Follicles Infected with Duck Tembusu Virus by Label-Free LC-MS. Front. Microbiol. 2016, 7, 463. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Huang, Y.; Li, L.; Dong, J.; Liao, M.; Sun, M. Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain. Int. J. Mol. Sci. 2020, 21, 2402. [Google Scholar] [CrossRef]
- Ma, D.Y.; Suthar, M.S. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr. Opin. Virol. 2015, 12, 26–37. [Google Scholar] [CrossRef]
- Chen, S.; Wu, Z.; Wang, M.; Cheng, A. Innate Immune Evasion Mediated by Flaviviridae Non-Structural Proteins. Viruses 2017, 9, 291. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, B.; Zeng, M.; Duan, Y.; Wu, Z.; Wu, Y.; Wang, T.; Wang, M.; Jia, R.; Zhu, D.; et al. Binding of Duck Tembusu Virus Nonstructural Protein 2A to Duck STING Disrupts Induction of Its Signal Transduction Cascade To Inhibit Beta Interferon Induction. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zeng, M.; Jiang, B.; Lu, T.; Guo, J.; Hu, T.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; et al. Amelioration of Beta Interferon Inhibition by NS4B Contributes to Attenuating Tembusu Virus Virulence in Ducks. Front. Immunol. 2021, 12, 671471. [Google Scholar] [CrossRef] [PubMed]
- Klema, V.J.; Padmanabhan, R.; Choi, K.H. Flaviviral Replication Complex: Coordination between RNA Synthesis and 5′-RNA Capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef] [PubMed]
- Daussy, C.F.; Espert, L. L’autophagie sélective au cours des infections virales. Virologie 2016, 20, 196–206. [Google Scholar] [CrossRef]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Randall, G. Dengue virus and autophagy. Viruses 2011, 3, 1332–1341. [Google Scholar] [CrossRef]
- Li, J.K.; Liang, J.J.; Liao, C.L.; Lin, Y.L. Autophagy is involved in the early step of Japanese encephalitis virus infection. Microbes Infect. 2012, 14, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Pan, Y.; Cheng, A.; Zhang, X.; Wang, M.; Chen, S.; Zhu, D.; Liu, M.; Yang, Q.; Wu, Y.; et al. Autophagy Is a Potential Therapeutic Target Against Duck Tembusu Virus Infection. Front. Cell. Infect. Microbiol. 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.D.; Mazzon, M.; Jacobs, M.; Amara, A. Pathogenesis of flavivirus infections: Using and abusing the host cell. Cell Host Microbe 2009, 5, 318–328. [Google Scholar] [CrossRef]
- Perera, N.; Miller, J.L.; Zitzmann, N. The role of the unfolded protein response in dengue virus pathogenesis. Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef]
- Blazquez, A.B.; Escribano-Romero, E.; Merino-Ramos, T.; Saiz, J.C.; Martin-Acebes, M.A. Stress responses in flavivirus-infected cells: Activation of unfolded protein response and autophagy. Front. Microbiol. 2014, 5, 266. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, J.; Han, K.; Liu, Q.; Wang, H.; Liu, Y.; Huang, X.; Zhang, L.; Li, Y. The unfolded protein response induced by Tembusu virus infection. BMC Vet. Res. 2019, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Kulprasertsri, S.; Aoshima, K.; Kobayashi, A.; Kimura, T. Minocycline prevents primary duck neurons from duck Tembusu virus-induced death. J. Vet. Med. Sci. 2021, 83, 734–741. [Google Scholar] [CrossRef]
- Zhu, Y.; Gu, X.; Zhang, M.; Lv, X.; Zhang, C.; Li, J.; Hu, Z.; Wu, Q.; Zhang, R.; Wei, J.; et al. Epigallocatechin-3-gallate exhibits antiviral effects against the duck Tembusu virus via blocking virus entry and upregulating type I interferons. Poult. Sci. 2021, 100, 100989. [Google Scholar] [CrossRef] [PubMed]
- Natsoulis, G.; Boeke, J.D. New antiviral strategy using capsid-nuclease fusion proteins. Nature 1991, 352, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.F.; Qin, E.; Yu, M.; Chen, S.P.; Jiang, T.; Deng, Y.Q.; Duan, H.Y.; Zhao, H. Therapeutic effects of dengue 2 virus capsid protein and staphylococcal nuclease fusion protein on dengue-infected cell cultures. Arch. Virol. 2005, 150, 659–669. [Google Scholar] [CrossRef]
- Pang, R.; He, D.N.; Zhou, B.; Liu, K.; Zhao, J.; Zhang, X.M.; Chen, P.Y. In vitro inhibition of Japanese encephalitis virus replication by capsid-targeted virus inactivation. Antiviral Res. 2013, 97, 369–375. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, R.; Pan, Y.; Wang, M.; Chen, S.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; et al. Therapeutic effects of duck Tembusu virus capsid protein fused with staphylococcal nuclease protein to target Tembusu infection in vitro. Vet. Microbiol. 2019, 235, 295–300. [Google Scholar] [CrossRef]
- Li, G.; Gao, X.; Xiao, Y.; Liu, S.; Peng, S.; Li, X.; Shi, Y.; Zhang, Y.; Yu, L.; Wu, X.; et al. Development of a live attenuated vaccine candidate against duck Tembusu viral disease. Virology 2014, 450–451, 233–242. [Google Scholar] [CrossRef]
- Sun, L.; Li, Y.; Zhang, Y.; Han, Z.; Xu, Y.; Kong, X.; Liu, S. Adaptation and attenuation of duck Tembusu virus strain Du/CH/LSD/110128 following serial passage in chicken embryos. Clin. Vaccine Immunol. 2014, 21, 1046–1053. [Google Scholar] [CrossRef][Green Version]
- Lin, J.; Liu, Y.; Wang, X.; Yang, B.; He, P.; Yang, Z.; Duan, H.; Xie, J.; Zou, L.; Zhao, J.; et al. Efficacy Evaluation of an Inactivated Duck Tembusu Virus Vaccine. Avian Dis. 2015, 59, 244–248. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.; Zhang, Q.; Sun, M.; Li, S.; Su, W.; Hu, X.; He, W.; Su, J. Efficacy assessment of an inactivated Tembusu virus vaccine candidate in ducks. Res. Vet. Sci. 2017, 110, 72–78. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Zhang, X.; Chen, L.; Tang, Y.; Diao, Y. Development of an attenuated live vaccine candidate of duck Tembusu virus strain. Vet. Microbiol. 2019, 231, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liang, T.; Lv, J.; Qu, S.; Meng, R.; Yang, B.; Feng, C.; Dai, W.; Wang, X.; Zhang, B.; et al. The Substantial Attenuation of Virulence of Tembusu Virus Strain PS Is Determined by an Arg at Residue 304 of the Envelope Protein. J. Virol. 2020, 95, e02331-20. [Google Scholar] [CrossRef]
- Chen, P.; Liu, J.; Jiang, Y.; Zhao, Y.; Li, Q.; Wu, L.; He, X.; Chen, H. The vaccine efficacy of recombinant duck enteritis virus expressing secreted E with or without PrM proteins of duck tembusu virus. Vaccine 2014, 32, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Liu, L.; Li, X.F.; Ye, Q.; Deng, Y.Q.; Qin, E.D.; Qin, C.F. In vitro and in vivo characterization of chimeric duck Tembusu virus based on Japanese encephalitis live vaccine strain SA14-14-2. J Gen. Virol. 2016, 97, 1551–1556. [Google Scholar] [CrossRef]
- Ma, T.; Liu, Y.; Cheng, J.; Fan, W.; Cheng, Z.; Niu, X.; Liu, J. Liposomes containing recombinant E protein vaccine against duck Tembusu virus in ducks. Vaccine 2016, 34, 2157–2163. [Google Scholar] [CrossRef]
- Tang, J.; Yin, D.; Wang, R.; Zhou, Q.; Zhou, X.; Xing, X.; Liu, H.M.; Liu, G.; Wang, G. A recombinant adenovirus expressing the E protein of duck Tembusu virus induces protective immunity in duck. J. Vet. Med. Sci. 2019, 81, 314–320. [Google Scholar] [CrossRef]
- Huang, J.; Shen, H.; Jia, R.; Wang, M.; Chen, S.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; et al. Oral Vaccination with a DNA Vaccine Encoding Capsid Protein of Duck Tembusu Virus Induces Protection Immunity. Viruses 2018, 10, 180. [Google Scholar] [CrossRef]
- Chen, H.; Yan, M.; Tang, Y.; Diao, Y. Evaluation of immunogenicity and protective efficacy of a CpG-adjuvanted DNA vaccine against Tembusu virus. Vet. Immunol. Immunopathol. 2019, 218, 109953. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Y.; Dong, J.; Zhang, J.; Zhang, C.; Qin, J.; Sun, M.; Xu, Z. Development of chimeric virus-like particles containing the E glycoprotein of duck Tembusu virus. Vet. Microbiol. 2019, 238, 108425. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef] [PubMed]
| Country/Location of Origin | Species | Year | References |
|---|---|---|---|
| Kuala Lumpur, Malaysia | Cx. tritaeniorhynchus | 1955 | [24] |
| Malaysia | Cx. vishnui | 1970 | [2] |
| Kamphaengphet Province, Thailand | Cx. gelidus Cx. tritaeniorhynchus Cx. vishnui | 1982 | [25] |
| Chiang Mai Province, Thailand | Cx. tritaeniorhynchus | 1992 | [1] |
| Kamphaeng Phet Province, Thailand | Cx. vishnui | 2005 | [37] |
| Shandong Province, China | Cx. pipiens | 2010–2012 | [38] |
| Yunnan Province, China | Cx. tritaeniorhynchus | 2012 | [39] |
| Shandong Province, China | Cx. spp | 2012 | [40] |
| Sing Buri Province, Thailand | Cx. tritaeniorhynchus | 2015 | [36] |
| Kanchanaburi Province, Thailand | Cx. quinquefasciatus | 2015 | [34] |
| Taipei, Taiwan | Cx. annulus | 2019 | [26] |
| Taichung, Taiwan | Cx. tritaeniorhynchus | 2019 | [26] |
| Country/Location of Origin | Species | Year | References |
|---|---|---|---|
| Perak State, Malaysia | Broiler chick | 2000 | [5] |
| Thailand | Duck | 2007 | [30] |
| Shandong Province, China | Human | 2010–2012 | [46] |
| Shanghai, China | Duck | 2010 | [47] |
| Shandong Province, China | Meat duck Layer duck | 2010 | [48] |
| Henan Province, China | Layer duck | 2010 | [48] |
| China | Pekin duck, Cherry Valley Pekin duck Shaoxing duck Breeder duck | 2010 | [4,23,49,50] |
| China | Goose | 2010 | [49] |
| China | Chicken | 2010 | [51] |
| Jiangsu Province, China | Goose | 2010 | [52,53] |
| Shandong Province, China | House sparrow (Passer domesticus) | 2010–2011 | [54] |
| Guangdong Province, China | Layer duck | 2011 | [55] |
| Guangxi Province, China | Layer duck | 2011 | [48] |
| Shandong Province, China | House sparrow | 2012 | [40] |
| Beijing Autonomous City, China | Pigeon | 2012 | [22] |
| Shandong Province, China | Goose | 2012 | [40,56] |
| Hebei Province, China | Duck | 2012 | [22] |
| Jiangsu Province, China | Egg-laying duck | 2012 | [48] |
| Shandong Province, China | Duck | 2012 | [48,56] |
| Fujian Province, China | Duck | 2012 | [57] |
| Malaysia | Pekin duck | 2012 | [3,58] |
| China | Duck | 2013 | [59] |
| Guangxi Province, China | Cherry Valley duck | 2013 | [60,61] |
| Shanghai Province, China | Pekin duck | 2013 | [62] |
| Shandong Province, China | Layer duck | 2013 | [40] |
| Anhui Province, China | Layer duck | 2013 | [40] |
| Thailand | Duck | 2013 | [63] |
| Thailand | Broiler chicken | 2013 | [32,64] |
| China | Chicken | 2013 | [40] |
| China | Goose | 2014–2015 | [65] |
| China | Chicken | 2014 | [66] |
| China | Layer duck | 2014 | [67] |
| China | Mallard (Anas platyrhynchos) | 2014 | [68] |
| China | Broiler duck | 2015 | [69] |
| China | Layer duck | 2015 | [70] |
| Guangdong Province, China | Muscovy duck | 2015 | [71] |
| Thailand | duck | 2015 | [72] |
| Shandong Province, China | Meat duck | 2016 | [40] |
| Inner Mongolia Autonomous Region, China | Meat duck | 2017 | [40] |
| Thailand | Free-grazing duck | 2018 | [42] |
| Name | Organism, Tissue | Type | Reference | |
|---|---|---|---|---|
| Avian | DF-1 | Chicken, embryo fibroblast | Cell line | [3,26,93] |
| HD11 | Chicken, bone marrow macrophage | Cell line | [97] | |
| DEF | Duck embryonic fibroblast | Primary cells from 9-day-old duck embryos | [10,98] | |
| goose PBMCs | Goose blood | Primary cell | [99] | |
| LSCC-BK3 | Chicken, B lymphocyte | Cell line | [5] | |
| Mammalian | MARC145 | Monkey kidney | Cell line | [5] |
| VERO | African green monkey kidney | Cell line | [3,5,26,58,95] | |
| BHK-21 | Hamster, kidney | Cell line | [10,22,26] | |
| CPK | Porcine, kidney | Cell line | [5] | |
| MEF | Mouse embryonic fibroblast | Primary cells | [98] | |
| A549 | Human, epithelial lung | Cell line | [95,96] | |
| HeLa | Human, epithelial cervix | Cell line | [95] | |
| SH-SY5Y | Human, epithelial bone marrow | Cell line | [95,96] | |
| HEK293T | Human, epithelial kidney | Cell line | [96,98] | |
| HUH7 | Human, liver | Cell line | [96,98] | |
| RD | Human rhabdomyosarcoma cell | Cell line | [96] | |
| SUP-T1 | Human T cell lymphoblastic lymphoma cell | Cell line | [96] | |
| Z-138 | Human B cell non-Hodgkin’s lymphoma cell | Cell line | [96] | |
| U937 | Human histiocytic lymphoma cell | Cell line | [96] | |
| imHC | Human hepatocyte-like cell | Derived cell from hiPSC | [96] | |
| Insect | C6/36 | Aedes albopictus, larva | Cell line | [22,26,58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamel, R.; Phanitchat, T.; Wichit, S.; Morales Vargas, R.E.; Jaroenpool, J.; Diagne, C.T.; Pompon, J.; Missé, D. New Insights into the Biology of the Emerging Tembusu Virus. Pathogens 2021, 10, 1010. https://doi.org/10.3390/pathogens10081010
Hamel R, Phanitchat T, Wichit S, Morales Vargas RE, Jaroenpool J, Diagne CT, Pompon J, Missé D. New Insights into the Biology of the Emerging Tembusu Virus. Pathogens. 2021; 10(8):1010. https://doi.org/10.3390/pathogens10081010
Chicago/Turabian StyleHamel, Rodolphe, Thipruethai Phanitchat, Sineewanlaya Wichit, Ronald Enrique Morales Vargas, Jiraporn Jaroenpool, Cheikh Tidiane Diagne, Julien Pompon, and Dorothée Missé. 2021. "New Insights into the Biology of the Emerging Tembusu Virus" Pathogens 10, no. 8: 1010. https://doi.org/10.3390/pathogens10081010
APA StyleHamel, R., Phanitchat, T., Wichit, S., Morales Vargas, R. E., Jaroenpool, J., Diagne, C. T., Pompon, J., & Missé, D. (2021). New Insights into the Biology of the Emerging Tembusu Virus. Pathogens, 10(8), 1010. https://doi.org/10.3390/pathogens10081010









