Culturable Seed Microbiota of Populus trichocarpa
Abstract
1. Introduction
2. Results
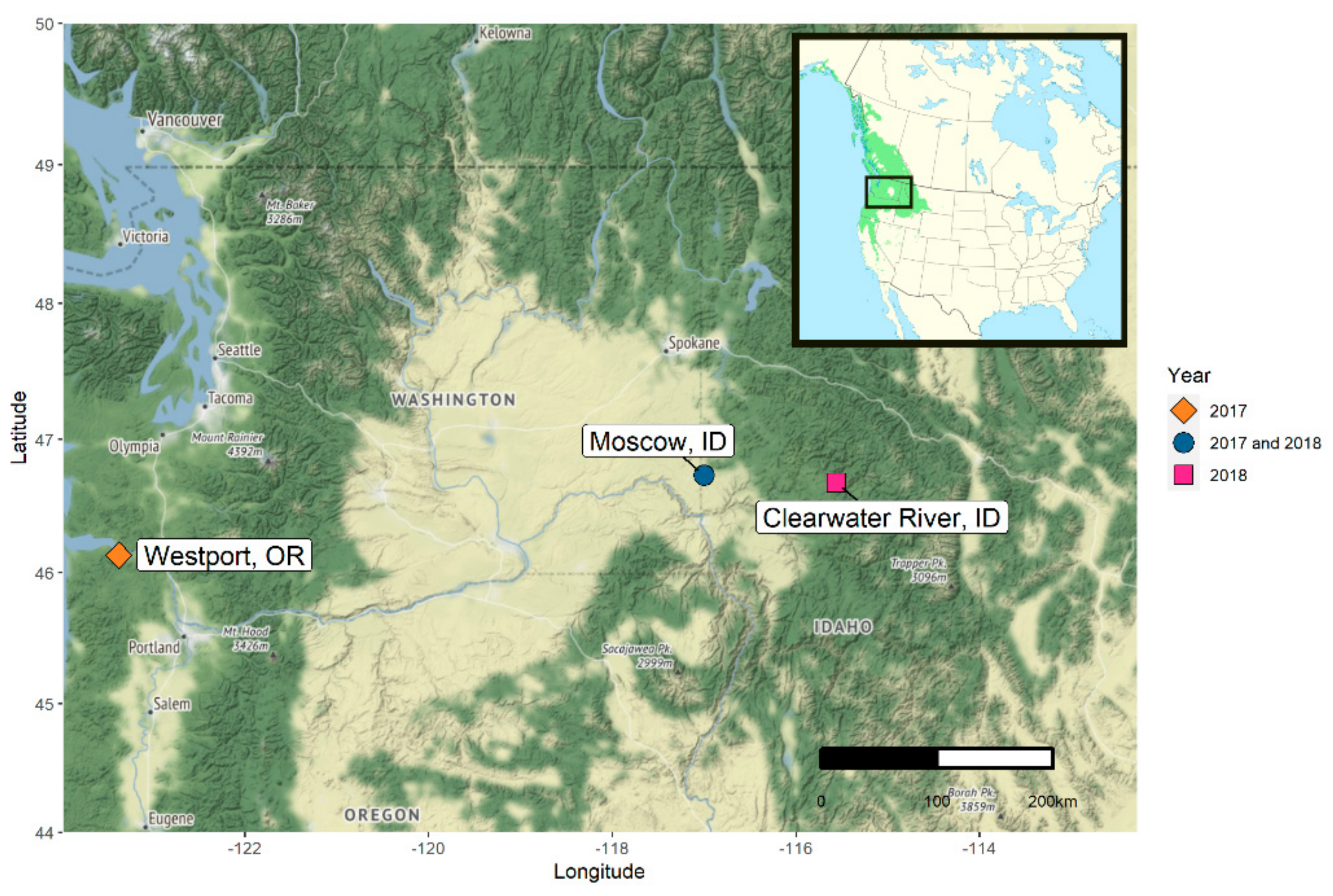
2.1. Description of Populus trichocarpa Seed Microbiota
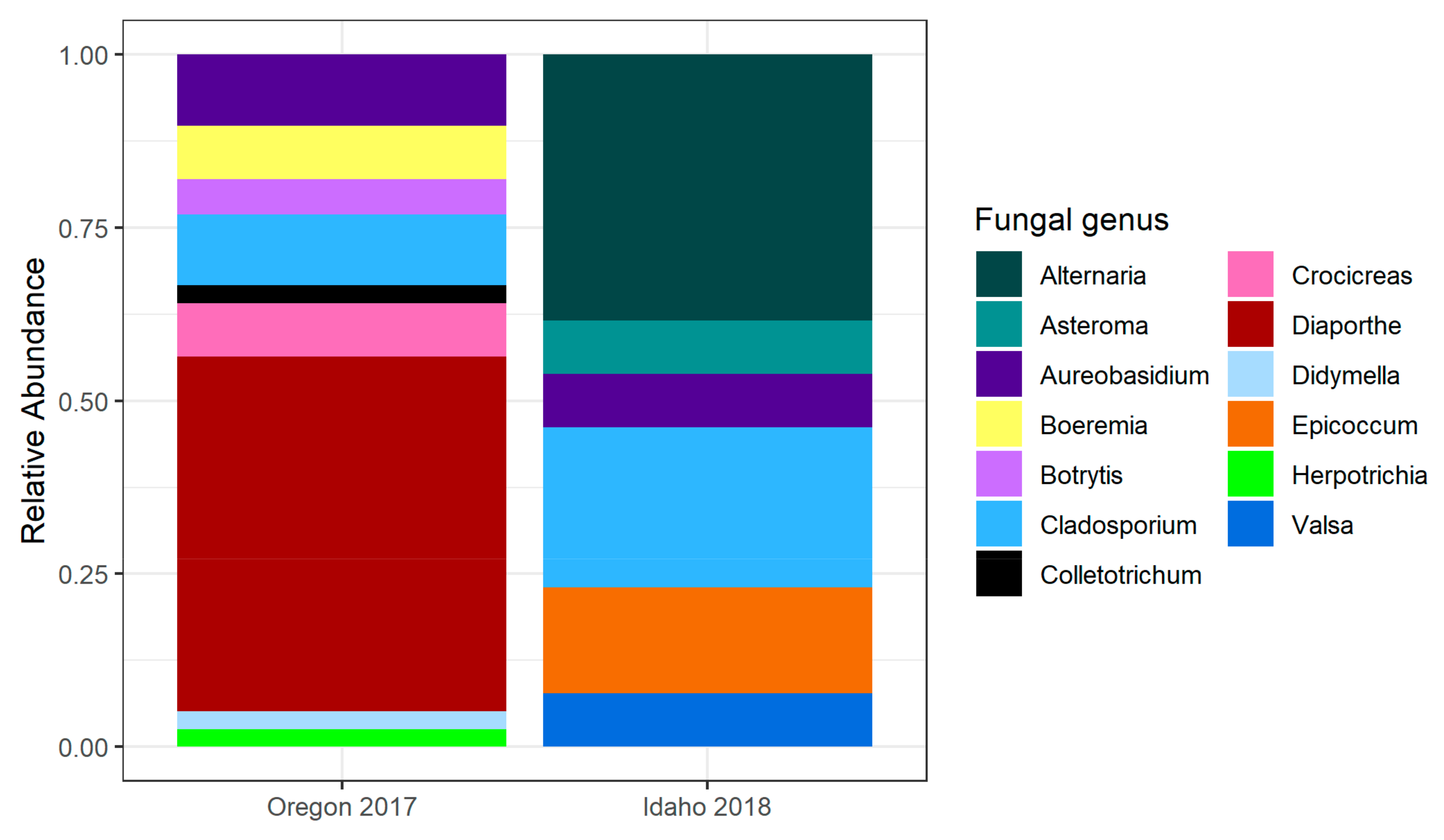
2.1.1. Oregon 2017
2.1.2. Idaho 2017
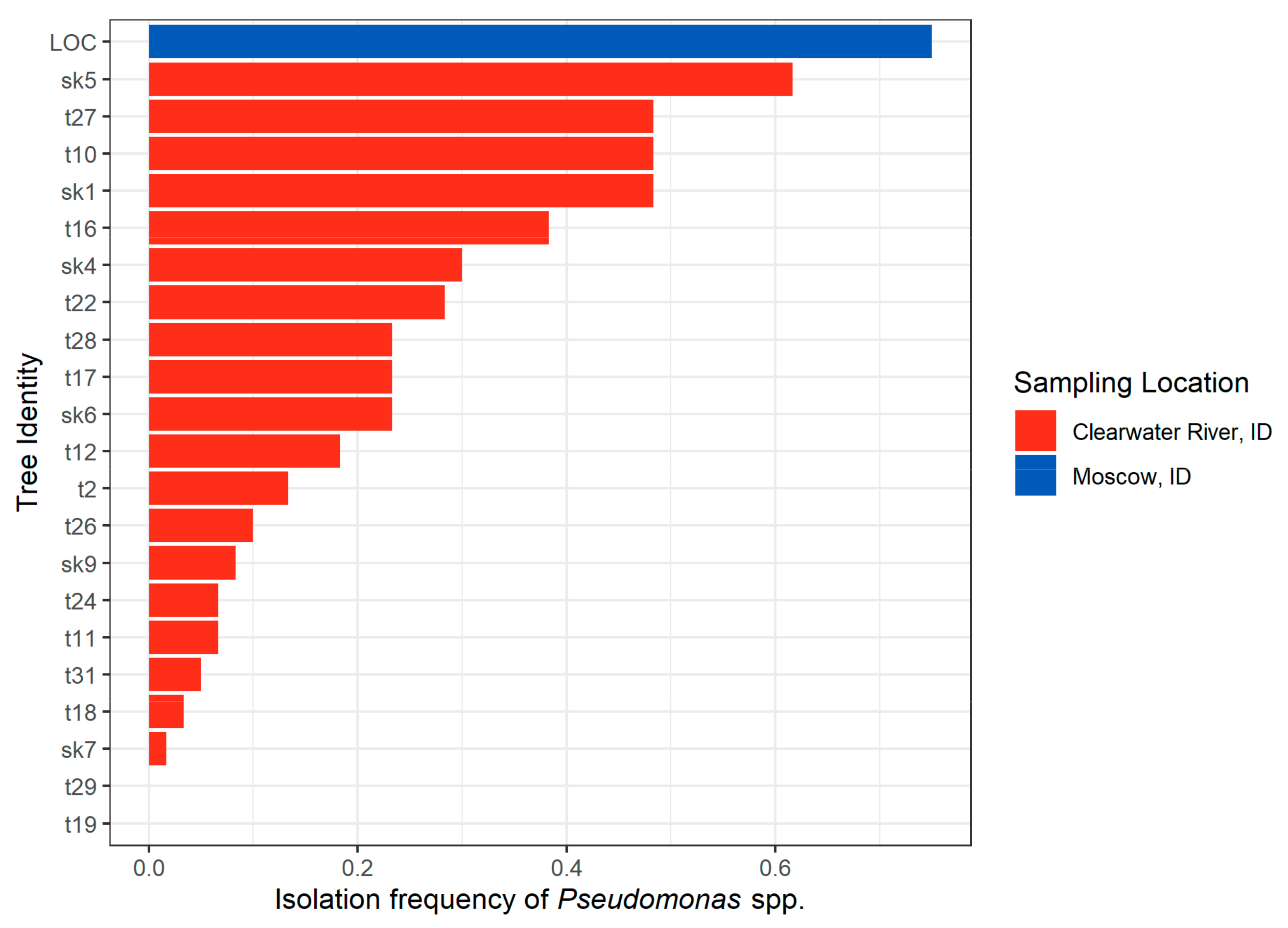
2.1.3. Idaho 2018
2.2. Seed and Seedling Mortality Experiment
3. Discussion
4. Materials and Methods
4.1. Sampling Populus trichocarpa Seeds
4.2. Seed Microbe Isolation
4.3. Seed Microbe Identification
4.4. Seedling Mortality Experiment
4.5. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Khan, Z.; Guelich, G.; Phan, H.; Redman, R.; Doty, S. Bacterial and Yeast Endophytes from Poplar and Willow Promote Growth in Crop Plants and Grasses. ISRN Agron. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Bever, J.D.; Mangan, S.A.; Alexander, H.M. Maintenance of Plant Species Diversity by Pathogens. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 305–325. [Google Scholar] [CrossRef]
- Fenner, M.; Thompson, K. The Ecology of Seeds; Cambridge University Press: Cambridge, UK, 2005; ISBN 9780511614101. [Google Scholar]
- Moles, A.T.; Westoby, M. Seedling survival and seed size: A synthesis of the literature. J. Ecol. 2004, 92, 372–383. [Google Scholar] [CrossRef]
- Czarnoleski, M.; Olejniczak, P.; Mikołajczak, P.; Lembicz, M.; Kozłowski, J. Fungal endophytes protect grass seedlings against herbivory and allow economical seed production. Evol. Ecol. Res. 2010, 12, 769–777. [Google Scholar]
- Bu, Y.; Guo, P.; Ji, Y.; Zhang, S.; Yu, H.; Wang, Z. Effects of Epichloë sinica on Roegneria kamoji seedling physiology under PEG-6000 simulated drought stress. Symbiosis 2019, 77, 123–132. [Google Scholar] [CrossRef]
- Clay, K. Effects of fungal endophytes on the seed and seedling biology of Lolium perenne and Festuca arundinacea. Oecologia 1987, 73, 358–362. [Google Scholar] [CrossRef]
- Verma, S.K.; White, J.F. Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J. Appl. Microbiol. 2018, 124, 764–778. [Google Scholar] [CrossRef]
- Morella, N.M.; Zhang, X.; Koskella, B. Tomato Seed-Associated Bacteria Confer Protection of Seedlings Against Foliar Disease Caused by Pseudomonas syringae. Phytobiomes J. 2019, 3, 177–190. [Google Scholar] [CrossRef]
- Vujanovic, V.; Nazrul, I.; Prasad, D.M. Transgenerational role of seed mycobiome-an endosymbiotic fungal composition as a prerequisite to stress resilience and adaptive phenotypes in Triticum. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Klaedtke, S.; Jacques, M.A.; Raggi, L.; Préveaux, A.; Bonneau, S.; Negri, V.; Chable, V.; Barret, M. Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 2016, 18, 1792–1804. [Google Scholar] [CrossRef]
- Adam, E.; Bernhart, M.; Müller, H.; Winkler, J.; Berg, G. The Cucurbita pepo seed microbiome: Genotype-specific composition and implications for breeding. Plant Soil 2016, 422, 35–49. [Google Scholar] [CrossRef]
- Bergna, A.; Cernava, T.; Grosch, R.; Zachow, C.; Berg, G. Tomato Seeds Preferably Transmit Plant Beneficial Endophytes. Phytobiomes J. 2018, 2, 183–193. [Google Scholar] [CrossRef]
- Van Overbeek, L.S.; Franke, A.C.; Nijhuis, E.H.M.; Groeneveld, R.M.W.; Nunes Da Rocha, U.; Lotz, L.A.P. Bacterial Communities Associated with Chenopodium album and Stellaria media Seeds from Arable Soils. Microb. Ecol. 2011, 62, 257–264. [Google Scholar] [CrossRef]
- Franíc, I.; Eschen, R.; Allan, E.; Hartmann, M.; Schneider, S.; Prospero, S. Drivers of richness and community composition of fungal endophytes of tree seeds. FEMS Microbiol. Ecol. 2020, 96, 166. [Google Scholar] [CrossRef]
- Fort, T.; Pauvert, C.; Zanne, A.E.; Ovaskainen, O.; Caignard, T.; Barret, M.; Compant, S.; Hampe, A.; Delzon, S.; Vacher, C. Maternal effects shape the seed mycobiome in Quercus petraea. New Phytol. 2021, 230, 1594–1608. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Schadt, C.W. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 2015, 205, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Barge, E.G.; Leopold, D.R.; Peay, K.G.; Newcombe, G.; Busby, P.E. Differentiating spatial from environmental effects on foliar fungal communities of Populus trichocarpa. J. Biogeogr. 2019, 46, 2001–2011. [Google Scholar] [CrossRef]
- Bonito, G.; Reynolds, H.; Robeson, M.S.; Nelson, J.; Hodkinson, B.P.; Tuskan, G.; Schadt, C.W.; Vilgalys, R. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol. Ecol. 2014, 23, 3356–3370. [Google Scholar] [CrossRef] [PubMed]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Saint-Vincent, P.M.B.; Ridout, M.; Engle, N.L.; Lawrence, T.J.; Yeary, M.L.; Tschaplinski, T.J.; Newcombe, G.; Pelletier, D.A. Isolation, characterization, and pathogenicity of two Pseudomonas syringae pathovars from Populus trichocarpa seeds. Microorganisms 2020, 8, 1137. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Haworth, R.H.; Spiers, A.G. Characterisation of bacteria from poplars and willows exhibiting leaf spotting and stem cankering in New Zealand. Eur. J. For. Pathol. 1988, 18, 426–436. [Google Scholar] [CrossRef]
- Ramstedt, M.; Rström, B.Å.; von Fircks, H.A. Dieback of poplar and willow caused by Pseudomonas syringae in combination with freezing stress. Eur. J. For. Pathol. 1994, 24, 305–315. [Google Scholar] [CrossRef]
- Raghavendra, A.K.H.; Newcombe, G.; Shipunov, A.; Baynes, M.; Tank, D. Exclusionary interactions among diverse fungi infecting developing seeds of Centaurea stoebe. FEMS Microbiol. Ecol. 2013, 84, 143–153. [Google Scholar] [CrossRef][Green Version]
- Mundt, O.J.; Hinkle, N.F. Ovules and Seeds. Appl. Environ. Microbiol. 1976, 32, 694–698. [Google Scholar] [CrossRef]
- Newcombe, G.; Harding, A.; Ridout, M.; Busby, P.E. A Hypothetical Bottleneck in the Plant Microbiome. Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Busby, P.E.; Peay, K.G.; Newcombe, G. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 2016, 209, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.S.C.; Okura, V.K.; Silveira, J.; Armanhi, L.; Jorrín, B.; Lozano, N.; José Da Silva, M.; González-Guerrero, M.; Migliorini De Araújo, L.; Verza, N.C.; et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef]
- Nelson, E.B. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 2004, 42, 271–309. [Google Scholar] [CrossRef]
- Jun, S.R.; Wassenaar, T.M.; Nookaew, I.; Hauser, L.; Wanchai, V.; Land, M.; Timm, C.M.; Lu, T.Y.S.; Schadt, C.W.; Doktycz, M.J.; et al. Diversity of Pseudomonas genomes, including populus-associated isolates, as revealed by comparative genome analysis. Appl. Environ. Microbiol. 2016, 82, 375–383. [Google Scholar] [CrossRef]
- Evans, L.M.; Slavov, G.T.; Rodgers-Melnick, E.; Martin, J.; Ranjan, P.; Muchero, W.; Brunner, A.M.; Schackwitz, W.; Gunter, L.; Chen, J.G.; et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 2014, 46, 1089–1096. [Google Scholar] [CrossRef]
- Ridout, M.; Houbraken, J.; Newcombe, G. Xerotolerance of Penicillium and Phialocephala fungi, dominant taxa of fine lateral roots of woody plants in the intermountain Pacific Northwest, USA. Rhizosphere 2017, 4, 94–103. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Stucky, B.J. Seqtrace: A graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech. 2012, 23, 90–93. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 13 April 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wright, K. Color Palettes, Colormaps, and Tools to Evaluate Them, version 1.7; R package pals; Github: San Francisco, CA, USA, 2021. [Google Scholar]
- Kahle, D.; Wickham, H. ggmap: Spatial Visualization with ggplot2. R J. 2013, 5, 144–161. [Google Scholar] [CrossRef]
- Wilke, C.O. Streamlined Plot Theme and Plot Annotations for “ggplot2”, version 1.1.1; R package cowplot; Github: San Francisco, CA, USA, 2020. [Google Scholar]
- Mcdonald, J.H. Handbook of Biological Statistics, 2nd ed.; Sparky House Publishing: Baltimore, MD, USA, 2009. [Google Scholar]
- Fisher, R.A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar]
| Total Incidence of Microbes | Percent of Seeds with Microbes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sampling Effort | Sampling Location | Number of Trees | Number of Seeds per Tree | Surface Sterilization | Fungi | Bacteria | Fungi and Bacteria | 0 | 1 | 2+ |
| Oregon 2017 | Westport, OR | 8 | 100 | Yes | 46 (5.8%) | 0 | 0 | 94.2% | 5.8% | 0% |
| Idaho 2017 | Moscow, ID | 1 | 1050 | No | 56 (5.3%) | 120 (11.4%) | 5 (0.05%) | 84% | 15.3% | 0.8% |
| Idaho 2018 | Moscow, ID | 1 | 60 | No | 15 (25%) | 46 (76.7%) | 9 (15%) | 18.3% | 63.3% | 18.3% |
| Idaho 2018 | Clearwater, ID | 21 | 60 | No | 260 (20.6%) | 353 (28%) | 51 (4.1%) | 56.7% | 37.9% | 5.3% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heitmann, S.; Bergmann, G.E.; Barge, E.; Ridout, M.; Newcombe, G.; Busby, P.E. Culturable Seed Microbiota of Populus trichocarpa. Pathogens 2021, 10, 653. https://doi.org/10.3390/pathogens10060653
Heitmann S, Bergmann GE, Barge E, Ridout M, Newcombe G, Busby PE. Culturable Seed Microbiota of Populus trichocarpa. Pathogens. 2021; 10(6):653. https://doi.org/10.3390/pathogens10060653
Chicago/Turabian StyleHeitmann, Sabrina, Gillian E. Bergmann, Edward Barge, Mary Ridout, George Newcombe, and Posy E. Busby. 2021. "Culturable Seed Microbiota of Populus trichocarpa" Pathogens 10, no. 6: 653. https://doi.org/10.3390/pathogens10060653
APA StyleHeitmann, S., Bergmann, G. E., Barge, E., Ridout, M., Newcombe, G., & Busby, P. E. (2021). Culturable Seed Microbiota of Populus trichocarpa. Pathogens, 10(6), 653. https://doi.org/10.3390/pathogens10060653







