Fungi Inhabiting the Wheat Endosphere
Abstract
1. Introduction
2. Isolation of Fungi from the Wheat Endosphere
3. Identification of Endophytic Fungi
4. Assortment and Role of Fungal Endophytes in Wheat
5. Molecular Interaction between Endophytic Fungi and Wheat
5.1. Epigenetic Control of Wheat–Fungi Interaction
5.1.1. DNA Methylation
5.1.2. Small RNAs
5.1.3. Long Non-Coding RNA (lncRNA)
6. Application of Endophytic Fungi in Modern Agronomy
7. New Perspectives and Research Needs
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grigoriev, I. Fungal genomics for energy and environment. In Genomics of Soil- and Plant-Associated Fungi: Soil Biology; Horwitz, B.A., Mukherjee, P.K., Mukherjee, M., Kubicek, C.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 36, pp. 11–27. [Google Scholar]
- Petrini, O. Fungal endophytes of tree leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Brock/Springer Series in Contemporary Bioscience; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Porras-Alfaro, A.; Bayman, P. Hidden fungi, emergent properties: Endophytes and microbiomes. Annu. Rev. Phytopathol. 2011, 49, 291–315. [Google Scholar] [CrossRef]
- Schardl, C.L.; Leuchtmann, A.; Spiering, M.J. Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 2004, 55, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttiläd, A.M.; Compante, S.; Campisanof, A.; Döringg, M.; Sessitsche, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Saikkonen, K.; Wali, P.R.; Helander, M. Genetic compatibility determines endophyte-grass combinations. PLoS ONE 2010, 5, e11395. [Google Scholar] [CrossRef]
- Kale, S.D.; Tyler, B.M. Entry of oomycete and fungal effectors into plant and animal host cells. Cell. Microbiol. 2011, 13, 1839–1848. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. N. Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Bacon, C.W.; White, J. (Eds.) Physiological adaptations in the evolution of endophytism in the Clavicipitaceae. In Microbial Endophytes; Taylor & Francis: New York, NY, USA, 2000; pp. 237–263. [Google Scholar]
- Saikkonen, K.I.D.; Gyllenberg, M. The persistence of vertically transmitted fungi in grass metapopulations. Proc. Biol. Sci. 2002, 269, 1397–1403. [Google Scholar] [CrossRef]
- Tadych, M.; Bergen, M.; Dugan, F.M.; White, J.F. Evaluation of the potential role of water in spread of conidia of the Neotyphodium endophyte of Poa ampla. Mycol. Res. 2007, 111, 466–472. [Google Scholar] [CrossRef]
- Gao, K.; Mendgen, K. Seed-transmitted beneficial endophytic Stagonospora sp can penetrate the walls of the root epidermis, but does not proliferate in the cortex, of Phragmites australis. Can. J. Bot. 2006, 84, 981–988. [Google Scholar] [CrossRef][Green Version]
- Redman, R.S.; Sheehan, K.B.; Stout, R.G.; Rodriguez, R.J.; Henson, J.M. Thermotolerance generated by plant/fungal symbiosis. Science 2002, 298, 1581. [Google Scholar] [CrossRef] [PubMed]
- Higgins, K.L.; Arnold, A.E.; Miadlikowska, J.; Sarvate, S.D.; Lutzoni, F. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol. Phylogenet. Evol. 2007, 42, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.C.; Shaw, A.J. Biogeographic and phylogenetic patterns in diversity of liverwort associated endophytes. Am. J. Bot. 2008, 95, 914–924. [Google Scholar] [CrossRef]
- Kumar, D.S.S.; Hyde, K.D. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004, 17, 69–90. [Google Scholar]
- O’Dell, T.E.; Massicotte, H.B.; Trappe, J.M. Root colonization of Lupinus latifolius Agardh. and Pinus contorta Dougl. by Phialocephala fortinii Wang & Wilcox. N. Phytol. 1993, 124, 93–100. [Google Scholar]
- Espinosa-Garcia, F.J.; Langenheim, J.H. The endophytic fungal community in leaves of a coastal redwood population diversity and spatial patterns. N. Phytol. 1990, 116, 89–97. [Google Scholar] [CrossRef]
- Li, W.C.; Zhou, J.; Guo, S.Y.; Guo, L.D. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers. 2007, 25, 69–80. [Google Scholar]
- Leuchtmann, A. Systematics, distribution, and host specificity of grass endophytes. Nat. Toxins 1993, 1, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Kai, W.; Zhiwei, Z. Occurrence of arbuscular mycorrhizas and dark septate endophytes in hydrophytes from lakes and streams in southwest China. Int. Rev. Hydrobiol. 2006, 91, 29–37. [Google Scholar] [CrossRef]
- Arnold, A.E.; Maynard, Z.; Gilbert, G.S.; Coley, P.D.; Kursar, T.A. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 2000, 3, 267–274. [Google Scholar] [CrossRef]
- Li, H.Y.; Shen, M.; Zhou, Z.P.; Li, T.; Wei, Y.L.; Lin, L.B. Diversity and cold adaptation of endophytic fungi from five dominant plant species collected from the Baima Snow Mountain, Southwest China. Fungal Divers. 2012, 54, 79–86. [Google Scholar] [CrossRef]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. N. Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef]
- Herre, E.A.; Mejía, L.C.; Kyllo, D.A.; Rojas, E.; Maynard, Z.; Butler, A.; Van Bael, S.A. Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology 2007, 88, 550–558. [Google Scholar] [CrossRef]
- Gao, F.K.; Dai, C.C.; Liu, X.Z. Mechanisms of fungal endophytes in plant protection against pathogens. Afr. J. Microbiol. Res. 2010, 4, 1346–1351. [Google Scholar]
- Hubbard, M.; Germida, J.; Vujanovic, V. Fungal endophytes improve wheat seed germination under heat and drought stress. Botany 2012, 90, 137–149. [Google Scholar] [CrossRef]
- Hubbard, M.; Germida, J.J.; Vujanovic, V. Fungal endophytes enhance wheat heat and drought tolerance in terms of grain yield and second-generation seed viability. J. Appl. Microbiol. 2014, 116, 109–122. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.L.; Kamran, M.; Hamayun, M.; Kang, S.M.; Kim, Y.H.; Lee, I.J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 2012, 17, 10754–10773. [Google Scholar] [CrossRef] [PubMed]
- © FAO. Crops and Livestock Products. 2021. Available online: http://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 6 October 2021).
- Mäkinen, H.; Kaseva, J.; Trnka, M.; Balek, J.; Kersebaum, K.C.; Nendel, C.; Gobin, A.; Olesen, J.E.; Bindi, M.; Ferrise, R.; et al. Sensitivity of European wheat to extreme weather. Field Crop. Res. 2018, 222, 209–217. [Google Scholar] [CrossRef]
- Araus, J.L.; Slafer, G.A.; Royo, C.; Serret, M.D. Breeding for yield potential and stress adaptation in cereals. Crit. Rev. Plant Sci. 2008, 27, 377–412. [Google Scholar] [CrossRef]
- Sieber, T.N. Endophytic fungi in forest trees: Are they mutualists? Fungal Biol. Rev. 2007, 21, 75–89. [Google Scholar] [CrossRef]
- Comby, M.; Lacoste, S.; Baillieul, F.; Profizi CDupont, J. Spatial and temporal variation of cultivable communities of co-occurring endophytes and pathogens in wheat. Front. Microbiol. 2016, 7, 403. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Gur, Y.; Ben-Moshe, S.; Sharon, O.; Kosman, E.; Mochli, E.; Sharon, A. Diversity of fungal endophytes in recent and ancient wheat ancestors Triticum dicoccoides and Aegilops sharonensis. FEMS Microbiol. Ecol. 2016, 92, 152. [Google Scholar] [CrossRef] [PubMed]
- Ripa, F.A.; Cao, W.D.; Tong, S.; Sun, J.G. Assessment of plant growth promoting and abiotic stress tolerance properties of wheat endophytic fungi. Biomed. Res. Int. 2019, 2019, 6105865. [Google Scholar] [CrossRef]
- Bouzouina, M.; Kouadria, R.; Lotmani, B. Fungal endophytes alleviate salt stress in wheat in terms of growth, ion homeostasis and osmoregulation. J. Appl. Microbiol. 2021, 130, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-X.; Zhou, H.; Xu, D.-Y.; Yu, C.-H.; Chen, Y.-Q.; Qu, L.-H. High diversity of endophytic fungi from the pharmaceutical plant, Heterosmilax japonica Kunth revealed by cultivation-independent approach. FEMS Microbiol. Lett. 2005, 249, 255–266. [Google Scholar] [CrossRef]
- Arnold, A.E.; Henk, D.A.; Eells, R.L.; Lutzoni, F.; Vilgalys, R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 2007, 99, 185–206. [Google Scholar] [CrossRef]
- Sun, X.; Kosman, E.; Sharon, O.; Ezrati, S.; Sharon, A. Significant host- and environment-dependent differentiation among highly sporadic fungal endophyte communities in cereal crops-related wild grasses. Environ. Microbiol. 2020, 20, 3357–3374. [Google Scholar] [CrossRef]
- Gamboa, M.A.; Laureano, S.; Bayman, P. Measuring diversity of endophytic fungi in leaf fragments: Does size matter? Mycopathologia 2003, 156, 41–45. [Google Scholar] [CrossRef]
- Larran, S.; Perello, A.; Simon, M.R.; Moreno, V. Isolation and analysis of endophytic microorganisms in wheat (Triticum aestivum L.) leaves. World J. Microbiol. Biotech. 2002, 18, 683–686. [Google Scholar] [CrossRef]
- Larran, S.; Perello, A.; Simon, M.R.; Moreno, V. The endophytic fungi from wheat (Triticum aestivum L.). World J. Microbiol. Biotechnol. 2007, 23, 565–572. [Google Scholar] [CrossRef]
- Cłapa, T.; Mikołajczak, K.; Błaszczyk, L.; Narożna, D. Development of high-resolution melting PCR (HRM-PCR) assay to identify native fungal species associated with the wheat endosphere. J. Appl. Genet. 2020, 61, 629–635. [Google Scholar] [CrossRef]
- Salamon, S.; Mikołajczak, K.; Błaszczyk, L.; Ratajczak, K.; Sulewska, H. Changes in root-associated fungal communities in Triticum aestivum ssp. spelta L. and Triticum aestivum ssp. vulgare L. under drought stress and in various soil processing. PLoS ONE 2020, 15, e0240037. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.C.; Jensen, B.; Jørgensen, H.J.; Latz, M.A.; Esteban, P.; Ding, Y.; Collinge, D.B. Selection of fungal endophytes with biocontrol potential against Fusarium head blight in wheat. Biol. Control. 2020, 144, 104222. [Google Scholar] [CrossRef]
- Sun, X.; Guo, L.-D. Endophytic fungal diversity: Review of traditional and molecular techniques. Mycology 2012, 3, 65–76. [Google Scholar]
- Torres, M.; White, J.F.; Tadych, M.; Bills, G. Isolation and Identification of Fungal Endophytes. Prospects and Applications for Plant-Associated Microbes. A laboratory Manual, Part B: Fungi; Paimo, BBI (Biobien Innovations): Turku, Finland, 2011; pp. 1–28. [Google Scholar]
- Comby, M.; Gacoin, M.; Robineau, M.; Rabenoelina, F.; Ptas, S.; Dupont, J.; Profizi, C.; Baillieul, F. Screening of wheat endophytes as biological control agents against Fusarium head blight using two different in vitro tests. Microbiol. Res. 2017, 202, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–662. [Google Scholar] [CrossRef]
- Geiser, D.M. Agriculture, and medicine. In Advances in Fungal Biotechnology for Industry; Springer: Berlin/Heidelberg, Germany, 2004; pp. 3–14. [Google Scholar]
- Lutzoni, F.; Kauff, F.; Cox, C.J.; McLaughlin, D.; Celio, G.; Dentinger, B.; Padamsee, M.; Hibbett, D.; James, T.Y.; Baloch, E.; et al. Assembling the fungal tree of life: Progress, classification, and evolution of subcellular traits. Am. J. Bot. 2004, 91, 1446–1480. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.W.; Petersen, R.H.; Lodge, D.J.; Bergemann, S.E.; Baumgartner, K.; Tulloss, R.E.; Lickey, E.; Cifuentes, J. Evolutionary consequences of putative intra-and interspecific hybridization in agaric fungi. Mycologia 2013, 105, 1577–1594. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Silar, P. Evolutionary Biology—Concepts, Molecular and Morphological Evolution; Pontarotti, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 317–328. [Google Scholar]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Letourneau, A.; Seena, S.; Marvanová, L.; Bärlocher, F. Potential use of barcoding to identify aquatic hyphomycetes. Fungal Divers. 2010, 40, 51–64. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Tailor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Ma, Z.; Michailides, T.J. A PCR-based technique for identification of Fusicoccum sp. from pistachio and various other hosts in California. Plant Dis. 2002, 86, 515–520. [Google Scholar] [CrossRef]
- Kuzdraliński, A.; Kot, A.; Szczerba, H.; Nowak, M.; Muszyńska, M. A review of Conventional PCR assays for the detection of selected phytopathogens of wheat. J. Mil. Microbiol. Biotechnol. 2017, 27, 175–189. [Google Scholar] [CrossRef]
- Sharma, R.; Polkade, A.V.; Shouche, Y.S. “Species Concept” in microbial taxonomy and systematics. Curr. Sci. 2015, 108, 1804–1814. [Google Scholar]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Hyde, K.D.; Pawłowska, J.; Ryberg, M.; Tedersoo, L.; Aas, A.B.; Alias, S.A.; Alves, A.; Anderson, C.L.; Antonelli, A.; et al. Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers. 2014, 67, 11–19. [Google Scholar] [CrossRef]
- Sun, X.; Guo, L.D.; Hyde, K.D. Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Divers. 2011, 47, 85–95. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Hirt, R.P.; Logsdon, J.M., Jr.; Healy, B.; Dorey, M.W.; Doolittle, W.F.; Embley, T.M. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. USA 1999, 19, 580–585. [Google Scholar] [CrossRef]
- Hong, S.B.; Go, S.J.; Shin, H.D.; Frisvad, J.C.; Samson, R.A. Polyhphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef] [PubMed]
- Maciá-Vicente, J.G.; Jansson, H.B.; Abdullah, S.K.; Descals, E.; Salinas, J.; Lopez-Llorca, L.V. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol. Ecol. 2008, 64, 90–105. [Google Scholar] [CrossRef]
- Brun, S.; Madrid, H.; Gerrits Van Den Ende, B.; Andersen, B.; Marinach-Patrice, C.; Mazier, D.; De Hoog, G.S. Multilocus phylogeny and MALDI-TOF analysis of the plant pathogenic species Alternaria dauci and relatives. Fungal Biol. 2013, 117, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Kulkarni, G.; Sonawane, M.S.; Shouche, Y.S. A new endophytic species of Chaetomium from Jatropha podagrica. Mycotaxon 2013, 124, 117–126. [Google Scholar] [CrossRef]
- Slippers, B.; Boissin, E.; Phillips, A.J.L.; Groenewald, J.Z.; Wingfield, M.J.; Postma, A.; Burgess, T.; Crous, P.W. Phylogenetic lineages in the Botryosphaeriales: A systematic an evolutionary framework. Stud. Mycol. 2013, 76, 31–49. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Dalling, J.W.; Gallery, R.E.; Maddison, D.R.; Davis, E.C.; Gibson, C.M.; Arnold, A.E. Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: A case study for analysing fungal environmental samples. Mycol. Res. 2009, 113, 432–449. [Google Scholar] [CrossRef]
- Gdanetz, K.; Trail, F. Wheat microbiome under four management strategies, and potential for endophytes in disease protection. Phytobiomes 2017, 1, 158–168. [Google Scholar] [CrossRef]
- Llorens, E.; Sharon, O.; Camañes, G.; García-Agustín, P.; Sharon, A. Endophytes from wild cereals protect wheat plants from drought by alteration of physiological responses of the plants to water stress. Environ. Microbiol. 2019, 21, 3299–3312. [Google Scholar] [CrossRef]
- Latz, M.A.; Jensen, B.; Collinge, D.B.; Jørgensen, H.J.L. Identification of two endophytic fungi that control Septoria tritici blotch in the field, using a structured screening approach. Biol. Control. 2020, 141, 104128. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Zhang, G.; Fu, C.; Liu, M.; Xing, Q.; et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Kraková, L.; Šoltys, K.; Otlewska, A.; Pietrzak, K.; Purkrtová, S.; Savická, D.; Puškárová, A.; Bučková, M.; Szemes, T.; Budiš, J.; et al. Comparison of methods for identification of microbial communities in book collections: Culture-dependent (sequencing and MALDI-TOF MS) and culture-independent (Illumina MiSeq). Int. Biodeterior. Biodegrad. 2018, 131, 51–59. [Google Scholar] [CrossRef]
- Mendoza, L.M.; Neef, A.; Vignolo, G.; Belloch, C. Yeast diversity during the fermentation of Andean chicha: A comparison of high-throughput sequencing and culturedependent approaches. Food Microbiol. 2017, 67, 1–10. [Google Scholar] [CrossRef]
- Malcolm, G.M.; Kuldau, G.A.; Gugino, B.K.; Jimenez-Gasco, M.D. Hidden host plant associations of soil borne fungal pathogens: Anecological perspective. Phytopathology 2013, 103, 538–544. [Google Scholar] [CrossRef]
- Zabalgogeazcoa, I. Fungal endophytes and their interaction with plant pathogens: A review. Span. J. Agric. 2008, 6, 138–146. [Google Scholar] [CrossRef]
- Dingle, J.; Mcgee, P.A. Some endophytic fungi reduce the density of pustules of Puccinia recondita f. sp. tritici in wheat. Mycol. Res. 2003, 107, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Zou, W.X.; Lu, H.; Tan, R.X. Antifungal activity of Artemisia annua endophyte cultures against phytopathogenic fungi. J. Biotechnol. 2001, 88, 277–282. [Google Scholar] [CrossRef]
- Rivera-Varas, V.V.; Freeman, T.A.; Gudmestad, N.C.; Secor, G.A. Mycoparasitism of Helminthosporium solani by Acremonium strictum. Phytopathology 2007, 97, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef]
- Card, S.D.; Faville, M.J.; Simpson, W.R.; Johnson, R.D.; Voisey, C.R.; de Bonth, A.C.M.; Hume, D.E. Mutualistic fungal endophytes in the Triticeae—Survey and description. FEMS Microbiol. Ecol. 2014, 88, 94–106. [Google Scholar] [CrossRef]
- Xu, K.G.; Jiang, Y.M.; Li, Y.K.; Xu, Q.Q.; Niu, J.S.; Zhu, X.X.; Li, Q.Y. Identification and pathogenicity of fungal pathogens causing black point in wheat on the north China plain. Indian J. Microbiol. 2018, 58, 159–164. [Google Scholar] [CrossRef]
- Zhan, G.; Tian, Y.; Wang, F.; Chen, X.; Guo, J.; Jiao, M.; Huang, L.; Kang, Z. A novel fungal hyperparasite of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS ONE 2014, 9, e111484. [Google Scholar] [CrossRef]
- Vujanovic, V.; Mavragani, D.; Hamel, C. Fungal communities associated with durum wheat production system: A characterization by growth stage, plant organ and preceding crop. Crop Prot. 2012, 37, 26–34. [Google Scholar] [CrossRef]
- Taheri, A.E.; Hamel, C.; Gan, Y. Cropping practices impact fungal endophytes and pathogens in durum wheat roots. Appl. Soil Ecol. 2016, 100, 104–111. [Google Scholar] [CrossRef]
- Al-Sadi, A.M. Bipolaris sorokiniana-induced black point, common root rot, and spot blotch diseases of wheat: A review. Front. Cell. Infect. Microbiol. 2021, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.-M.; Wang, M.; Gong, W.-F.; Zhang, L.-O. The screening and identification of the biological control fungi Chaetomium spp. against wheat common root rot. FEMS Microbiol. Lett. 2018, 365, fny242. [Google Scholar] [CrossRef]
- Istifadah, N.; McGee, P.A. Endophytic Chaetomium globosum reduces development of tan spot in wheat caused by Pyrenophora tritici-repentis. Australas. Plant Pathol. 2006, 35, 411–418. [Google Scholar] [CrossRef]
- Roberti, R.; Veronesi, A.R.; Cesari, A.; Cascone, A.; Di Berardina, I.; Bertini, L.; Caruso, C. Induction of PR proteins and resistance by the biocontrol agent Clonostachys rosea in wheat plants infected with Fusarium culmorum. Plant Sci. 2008, 175, 339–347. [Google Scholar] [CrossRef]
- Banerjee, A.; Mittra, B. Morphological modification in wheat seedlings infected by Fusarium oxysporum. Eur. J. Plant Pathol. 2018, 152, 521–524. [Google Scholar] [CrossRef]
- Alkan, M.; Özer, G.; İmren, M.; Özdemir, F.; Morgounov, A.; Dababat, A.A. First Report of Fusarium culmorum and Microdochium bolleyi Causing Root Rot on Triticale in Kazakhstan. Plant Dis. 2021, 1. [Google Scholar] [CrossRef]
- Littlefield, L.J. Biology of the Plant Rust: An Introduction; Iowa State University Press Ames: Iowa City, IA, USA, 1981; p. 103. [Google Scholar]
- Bartosiak, S.F.; Arseniuk, E.; Szechyńska-Hebda, M.; Bartosiak, E. Monitoring of natural occurrence and severity of leaf and glume blotch diseases of winter wheat and winter triticale incited by necrotrophic fungi Parastagonospora spp. and Zymoseptoria tritici. Agronomy 2021, 11, 967. [Google Scholar] [CrossRef]
- Abdel-Kareem, M.M.; Zohri, A.N.A.; Nasr, S.A.E.E. Novel marine yeast strains as plant growth-promoting agents improve defense in wheat (Triticum aestivum) against Fusarium oxysporum. J. Plant Dis. Prot. 2021, 128, 973–988. [Google Scholar] [CrossRef]
- Ponomarenko, A.; Goodwin, S.B.; Kema, G.H.J. Septoria tritici blotch (STB) of wheat. Plant Health Instr. 2011. [Google Scholar] [CrossRef]
- Błaszczyk, L.; Basińska-Barczak, A.; Ćwiek-Kupczyńska, H.; Gromadzka, K.; Popiel, D.; Stępień, Ł. Suppressive effect of Trichoderma spp. on toxigenic Fusarium species. Pol. J. Microbiol. 2017, 66, 85–100. [Google Scholar] [CrossRef]
- Rascovan, N.; Carbonetto, B.; Perrig, D.; Diaz, M.; Canciani, W.; Abalo, M.; Alloati, J.; Gonzalez-Ant, G.; Vazquez, M.P. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Karlsson, I.; Friberg, H.; Kolseth, A.K.; Steinberg, C.; Persson, P. Organic farming increases richness of fungal taxa in the wheat phyllosphere. Mol. Ecol. 2017, 26, 3424–3436. [Google Scholar] [CrossRef]
- Sapkota, R.; Jørgensen, L.N.; Nicolaisen, M. Spatiotemporal variation and networks in the mycobiome of the wheat canopy. Front. Plant Sci. 2017, 8, 1357. [Google Scholar] [CrossRef]
- Vujanovic, V.; Kim, S.H.; Lahlali, R.; Karunakaran, C. Spectroscopy and SEM imaging reveal endosymbiont-dependent components changes in germinating kernel through direct and indirect coleorhiza-fungus interactions under stress. Sci. Rep. 2019, 9, 1665. [Google Scholar] [CrossRef]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci. 2017, 8, 132. [Google Scholar] [CrossRef]
- Ofek, M.; Voronov-Goldman, M.; Hadar, Y.; Minz, D. Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities. Environ. Microbiol. 2013, 16, 2157–2167. [Google Scholar] [CrossRef]
- Yin, C.; Mueth, N.; Hulbert, S.; Schlatter, D.; Paulitz, T.C.; Schroeder, K.; Prescott, A.; Dhingra, A. Bacterial communities on wheat grown under long-term conventional tillage and no-tiol in the Pacific Northwest of the United States. Phytobiomes 2017, 1, 83–90. [Google Scholar] [CrossRef]
- Granzow, S.; Kaiser, K.; Wemheuer, B.; Pfeiffer, B.; Daniel, R.; Vidal, S.; Wemheuer, F. The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front. Microbiol. 2017, 8, 902. [Google Scholar] [CrossRef]
- Huang, Y.; Kuang, Z.; Wang, W.; Cao, L. Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control. 2016, 98, 27–33. [Google Scholar] [CrossRef]
- Kothe, E.; Turnau, K. Mycorrhizosphere communication: Mycorrhizal fungi and endophytic fungus-plant interactions. Front. Microbiol. 2018, 9, 3015. [Google Scholar] [CrossRef] [PubMed]
- Nicolaisen, M.; Justesen, A.F.; Knorr, K.; Wang, J.; Pinnschmidt, H.O. Fungal communities in wheat grain show significant co-existence patterns among species. Fungal Ecol. 2014, 11, 145–153. [Google Scholar] [CrossRef]
- Karlsson, I.; Friberg, H.; Steinberg, C.; Persson, P. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS ONE 2014, 9, e0111786. [Google Scholar] [CrossRef]
- Sapkota, R.; Knorr, K.; Jørgensen, L.N.; O’Hanlon, K.A.; Nicolaisen, M. Host genotype is an important determinant of the cereal phyllosphere mycobiome. N. Phytol. 2015, 207, 1134–1144. [Google Scholar] [CrossRef]
- Hertz, M.; Jensen, I.R.; Jensen, L.Ø.; Thomsen, S.N.; Winde, J.; Dueholm, M.S.; Sørensen, L.H.; Wollenberg, R.D.; Sørensen, H.O.; Sondergaard, T.E.; et al. The fungal community changes over time in developing wheat heads. Int. J. Food Microbiol. 2016, 222, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, E.; Savova-Bianchi, D.; Niculita-Hirzel, H. Major differences in the diversity of mycobiomes associated with wheat processing and domestic environments: Significant findings from high-throughput sequencing of fungal barcode ITS1. Int. J. Environ. Res. Public Health 2019, 16, 2335. [Google Scholar] [CrossRef] [PubMed]
- Schiro, G.; Colangeli, P.; Müller, M.E.H. A metabarcoding analysis of the mycobiome of wheat ears across a topographically heterogeneous field. Front Microbiol. 2019, 10, 2095. [Google Scholar] [CrossRef] [PubMed]
- Knorr, K.; Jørgensen, L.N.; Nicolaisen, M. Fungicides have complex effects on the wheat phyllosphere mycobiome. PLoS ONE 2019, 14, e0213176. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Mendes, R.; Bargaz, A.; Mauchline, T.H. Defining the wheat microbiome: Towards microbiome-facilitated crop production. Comput. Struct. Biotechnol. 2021, 19, 1200. [Google Scholar] [CrossRef] [PubMed]
- Simonin, M.; Dasilva, C.; Terzi, V.; Ngonkeu, E.L.M.; Diouf, D.; Kane, A.; Béna, G.; Moulin, L. Influence of plant genotype and soil on the wheat rhizosphere microbiome: Evidences for a core microbiome across eight African and European soils. FEMS Microbiol. Ecol. 2020, 96, fiaa067. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Yin, C.; Hulbert, S.; Paulitz, T.C. Core rhizosphere microbiomes of dry land wheat are influenced by location and land use history. Appl. Environ. Microbiol. 2019, 86, e02135-19. [Google Scholar]
- Rossmann, M.; Pérez-Jaramillo, J.E.; Kavamura, V.N.; Chiaramonte, J.B.; Dumack, K.; Fiore-Donno, A.M.; Mendes, L.W.; Ferreira, M.M.C.; Bonkowski, M.; Raaijmakers, J.M.; et al. Multitrophic interactions in the rhizosphere microbiome of wheat: From bacteria and fungi to protists. FEMS Microbiol. Ecol. 2020, 96, fiaa032. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247. [Google Scholar] [CrossRef]
- Fiorilli, V.; Belmondo, S.; Khouja, H.R.; Abbà, S.; Faccio, A.; Daghino, S.; Lanfranco, L. RiPEIP1, a gene from the arbuscular mycorrhizal fungus Rhizophagus irregularis, is preferentially expressed in planta and may be involved in root colonization. Mycorrhiza 2016, 26, 609–621. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under longterm organic and conventional farming. ISME J. 2014, 9, 1177–1194. [Google Scholar] [CrossRef]
- Martienssen, R.A.; Colot, V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 2001, 293, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jacobsen, S.E.; Reik, W. Epigenetic reprogramming in plant and animal development. Science 2010, 330, 622–627. [Google Scholar] [CrossRef]
- Asgari, S. Epigenetic modifications underlying symbiont–host interactions. Adv. Genet. 2014, 86, 253–276. [Google Scholar] [PubMed]
- Varga, S.; Soulsbury, C.D. Arbuscular mycorrhizal fungi change host plant DNA methylation systemically. Plant Biol. 2019, 21, 278–283. [Google Scholar] [CrossRef]
- Ichida, H.; Matsuyama, T.; Abe, T.; Koba, T. DNA adenine methylation changes dramatically during establishment of symbiosis. FEBS J. 2007, 274, 951–962. [Google Scholar] [CrossRef][Green Version]
- Kinoshita, T.; Motoaki, S. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef]
- Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. Plant Sci. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Khurana, J.P. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L. International Wheat Genome Sequencing Consortium. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Saripalli, G.; Sharma, C.; Gautam, T.; Singh, K.; Jain, N.; Prasad, P.; Roy, J.K.; Sharma, J.B.; Sharma, P.K.; Prabhu, K.V.; et al. Complex relationship between DNA methylation and gene expression due to Lr28 in wheat-leaf rust pathosystem. Mol. Biol. Rep. 2020, 47, 1339–1360. [Google Scholar] [CrossRef]
- Geng, S.; Kong, X.; Song, G.; Jia, M.; Guan, J.; Wang, F.; Qin, Z.; Wu, L.; Lan, X.; Li, A.; et al. DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. N. Phytol. 2019, 221, 1023–1035. [Google Scholar] [CrossRef]
- Sun, X.; Lin, L.; Sui, N. Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol. Biol. Rep. 2019, 46, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Wang, Y.; Yao, Y.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Gupta, O.P.; Permar, V.; Koundal, V.; Singh, U.D.; Praveen, S. MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f. sp. tritici infection. Mol. Biol. Rep. 2012, 39, 817–824. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, D.; Kanodia, P.; Prabhu, K.V.; Kumar, M.; Mukhopadhyay, K. Discovery of novel leaf rust responsive microRNAs in wheat and prediction of their target genes. J. Nucleic Acids. 2014, ID 570176. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Jin, H. A safe ride in extracellular vesicles—Small RNA trafficking between plant hosts and pathogens. Curr. Opin. Plant Biol. 2019, 52, 140–148. [Google Scholar] [CrossRef]
- Lauressergues, D.; Delaux, P.M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.P. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.-M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.-P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef]
- Jiao, J.; Peng, D. Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J. Plant Interact. 2018, 13, 514–521. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, W.; Hao, J.; Lv, S.; Wang, C.; Tong, W.; Wang, Y.; Wang, Y.; Liu, X.; Ji, W. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genom. 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Sinha, N.; Krishna, H.; Singh, P.K.; Gautam, T.; Prasad, P.; Gupta, P.K. A study of miRNAs and lncRNAs during Lr28-mediated resistance against leaf rust in wheat (Triticum aestivum L.). Physiol. Mol. Plant Pathol. 2020, 112, 101552. [Google Scholar] [CrossRef]
- Han, G.; Cheng, C.; Zheng, Y.; Wang, X.; Xu, Y.; Wang, W.; Zhu, S.; Cheng, B. Identification of long non-coding RNAs and the regulatory network responsive to arbuscular mycorrhizal fungi colonization in maize roots. Int. J. Mol. Sci. 2019, 20, 4491. [Google Scholar] [CrossRef] [PubMed]
- World Population Prospects 2019, Volume I: Comprehensive Tables; United Nations Department of Economic and Social Affairs/Population Division, United Nations New York: New York, NY, USA, 2019; ISBN 978-92-1-148327-7. eISBN: 978-92-1-004642-8.
- Larran, S.; Simon, M.R.; Moreno, M.V.; Siurana, M.S.; Perello, A. Endophytes from wheat as biocontrol agents against tan spot disease. Biol. Control. 2016, 92, 17–23. [Google Scholar] [CrossRef]
- Fernández-Pastor, I.; González-Menéndez, V.; Annang, F.; Toro, C.; Mackenzie, T.A.; Bosch-Navarrete, C.; Reyes, F. Pipecolisporin, a novel cyclic peptide with antimalarial and antitrypanosome activities from a wheat endophytic nigrospora oryzae. Pharmaceuticals 2021, 14, 268. [Google Scholar] [CrossRef]
- Reddy, G.V.P.; Tangtrakulwanich, K.; Wu, S.; Miller, J.H.; Ophus, V.L.; Prewett, J.; Jaronski, S.T. Evaluation of the effectiveness of entomopathogens for the management of wireworms (Coleoptera. Elateridae) on spring wheat. J. Invertebr. Pathol. 2014, 120, 43–49. [Google Scholar] [CrossRef]
- Keyser, C.A.; Jensen, B.; Meyling, N.V. Dual effects of Metarhizium spp. and Clonostachys rosea against an insect and a seed-borne pathogen in wheat. Pest Manag. Sci. 2015, 72, 517–526. [Google Scholar] [CrossRef]
- Herreraa, S.D.; Grossia, C.; Zawoznika, M.; Groppaa, M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 2016, 186–187, 37–43. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y.; Bonini, P.; Cardarelli, M. Coating seeds with endophytic fungi enhance growth, nutrient uptake, yield, and grain quality of winter wheat. Int. J. Plant Prot. 2015, 9, 171–189. [Google Scholar]
- Serfling, A.; Wirsel, S.G.R.; Lind, V.; Deising, H.B. Performance of the BioControl fungus piriformospora indica on wheat under greenhouse and field conditions. Phytopathology 2007, 97, 523–531. [Google Scholar] [CrossRef]
- Malik, A.; Butt, T.A.; Naqvi, S.T.A.; Yousaf, S.; Qureshi, M.K.; Zafar, M.I.; Farooq, G.; Nawaz, I.; Iqbal, M. Lead tolerant endophyte Trametes Hirsuta improved the growth and lead accumulation in the vegetative parts of Triticum aestivum L. Heliyon 2020, 6, e04188. [Google Scholar] [CrossRef] [PubMed]
- Ikram, M.; Ali, N.; Jan, G.; Jan, F.G.; Rahman, I.U.; Iqbal, A.; Hamayun, M. IAA producing fungal endophyte Penicillium roqueforti Thom., enhances stress tolerance and nutrients uptake in wheat plants grown on heavy metal contaminated soils. PLoS ONE 2018, 13, e0208150. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Junaid, J.M.; Dar, N.A.; Bhat, T.A.; Bhat, A.H.; Bhat, M.A. Commercial biocontrol agents and their mechanism of action in the management of plant pathogens. Int. J. Mod. Plant Anim. Sci. 2013, 1, 39–57. [Google Scholar]
- Kiss, L. A review of fungal antagonists of powdery mildews and their potential as biocontrol agents. Pestic. Manag. Sci. 2003, 59, 475–483. [Google Scholar] [CrossRef]
- Butt, T.; Jackson, C.; Magan, N. Fungi as Biocontrol Agents: Progress, Problems and Potential; CABI: Wallingford, UK, 2001; p. 390. [Google Scholar]
- Degenkolb, T.; Fog-Nielsen, K.; Dieckmann, R.; Branco-Rocha, F.; Chaverri, P.; Samuels, G.J.; Thrane, U.; von Dohren, H.; Vilcinskas, A.; Bruckner, H. Peptaibol, secondary metabolite, and hydrophobin pattern of commercial biocontrol agents formulated with species of the Trichoderma harzianum complex. Chem. Biodivers. 2015, 12, 662–684. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef]
- Simon, J.C.; Marchesi, J.R.; Mougel, C.; Selosse, M.A. Host-microbiota interactions: From holobiont theory to analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Geng, S.; Li, A.; Mao, Y.; Mao, L. RNAi technology for plant protection and its application in wheat. aBIOTECH 2021. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]
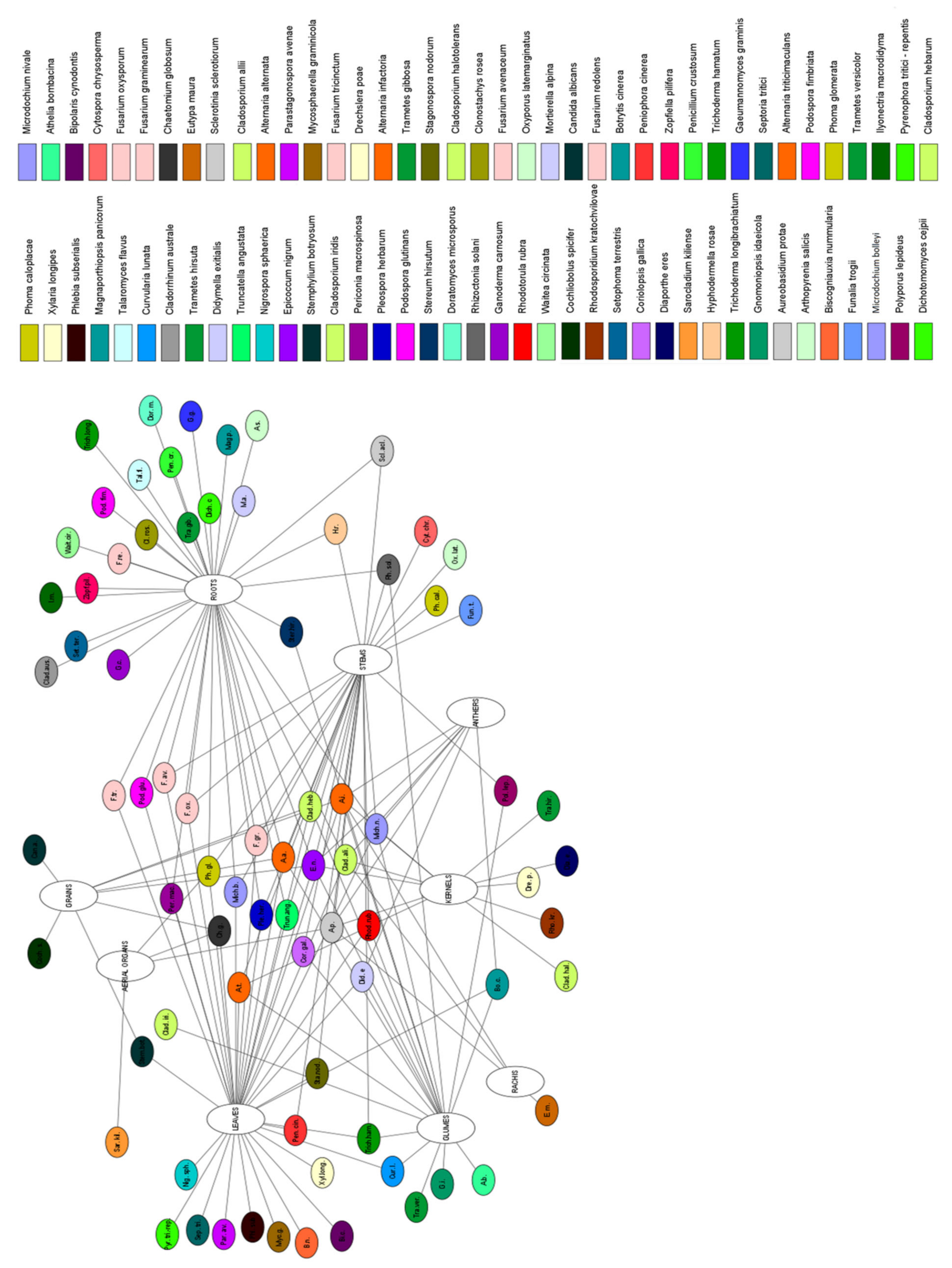
| Species 1 | Tissue Type | Role 2 | Localization | Wheat | References |
|---|---|---|---|---|---|
| Alternaria alternata | roots, stems, leaves | saprophyte/ pathogen | South Africa | Triticum aestivum | [46,47,90,91] |
| leaves | Argentina | ||||
| leaves, stems, glumes, grains | |||||
| Alternaria infectoria | rachis, leaves, glumes, anthers, stems, grains | pathogen | France | [38] | |
| leaves, glumes, grains | Argentina | [53] | |||
| Alternaria triticimaculans | leaves, glumes, stems, grains | pathogen | France | [38] | |
| Arthopyrenia salicis | roots | unrecognized | Poland | [49] | |
| Athelia bombacina | glumes | unrecognized | France | [38] | |
| Aureobasidium proteae | aerial organs | unrecognized | [92] | ||
| rachis, anthers, stems, grains | [38] | ||||
| Bipolaris cynodontis | leaves | unrecognized | Argentina | [47] | |
| Bipolaris sorokiniana | roots, stems, leaves, crowns | pathogen | Canada | Triticum durum | [47,53,93,94] |
| leaves | Argentina | Triticum aestivum | |||
| stems, grains | |||||
| Biscogniauxia nummularia | leaves | unrecognized | France | [38] | |
| Botrytis cinerea | leaves, glumes, anthers | pathogen | [38] | ||
| Candida albicans | grains | unrecognized | Argentina | [53] | |
| Chaetomium globosum | leaves | unrecognized mycoparasites | France | [38,47,53,92,95,96] | |
| leaves | Argentina | ||||
| aerial organs | France | ||||
| leaves, grains | Argentina | ||||
| Cladorrhinum australe | roots | unrecognized | Poland | [49] | |
| Cladosporium allii | grains, rachis, roots, leaves, anthers | unrecognized | France | [38] | |
| Cladosporium cladosporoides | roots, stems, leaves, awns, crowns | unrecognized mycoparasite | Canada | Triticum durum | [93] |
| Cladosporium halotolerans | grains | unrecognized | France | Triticum aestivum | [38,92] |
| grains | |||||
| Cladosporium herbarum | leaves | saprophyte/ pathogen | Argentina | [47] | |
| leaves, stems glumes, grains | [53] | ||||
| Cladosporium iridis | glumes | unrecognized | France | [38] | |
| Cladosporium minourae | roots, stems, leaves, awns, crowns | unrecognized | Canada | Triticum durum | [93] |
| Clonostachys rosea | roots | unrecognized mycoparasite | France | Triticum aestivum | [38,92,97] |
| Cochliobolus sativus (Bipolaris sorokiniana) | pathogen | Canada | Triticum durum | [93] | |
| Cochliobolus spicifier (Curvularia spicifera) | grains | pathogen | Argentina | Triticum aestivum | [53,90] |
| Coriolopsis gallica | glumes, stems | unrecognized | France | [38] | |
| Curvularia lunata | leaves, glumes | pathogen | Argentina | [53] | |
| Cytospora chrysosperma | stems | unrecognized | France | [38] | |
| Diaporthe eres (Phomopsis velata) | grains | unrecognized | |||
| Dichotomomyces cejpii (Aspergillus cejpii) | roots | unrecognized | |||
| Didymella exitialis (Neoascochyta exitialis) | leaves, glumes, anthers | pathogen | |||
| Doratomyces microsporus (Cephalotrichum microsporum) | roots | unrecognized | |||
| Drechslera poae (Pyrenophora poae) | grains | pathogen | |||
| Epicoccum nigrum | roots, stems, leaves | saprophyte/ pathogen | South Africa | [38,46,47,53] | |
| leaves, anthers, grains | pathogen | France | |||
| leaves | saprophyte/ pathogen | Argentina Argentina | |||
| leaves, stems, glumes, grains | |||||
| Eutypa maura | rachis | unrecognized | France | [38] | |
| Funalia trogii (Trametes trogii) | stems | unrecognized | |||
| Fusarium tricinctum | roots, stems, awns, crowns | pathogen | Canada | Triticum durum | [93] |
| Fusarium acuminatum | roots | pathogen | |||
| Fusarium avenaceum | roots, stems, leaves, awns, crowns | pathogen | [46,49,93] | ||
| roots, stems, leaves | pathogen | South Africa | Triticum aestivum | ||
| roots | pathogen | Poland | Triticum aestivum | ||
| Fusarium culmorum | pathogen | Canada | Triticum durum | [94] | |
| Fusarium graminearum | stems | pathogen | France | Triticum aestivum | [38,49,53] |
| leaves, stems | Argentina | ||||
| roots | Poland | Triticum aestivum spp. spelta | |||
| Fusarium oxysporum | leaves, stems | pathogen | Argentina | Triticum aestivum | [49,53,98] |
| roots | Poland | ||||
| Fusarium redolens | pathogen | France | [38,49] | ||
| Poland | |||||
| Fusarium reticulatum | roots, stems, leaves, awns, crowns | pathogen | Canada | Triticum durum | [93] |
| Fusarium torulosum | pathogen | ||||
| Fusarium tricinctum | leaves | pathogen | France | Triticum aestivum | [38,49] |
| roots | Poland | ||||
| Gaeumannomyces graminis | pathogen | Canada | Triticum durum | [93] | |
| France | Triticum aestivum | [38] | |||
| Ganoderma carnosum | unrecognized | ||||
| Geomyces pannorum (Pseudogymnoascus pannorum) | roots, stems, leaves, awns, crowns | unrecognized | Canada | Triticum durum | [93] |
| Gnomoniopsis idaeicola | glumes | unrecognized | France | Triticum aestivum | [38] |
| Hyphodermella rosae | roots, stems | unrecognized | |||
| Ilyonectria macrodidyma (Dactylonectria macrodidyma) | roots | unrecognized | |||
| Magnaporthiopsis panicorum | roots | unrecognized | Poland | Triticum aestivum spp. spelta | [49] |
| Microdochium bolleyi | roots, stems, leaves | unrecognized pathogen | South Africa | Triticum aestivum | [38,46,49,92,99] |
| roots | France | ||||
| Poland | |||||
| Microdochium nivale | roots, leaves, glumes, stems, anthers, grains | pathogen mycoparasite | France | [38,100] | |
| Mortierella hyalina | awns | unrecognized | Canada | Triticum durum | [93] |
| Mortierella alpina | roots | unrecognized | France | Triticum aestivum | [38] |
| Mycosphaerella graminicola (Zymoseptoria tritici) | leaves | pathogen | |||
| Nigrospora sphaerica (Nigrospora oryzae) | unrecognized | South Africa | [46] | ||
| Oxyporus latemarginatus | stems | France | [38,101] | ||
| Parastagonospora avenae | leaves | unrecognized/ pathogen | |||
| Penicillium aurantiogriseum | roots, stems, leaves, awns, crowns | Canada | Triticum durum | [93] | |
| Penicillium crustosum (Penicillium solitum) | roots | unrecognized | Poland | Triticum aestivum spp. vulgare | [49] |
| Penicillium griseofulvum | roots stems, leaves, awns, crowns | Canada | Triticum durum | [93] | |
| Peniophora cinerea | leaves, stems | France | Triticum aestivum | [38] | |
| Periconia macrospinosa | roots | Canada | Triticum durum | [38,49,93] | |
| roots, leaves | France | Triticum aestivum | |||
| roots | Poland | ||||
| Phlebia subserialis | leaves | France | [38] | ||
| Phoma caloplacae (Diederichomyces caloplacae) | stems | ||||
| Phoma glomerata (Didymella glomerata) | roots, stems, leaves | saprophyte/ pathogen | South Africa | [46,92] | |
| aerial organs | France | ||||
| Pleospora herbarum (Stemphylium vesicarium) | roots, stems, leaves | saprophyte/ pathogen | South Africa | [46,47,53] | |
| leaves | Argentina | ||||
| Podospora fimbriata (Schizothecium fimbriatum) | roots | unrecognized | France | [38] | |
| Podospora glutinans (Schizothecium glutinans) | roots, leaves | ||||
| Polyporus lepideus | glumes, stems | ||||
| Pyrenophora tritici-repentis | leaves, awns | pathogen | Canada | Triticum durum | [93] |
| leaves | France | Triticum aestivum | [38] | ||
| Rhizoctonia solani | stems, roots, glumes | ||||
| roots | Poland | [49] | |||
| Rhodosporidium kratochvilovae (Rhodotorula kratochvilovae) | grains | unrecognized | France | [38] | |
| Rhodotorula rubra (Rhodotorula mucilaginosa) | leaves | unrecognized/ mycoparasites | Argentina | [47,53,102] | |
| leaves, stems, glumes | |||||
| Sarocladium kiliense | stems | unrecognized | France | [38,92] | |
| aerial organs | |||||
| Sclerotinia sclerotiorum | roots, stems | pathogen | [38] | ||
| Septoria tritici (Zymoseptoria tritici) | leaves | unrecognized/ pathogen | Argentina | [53,103] | |
| Setophoma terrestris | roots | unrecognized | Poland | [49] | |
| Stagonospora nodorum (Parastagonospora nodorum) | stems, leaves | pathogen | South Africa | [46] | |
| Stemphylium botryosum | leaves, grains | unrecognized | Argentina | [53] | |
| Stereum hirsutum | roots, glumes | unrecognized | France | [38] | |
| Talaromyces flavus | roots | unrecognized | |||
| Thielavia hyalocarpa (Cladorrhinum hyalocarpum) | awns | unrecognized | Canada | Triticum durum | [93] |
| Trametes gibbosa | roots | unrecognized | France | Triticum aestivum | [38] |
| Trametes hirsuta | grains | unrecognized | |||
| Trametes versicolor | glumes | unrecognized | |||
| Trichoderma harzianum | roots | mycoparasites | Canada | Triticum durum | [93,104] |
| Trichoderma hamatum | leaves, stems, glumes | unrecognized/ mycoparasite | Argentina | Triticum aestivum | [53,104] |
| Trichoderma longibrachiatum | roots | unrecognized/ mycoparasite | Poland | [49,104] | |
| Truncatella angustata | roots, stems, leaves | unrecognized | South Africa | [46] | |
| Xylaria longipes | leaves | unrecognized | France | [38] | |
| Waitea circinata | roots | unrecognized | Poland | [49] | |
| Zopfiella pilifera | roots | unrecognized |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Błaszczyk, L.; Salamon, S.; Mikołajczak, K. Fungi Inhabiting the Wheat Endosphere. Pathogens 2021, 10, 1288. https://doi.org/10.3390/pathogens10101288
Błaszczyk L, Salamon S, Mikołajczak K. Fungi Inhabiting the Wheat Endosphere. Pathogens. 2021; 10(10):1288. https://doi.org/10.3390/pathogens10101288
Chicago/Turabian StyleBłaszczyk, Lidia, Sylwia Salamon, and Katarzyna Mikołajczak. 2021. "Fungi Inhabiting the Wheat Endosphere" Pathogens 10, no. 10: 1288. https://doi.org/10.3390/pathogens10101288
APA StyleBłaszczyk, L., Salamon, S., & Mikołajczak, K. (2021). Fungi Inhabiting the Wheat Endosphere. Pathogens, 10(10), 1288. https://doi.org/10.3390/pathogens10101288






