Nucleosome Positioning on Episomal Human Papillomavirus DNA in Cultured Cells
Abstract
1. Introduction
2. Results
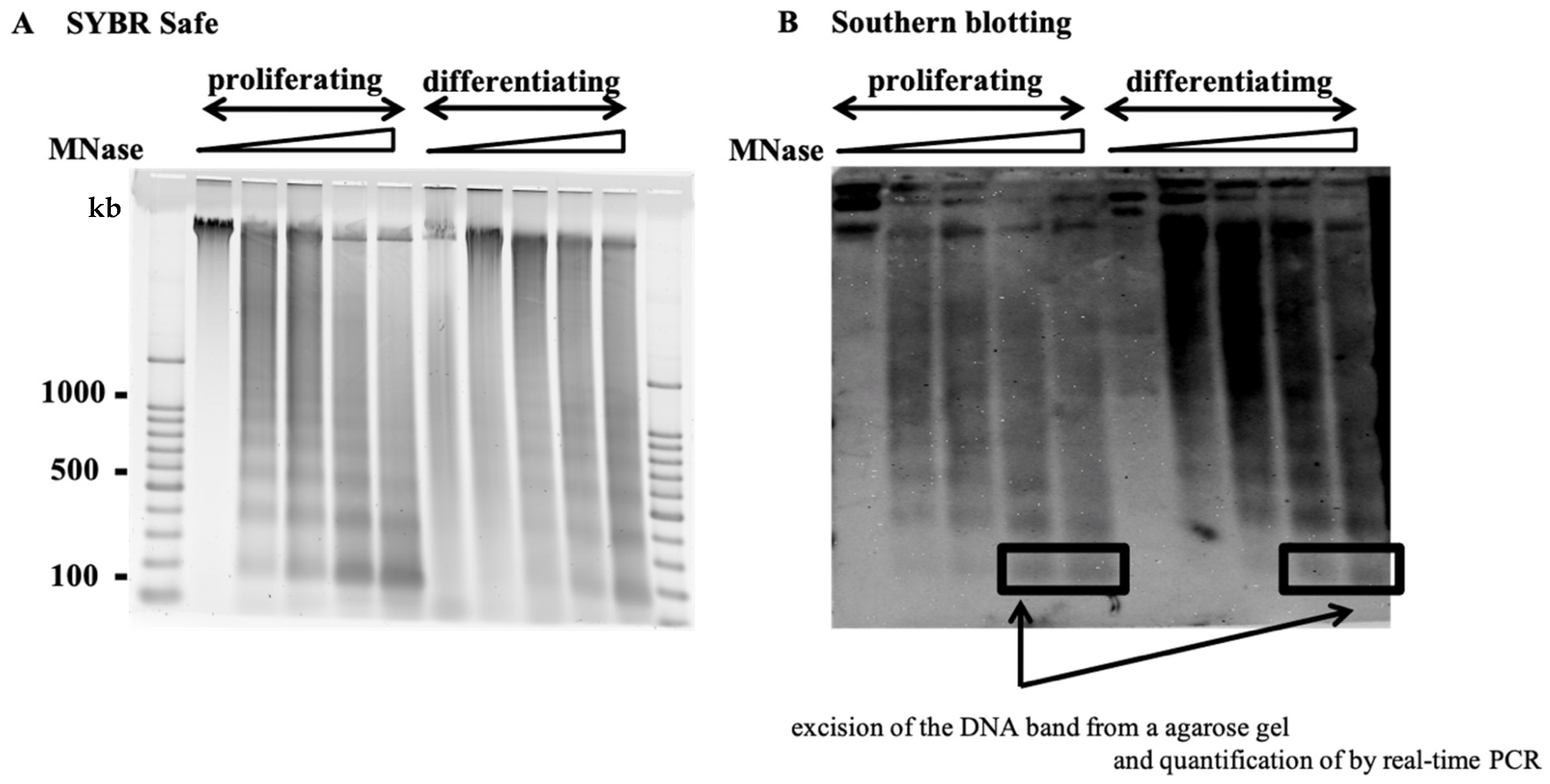
2.1. Nucleosome Organization of the HPV Genome and Host Genome
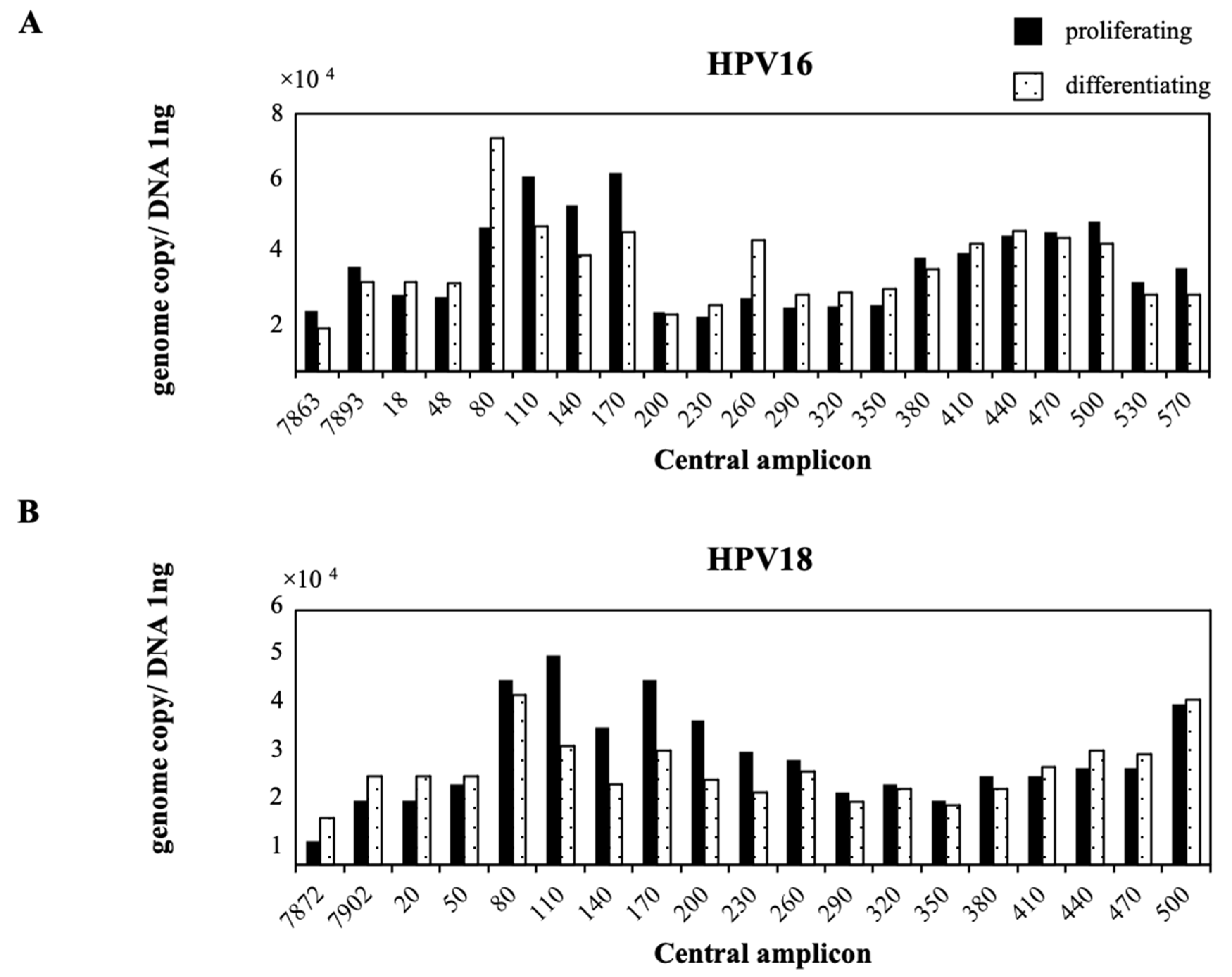
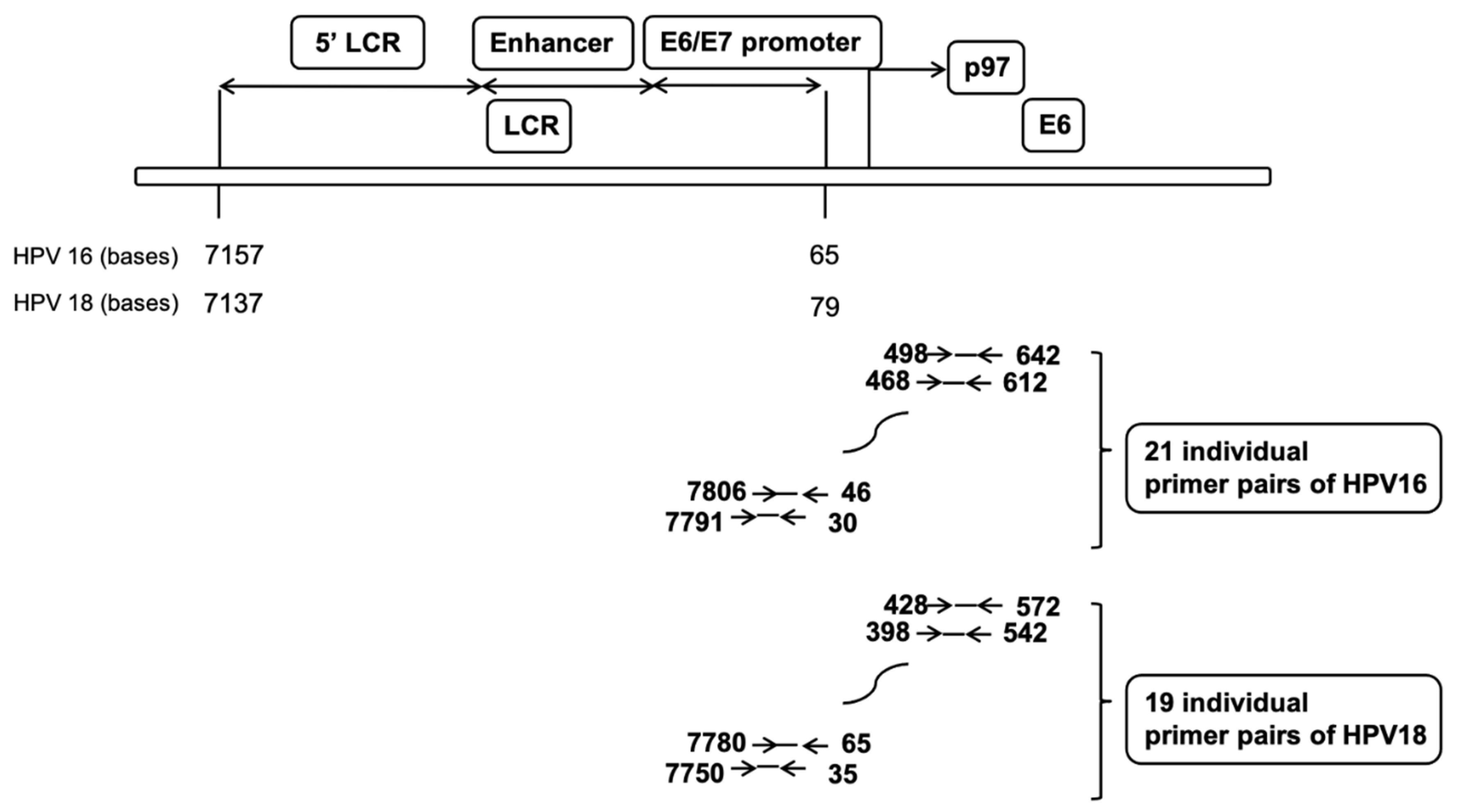
2.2. Nucleosome Positioning on Episomal HPV DNA in Cultured Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of NIKS Cell Lines Containing HPV Genomes
4.3. Nuclei Preparation
4.4. MNase Digestion of Chromatin and Purified DNA
4.5. Southern Blotting
4.6. Nucleosome Scanning Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Doorslaer, K. Evolution of the papillomaviridae. Virology 2013, 445, 11–20. [Google Scholar] [CrossRef]
- Bravo, I.G.; Felez-Sanchez, M. Papillomaviruses: Viral evolution, cancer and evolutionary medicine. Evol. Med. Public Health 2015, 2015, 32–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; DeSalle, R.; Schiffman, M.; Herrero, R.; Wood, C.E.; Ruiz, J.C.; Clifford, G.M.; Chan, P.K.S.; Burk, R.D. Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog. 2018, 14, e1007352. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Cosma, M.P. Ordered recruitment: Gene-specific mechanism of transcription activation. Mol. Cell 2002, 10, 227–236. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Fan, H.-Y.; Kingston, R.E. Cooperation between Complexes that Regulate Chromatin Structure and Transcription. Cell 2002, 108, 475–487. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 6, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Archer, T.K.; Cordingley, M.G.; Wolford, R.G.; Hager, G.L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol. 1991, 11, 688–698. [Google Scholar] [CrossRef]
- Truss, M.; Bartsch, J.; Schelbert, A.; Hache, R.J.; Beato, M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995, 14, 1737–1751. [Google Scholar] [CrossRef]
- Batson, S.C.; Rimsky, S.; Sundseth, R.; Hansen, U. Association of nucleosome-free regions and basal transcription factors with in vivo-assembled chromatin templates active in vitro. Nucleic Acids Res. 1993, 21, 3459–3468. [Google Scholar] [PubMed]
- Kondoleon, S.K.; Kurkinen, N.A.; Hallick, L.M. The SV40 nucleosome-free region is detected throughout the virus life cycle. Virology 1989, 173, 129–135. [Google Scholar] [CrossRef]
- Jeong, S.W.; Stein, A. DNA sequence affects nucleosome ordering on replicating plasmids in transfected COS-1 cells and in vitro. J. Biol. Chem. 1994, 269, 2197–2205. [Google Scholar] [CrossRef]
- Knezetic, J.A.; Luse, D.S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell 1986, 45, 95–104. [Google Scholar] [CrossRef]
- Li, B.; Adams, C.; Workman, J. Nucleosome binding by the constitutive transcription factor Sp1. J. Biol. Chem. 1994, 269, 7756–7763. [Google Scholar] [CrossRef]
- Alexiadis, V.; Varga-Weisz, P.D.; Bonte, E.; Becker, P.B.; Gruss, C. In vitro chromatin remodeling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998, 17, 3428–3438. [Google Scholar] [CrossRef]
- Ramsperger, U.; Stahl, H. Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J. 1995, 14, 3215–3225. [Google Scholar] [CrossRef]
- Rösl, F.; Waldeck, W.; Zentgraf, H.; Sauer, G. Properties of intracellular bovine papillomavirus chromatin. J. Virol. 1986, 58, 500–507. [Google Scholar] [CrossRef]
- Waldeck, W.; Rösl, F.; Zentgraf, H. Origin of replication in episomal bovine papilloma virus type 1 DNA isolated from transformed cells. EMBO J. 1984, 3, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.M.; Storgaard, L.; Fey, S.J. Proteins present in bovine papillomavirus particles. J. Virol. 1987, 61, 3596–3601. [Google Scholar] [CrossRef]
- Peña, L.D.M.; Laimins, L.A. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. J. Virol. 2001, 75, 10005–10013. [Google Scholar] [CrossRef]
- Cornet, I.; Gheit, T.; Franceschi, S.; Vignat, J.; Burk, R.; Sylla, B.; Tommasino, M.; Clifford, G. Human pappilomavirus type 16 genetic variants: Phylogeny and classification based on E6 and LCR. J. Virol. 2012, 86, 6855–6861. [Google Scholar] [CrossRef]
- Poumay, Y.; Pittelkow, M. Cell Density and Culture Factors Regulate Keratinocyte Commitment to Differentiation and Expression of Suprabasal Kl/Kl0 Keratins. J. Investig. Dermatol. 1995, 104, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Murakami, I.; Egawa, N.; Griffin, H.; Yin, W.; Kranjec, C.; Nakahara, T.; Kiyono, T.; Doorbar, J. Roles for E1-independent replication and E6-mediated p53 degradation during low-risk and high-risk human papillomavirus genome maintenance. PLoS Pathog. 2019, 15, e1007755. [Google Scholar] [CrossRef]
- Simpson, R.T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 1990, 343, 387–389. [Google Scholar] [CrossRef]
- Meersseman, G.; Pennings, S.; Bradbury, E. Mobile nucleosomes--a general behavior. EMBO J. 1992, 11, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Pazin, M.J.; Bhargava, P.; Geiduschek, E.P.; Kadonaga, J.T. Nucleosome mobility and the maintenance of nucleosome positioning. Science 1997, 276, 809–812. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, P.C.; Simpson, R.T. Effects of sequence alterations in a DNA segment containing the 5S RNA gene from Lythechinus variegatus on positioning of a nucleosome core particle in vitro. J. Biol. Chem. 1985, 260, 15318–15324. [Google Scholar] [CrossRef]
- Hayes, J.J.; Tullius, T.D.; Wolffe, A.P. The structure of DNA in a nucleosome. Proc. Natl. Acad. Sci. USA 1990, 87, 7405–7409. [Google Scholar] [CrossRef] [PubMed]
- Chavez, S.; Beato, M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc. Natl. Acad. Sci. USA 1997, 94, 2885–2890. [Google Scholar] [CrossRef] [PubMed]
- Richard-Foy, H.; Hager, G.L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987, 6, 2321–2328. [Google Scholar] [CrossRef]
- Sera, T.; Wolffe, A.P. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus 5S rRNA genes. Mol. Cell. Biol. 1998, 18, 3668–3680. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.R.; Hoffmannb, B.L.A.-; Leea, D.; Sattler, C.A.; Lambert, P.F. Establishment of the Human Papillomavirus Type 16 (HPV-16) Life Cycle in an Immortalized Human Foreskin Keratinocyte Cell Line. Virology 1999, 262, 344–354. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Egawa, N.; Wang, Q.; Griffin, H.M.; Murakami, I.; Jackson, D.; Mahmood, R.; Doorbar, J. HPV16 and 18 genome amplification show different E4-dependence, with 16E4 enhancing E1 nuclear accumulation and replicative efficiency via its cell cycle arrest and kinase activation functions. PLoS Pathog. 2017, 13, e1006282. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murakami, I.; Iwata, T.; Morisada, T.; Tanaka, K.; Aoki, D. Nucleosome Positioning on Episomal Human Papillomavirus DNA in Cultured Cells. Pathogens 2021, 10, 772. https://doi.org/10.3390/pathogens10060772
Murakami I, Iwata T, Morisada T, Tanaka K, Aoki D. Nucleosome Positioning on Episomal Human Papillomavirus DNA in Cultured Cells. Pathogens. 2021; 10(6):772. https://doi.org/10.3390/pathogens10060772
Chicago/Turabian StyleMurakami, Isao, Takashi Iwata, Tohru Morisada, Kyoko Tanaka, and Daisuke Aoki. 2021. "Nucleosome Positioning on Episomal Human Papillomavirus DNA in Cultured Cells" Pathogens 10, no. 6: 772. https://doi.org/10.3390/pathogens10060772
APA StyleMurakami, I., Iwata, T., Morisada, T., Tanaka, K., & Aoki, D. (2021). Nucleosome Positioning on Episomal Human Papillomavirus DNA in Cultured Cells. Pathogens, 10(6), 772. https://doi.org/10.3390/pathogens10060772





