Nanopore-Sequencing Characterization of the Gut Microbiota of Melolontha melolontha Larvae: Contribution to Protection against Entomopathogenic Nematodes?
Abstract
1. Introduction
2. Results
2.1. Nanopore Sequencing Results
2.2. Bacterial Species Richness and α-Diversity
2.3. Diversity of Bacterial Microbiota of Control Group of M. melolontha Larvae
2.4. Selection of EPN-Resistant Insects
2.5. Diversity of the Bacterial Microbiota of the EPN-Resistant Group of Insects
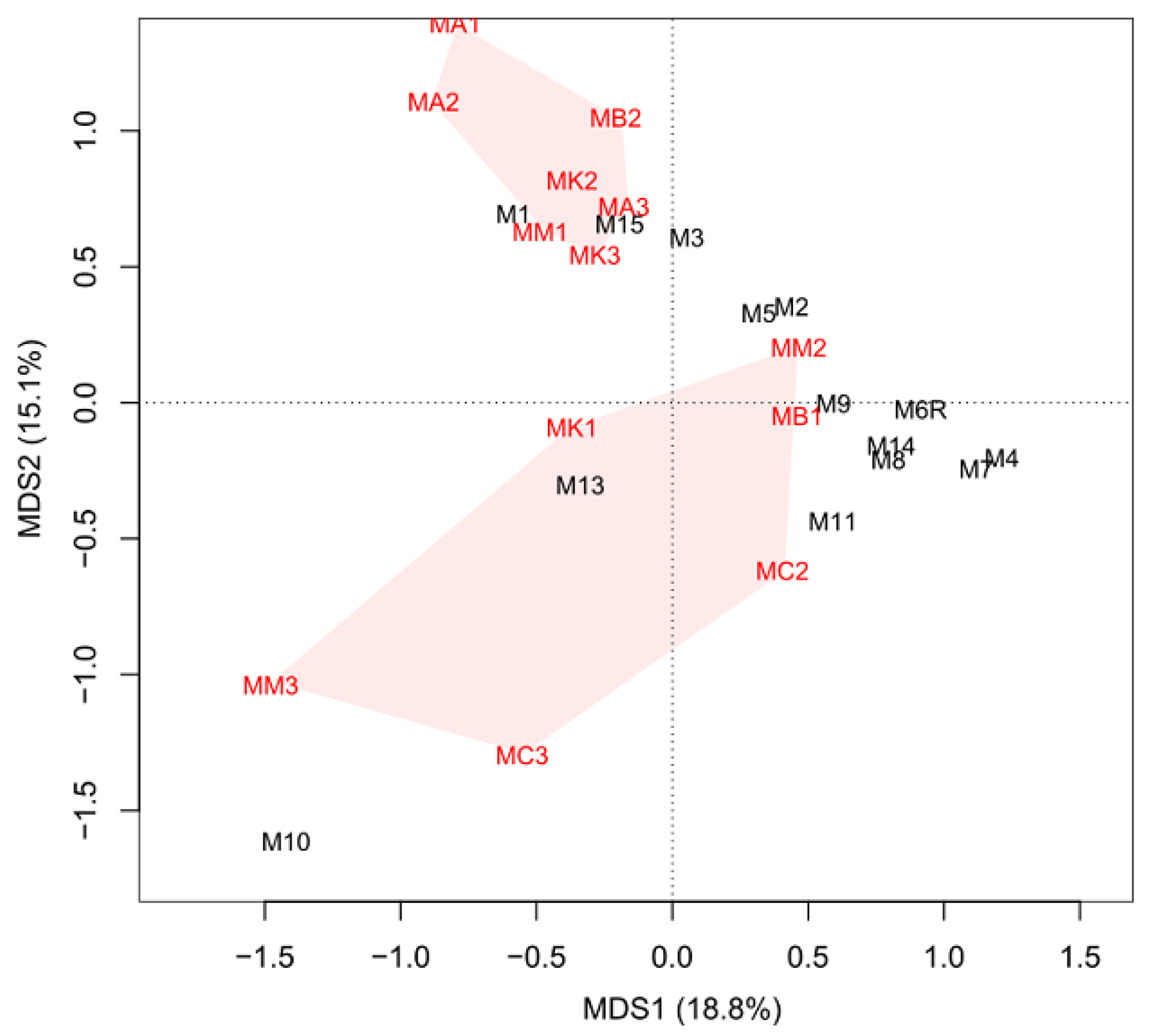
2.6. Comparison of the Midgut Microbiota between the Control and EPN-Resistantt Group of Insects
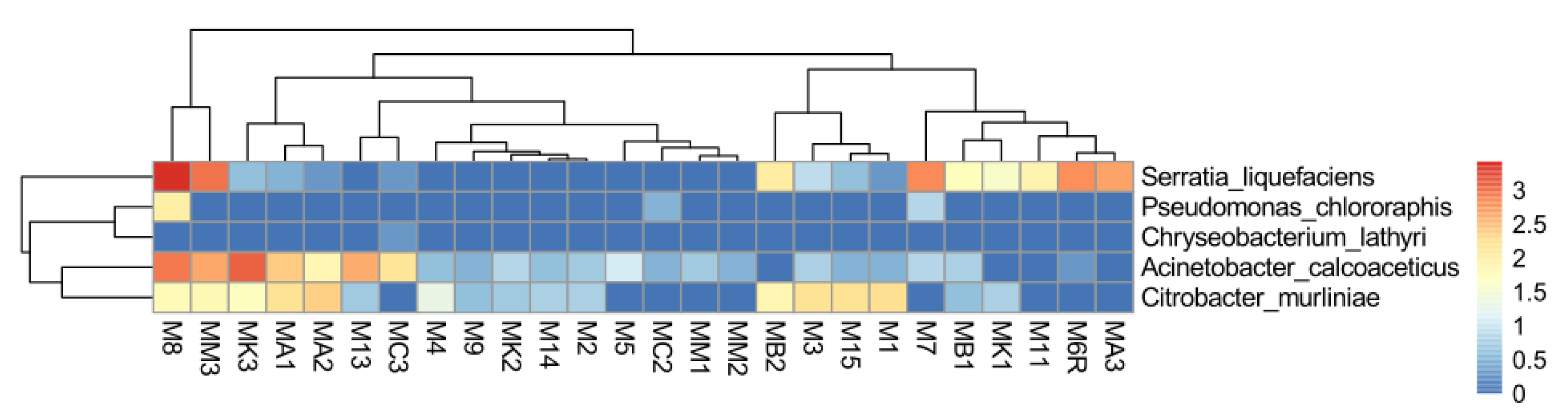
2.7. Abundance of EPN Symbionts and Their Antagonists in the Midgut Communities
3. Discussion
3.1. Natural Gut Microbiota of the Common Cockchafer
3.2. Alterations in Gut Microbial Composition in EPN-Resistant Insects
3.3. Presence of Xenorhabdus and Photorhabdus Entomopathogens and Their Antagonists in the M. melolontha Midgut Microbiota
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. DNA Extraction
4.3. Nanopore High Throughput Sequencing
4.4. Exploratory Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| EPN | entomopathogenic nematodes |
| IJ | infective juveniles |
| NCBI | National Centre for Biotechnology Information |
| PCoA | Principal Component Analysis |
| RA | relative abundance |
References
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Gurung, K.; Wertheim, B.; Falcao Salles, J. The microbiome of pest insects: It is not just bacteria. Entomol. Exp. Appl. 2019, 167, 156–170. [Google Scholar] [CrossRef]
- Malacrinò, A. Meta-omics tools in the world of insect-microorganism interactions. Biology 2018, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Colman, D.R.; Toolson, E.C.; Takacs-Vesbach, C.D. Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 2012, 21, 5124–5137. [Google Scholar] [CrossRef]
- Augustinos, A.A.; Tsiamis, G.; Cáceres, C.; Abd-Alla, A.M.M.; Bourtzis, K. Taxonomy, diet, and developmental stage contribute to the structuring of gut-associated bacterial communities in tephritid pest species. Front. Microbiol. 2019, 10, 2004. [Google Scholar] [CrossRef] [PubMed]
- Kucuk, R.A. Gut bacteria in the holometabola: A review of obligate and facultative symbionts. J. Insect Sci. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Oliver, K.M.; Perlman, S.J. Toxin-mediated protection against natural enemies by insect defensive symbionts. Adv. Insect Physiol. 2020, 58, 277–303. [Google Scholar] [CrossRef]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef]
- Flórez, L.V.; Scherlach, K.; Miller, I.J.; Rodrigues, A.; Kwan, J.C.; Hertweck, C.; Kaltenpoth, M. An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat. Commun. 2018, 9, 2478. [Google Scholar] [CrossRef]
- Wang, Y.; Rozen, D.E. Gut microbiota in the burying beetle, Nicrophorus vespilloides, provide colonization resistance against larval bacterial pathogens. Ecol. Evol. 2018, 8, 1646–1654. [Google Scholar] [CrossRef] [PubMed]
- Heise, P.; Liu, Y.; Degenkolb, T.; Vogel, H.; Schäberle, T.F.; Vilcinskas, A. Antibiotic-producing beneficial bacteria in the gut of the burying beetle Nicrophorus vespilloides. Front. Microbiol. 2019, 10, 1178. [Google Scholar] [CrossRef]
- Huang, S.-W.; Zhang, H.-Y.; Marshall, S.; Jackson, T.A. The scarab gut: A potential bioreactor for bio-fuel production. Insect. Sci. 2010, 17, 175–183. [Google Scholar] [CrossRef]
- Krishnan, M.; Bharathiraja, C.; Pandiarajan, J.; Prasanna, V.A.; Rajendhran, J.; Gunasekaran, P. Insect gut microbiome—An unexploited reserve for biotechnological application. Asian Pac. J. Trop. Biomed. 2014, 4 (Suppl. 1), S16–S21. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Kikuchi, Y. Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 2019, 41, 1–7. [Google Scholar] [CrossRef]
- Huiting, H.F.; Moral, L.G.; Griepink, F.C.; Ester, A. Biology, control and luring of the cockchafer, Melolontha melolontha. Appl. Plant Res. 2006, 1–34. [Google Scholar]
- Jackson, T.A.; Klein, M.G. Scarabs as pests: A continuing problem. Coleopt. Soc. Monogr. 2006, 5, 102–119. [Google Scholar] [CrossRef]
- Polavarapu, S.; Koppenhöfer, A.M.; Barry, J.D.; Holdcraft, R.J.; Fuzy, F.M. Entomopathogenic nematodes and neonicotinoids for remedial control of oriental beetle, Anomala orientalis (Coleoptera: Scarabaeidae), in high bush blueberry. Crop Prot. 2007, 26, 1266–1271. [Google Scholar] [CrossRef]
- Marianelli, L.; Paoli, F.; Torrini, G.; Mazza, G.; Benvenuti, C.; Binazzi, F.; Sabbatini Peverieri, G.; Bosio, G.; Venanzio, D.; Giacometto, E.; et al. Entomopathogenic nematodes as potential biological control agents of Popillia japonica (Coleoptera, Scarabaeidae) in Piedmont Region (Italy). J. Appl. Entomol. 2017, 142, 311–318. [Google Scholar] [CrossRef]
- Torrini, G.; Paoli, F.; Mazza, G.; Simoncini, S.; Benvenuti, C.; Strangi, A.; Tarasco, E.; Barzanti, G.P.; Bosio, G.; Cutino, I.; et al. Evaluation of indigenous entomopathogenic nematodes as potential biocontrol agents against Popillia japonica (Coleoptera: Scarabaeidae) in Northern Italy. Insects 2020, 11, 804. [Google Scholar] [CrossRef]
- Adams, B.J.; Fodor, A.; Koppenhöfer, H.S.; Stackebrandt, E.; Stock, S.P.; Klein, M.G. Biodiversity and systematics of nematode–bacterium entomopathogens, Biol. Control 2006, 37, 32–49. [Google Scholar] [CrossRef]
- Sajnaga, E.; Kazimierczak, W. Evolution and taxonomy of nematode-associated entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus: An overview. Symbiosis 2020, 80, 1–13. [Google Scholar] [CrossRef]
- Stock, S.P. Diversity, biology and evolutionary relationships. In Nematode Pathogenesis of Insects and Other Pests: Ecology and Applied Technologies for Sustainable Plant and Crop Protection; Campos-Herrera, R., Ed.; Springer International Publishing: Neuchâtel, Switzerland, 2015; pp. 3–27. [Google Scholar]
- Tobias, N.J.; Wolff, H.; Djahanschiri, B.; Grundmann, F.; Kronenwerth, M.; Shi, Y.M.; Simonyi, S.; Grün, P.; Shapiro-Ilan, D.; Pidot, S.J.; et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2017, 2, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Skowronek, M.; Sajnaga, E.; Pleszczyńska, M.; Kazimierczak, W.; Lis, M.; Wiater, A. Bacteria from the midgut of common cockchafer (Melolontha melolontha L.) larvae exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes: Isolation and molecular identification. Int. J. Mol. Sci. 2020, 21, 580. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Reese, J.M.; Casanova-Torres, A.M.; Goodrich-Blair, H.; Forst, S. Microbial population dynamics in the hemolymph of Manduca sexta infected with Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl. Environ. Microbiol. 2014, 80, 4277–4285. [Google Scholar] [CrossRef] [PubMed]
- Cambon, M.C.; Lafont, P.; Frayssinet, M.; Lanois, A.; Ogier, J.-C.; Pagès, S.; Parthuisot, N.; Ferdy, J.-B.; Gaudriault, S. Bacterial community profile after the lethal infection of Steinernema–Xenorhabdus pairs into soil-reared Tenebrio molitor larvae. FEMS Microbiol. Ecol. 2020, 96, fiaa009. [Google Scholar] [CrossRef]
- Wollenberg, A.C.; Jagdish, T.; Slough, G.; Hoinville, M.E.; Wollenberg, M.S. Death becomes them: Bacterial community dynamics and stilbene antibiotic production in cadavers of Galleria mellonella killed by Heterorhabditis and Photorhabdus spp. Appl. Environ. Microbiol. 2016, 82, 5824–5837. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based fourth-generation DNA sequencing technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef]
- Egert, M.; Stingl, U.; Bruun, L.D.; Brune, A.; Friedrich, M.W.; Pommerenke, B. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 2005, 71, 4556–4566. [Google Scholar] [CrossRef]
- Arias-Cordero, E.; Ping, L.; Reichwald, K.; Delb, H.; Platzer, M.; Boland, W. Comparative evaluation of the gut microbiota associated with the below- and above-ground life stages (larvae and beetles) of the forest cockchafer, Melolontha hippocastani. PLoS ONE 2012, 7, e51557. [Google Scholar] [CrossRef]
- Franzini, P.Z.; Ramond, J.B.; Scholtz, C.H.; Sole, C.L.; Ronca, S.; Cowan, D.A. The gut microbiomes of two Pachysoma MacLeay desert dung beetle species (Coleoptera: Scarabaeidae: Scarabaeinae) feeding on different diets. PLoS ONE 2016, 11, e0161118. [Google Scholar] [CrossRef]
- Bozorov, T.A.; Rasulov, B.A.; Zhang, D. Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera: Buprestidae) using high-throughput sequencing: Uncovering plant cell-wall degrading bacteria. Sci. Rep. 2019, 9, 4923. [Google Scholar] [CrossRef]
- Luo, C.; Li, Y.; Chen, Y.; Fu, C.; Long, W.; Xiao, X.; Liao, H.; Yang, Y. Bamboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti. Biotechnol. Biofuels 2019, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.Z.; Qi, F.H.; Wang, Z.Y. Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Andert, J.; Marten, A.; Brandl, R.; Brune, A. Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.). FEMS Microbiol. Ecol. 2010, 74, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Lupatini, M.; Suleiman, A.; Jacques, R.; Antoniolli, Z.; de Siqueira Ferreira, A.; Kuramae, E.; Roesch, L. Network topology reveals high connectance levels and few key microbial genera within soils. Front. Environ. Sci. 2014, 2, 10. [Google Scholar] [CrossRef]
- Venturelli, O.S.; Carr, A.C.; Fisher, G.; Hsu, R.H.; Lau, R.; Bowen, B.P.; Hromada, S.; Northen, T.; Arkin, A.P. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 2018, 14, e8157. [Google Scholar] [CrossRef]
- Råberg, L.; Sim, D.; Read, A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 2007, 318, 812–814. [Google Scholar] [CrossRef]
- Dillon, R.J.; Vennard, C.T.; Buckling, A.; Charnley, A.K. Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 2005, 8, 1291–1298. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Hernández-Martínez, P.; Naseri, B.; Navarro-Cerrillo, G.; Escriche, B.; Ferré, J.; Herrero, S. Increase in midgut microbiota load induces an apparent immune priming and increases tolerance to Bacillus thuringiensis. Environ. Microbiol. 2010, 12, 2730–2737. [Google Scholar] [CrossRef]
- Dubovskiy, I.M.; Grizanova, E.V.; Whitten, M.M.; Mukherjee, K.; Greig, C.; Alikina, T.; Kabilov, M.; Vilcinskas, A.; Glupov, V.V.; Butt, T.M. Immuno-physiological adaptations confer wax moth Galleria mellonella resistance to Bacillus thuringiensis. Virulence 2016, 7, 860–870. [Google Scholar] [CrossRef]
- Tetreau, G.; Grizard, S.; Patil, C.D.; Tran, F.H.; Tran Van, V.; Stalinski, R.; Laporte, F.; Mavingui, P.; Després, L.; Valiente Moro, C. Bacterial microbiota of Aedes aegypti mosquito larvae is altered by intoxication with Bacillus thuringiensis israelensis. Parasites Vectors 2018, 11, 121. [Google Scholar] [CrossRef]
- Xia, X.; Zheng, D.; Zhong, H.; Qin, B.; Gurr, G.M.; Vasseur, L.; Lin, H.; Bai, J.; He, W.; You, M. DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS ONE 2013, 8, e68852. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Xia, X.; Gurr, G.M.; Vasseur, L.; Zheng, D.; Zhong, H.; Qin, B.; Li, J.; Wang, Y.; Song, F.; Li, Y.; et al. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef]
- Wei, G.; Lai, Y.; Wang, G.; Chen, H.; Li, F.; Wang, S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. USA. 2017, 114, 5994–5999. [Google Scholar] [CrossRef]
- Lu, D.; Macchietto, M.; Chang, D.; Barros, M.M.; Baldwin, J.; Mortazavi, A.; Dillman, A.R. Activated entomopathogenic nematode infective juveniles release lethal venom proteins. PLoS Pathog. 2017, 13, e1006302. [Google Scholar] [CrossRef] [PubMed]
- Dierking, K.; Yang, W.; Schulenburg, H. Antimicrobial effectors in the nematode Caenorhabditis elegans: An outgroup to the Arthropoda. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016, 371, 20150299. [Google Scholar] [CrossRef] [PubMed]
- Midha, A.; Janek, K.; Niewienda, A.; Henklein, P.; Guenther, S.; Serra, D.O.; Schlosser, J.; Hengge, R.; Hartmann, S. The Intestinal roundworm Ascaris suum releases antimicrobial factors which interfere with bacterial growth and biofilm formation. Front. Cell Infect. Microbiol. 2018, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Longdon, B.; Bauer, S.; Chan, Y.S.; Miller, W.J.; Bourtzis, K.; Teixeira, L.; Jiggins, F.M. Symbionts commonly provide broad spectrum resistance to viruses in insects: A comparative analysis of Wolbachia strains. PLoS Pathog. 2014, 10, e1004369. [Google Scholar] [CrossRef] [PubMed]
- Dahan, D.; Preston, G.; Sealey, J.; King, K.C. Impacts of a novel defensive symbiosis on the nematode host microbiome. BMC Microbiol. 2020, 20, 159. [Google Scholar] [CrossRef]
- Caccia, S.; Di Lelio, I.; La Storia, A.; Marinelli, A.; Varricchio, P.; Franzetti, E.; Banyuls, N.; Tettamanti, G.; Casartelli, M.; Giordana, B.; et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 9486–9491. [Google Scholar] [CrossRef]
- Bando, H.; Okado, K.; Guelbeogo, W.M.; Badolo, A.; Aonuma, H.; Nelson, B.; Fukumoto, S.; Xuan, X.; Sagnon, N.; Kanuka, H. Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci. Rep. 2013, 3, 1641. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Wang, L.; Vega-Rodríguez, J.; Wang, G.; Wang, S. A gut symbiotic bacterium Serratia marcescens renders mosquito resistance to Plasmodium Infection through activation of mosquito immune responses. Front. Microbiol. 2019, 10, 1580. [Google Scholar] [CrossRef] [PubMed]
- Osimani, A.; Milanović, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Pasquini, M.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Loreto, N.; et al. The bacterial biota of laboratory-reared edible mealworms (Tenebrio molitor L.): From feed to frass. Int. J. Food Microbiol. 2018, 272, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, S.; Dominelli, N.; Brachmann, A.; Heermann, R. Phenotypic heterogeneity of the insect pathogen Photorhabdus luminescens: Insights into the fate of secondary cells. Appl. Environ. Microbiol. 2019, 85, e01910–e01919. [Google Scholar] [CrossRef]
- Koppenhöfer, A.M.; Fuzy, E. Effect of white grub developmental stage on susceptibility to entomopathogenic nematodes. J. Econ. Entomol. 2004, 97, 1842–1849. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996, 62, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA. 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Lagendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package R Version 2.5-6. 2019. Available online: https://cran.r-project.org (accessed on 1 December 2020).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Paleontol. Electron. 2001, 4, 9. [Google Scholar]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Constructing and analyzing microbiome networks in R. Methods Mol. Biol. 2018, 1849, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed]
| Sample Name | Developmental Stage of Larvae | Geographic Origin | Geographic Origin Code | EPN Exposure/EPN (Bacterial Symbiont) Species |
|---|---|---|---|---|
| M1, M2 | L2 | Forest 50°54′41.9″ N 22°19′27.1″ E | FK | none |
| M3, M4 | L3 | Forest 50°54′41.9″ N 22°19′27.1″ E | FK | none |
| M5, M6 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | none |
| M7 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | none |
| M8, M9 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | none |
| M10, M11, M12 | L2 | Forest 51°23′45.9″ N 22°47′43.2″ E | ZF | none |
| M13, M14 | L2 | Field 51°18′44.8″ N 22°24′33.7″ E | PF | none |
| M15 | L3 | Field 51°18′44.8″ N 22°24′33.7″ E | PF | none |
| MM1, MM2 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Heterorhabditis megidis (Photorhabdus temperata) |
| MM3 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Heterorhabditis megidis (Photorhabdus temperata) |
| MA1, MA2, MA3 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema arenarium (Xenorhabdus kozodoii) |
| MC1, MC2 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema carpocapse (Xenorhabdus nematophila) |
| MC3 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema carpocapsae (Xenorhabdus nematophila) |
| MB1, MB2 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema bicornutum (Xenorhabdus budapestensis) |
| MB3 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema bicornutum (Xenorhabdus budapestensis) |
| MK1, MK2 | L2 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema kraussei (Xenorhabdus bovienii) |
| MK3 | L3 | Forest nursery 51°23′37.0″ N 22°29′44.0″ E | KF | Steinernema kraussei (Xenorhabdus bovienii) |
| Taxname | Relative Abundance (Mean in %) and 95% Confidence Interval | |
|---|---|---|
| Control Larvae | EPN-Resistant Larvae | |
| Phylum level | ||
| Firmicutes | 72.85 [63.16–82.54] | 48.45 [33.59–63.31] |
| Proteobacteria | 12.15 [5.61–18.69] | 20.17 [12.68–27.65] |
| Actinobacteria | 5.85 [2.42–9.27] | 19.35 [10.74–27.96] |
| Bacteroidetes | 6.75 [3.03–10.47] | 6.18 [2.33–10.03] |
| Tenericutes | 0.32 [0.05–0.58] | 1.77 [0.10–3.43] |
| Verrucomicrobia | 0.26 [0.02–0.50] | 1.01 [0.34–61.68] |
| Class level | ||
| Clostridia | 49.43 [33.29–65.58] | 31.95 [18.85–45.06] |
| Actinobacteria | 5.85 [2.42–9.27] | 19.35 [10.74–27.96] |
| Bacilli | 12.74 [4.21–21.27] | 5.99 [3.44–8.53] |
| γ-Proteobacteria | 6.89 [1.70–12.08] | 10.09 [3.67–16.50] |
| Erysipelotrichia | 7.81 [0–19.04] | 8.54 [0–17.65] |
| Bacteroidia | 6.55 [2.74–10.36] | 5.70 [1.82–9.57] |
| α-Proteobacteria | 3.14 [0.03–6.24] | 7.75 [3.95–11.54] |
| β-Proteobacteria | 1.07 [0.32–1.81] | 1.01 [0–2.07] |
| Mollicutes | 0.32 [0.05–0.58] | 1.77 [0.10–3.43] |
| Taxname | Relative Abundance (Mean in %) and 95% Confidence Interval | |
|---|---|---|
| Control Larvae | EPN-Resistant Larvae | |
| Lachnospiraceae | 21.44 [11.31–31.56] | 14.11 [5.80–22.42] |
| Ruminococcaceae | 18.04 [10.72–25.36] | 13.27 [2.47–24.07] |
| Erysipelotrichaceae | 7.81 [0.25–19.04] | 8.54 [0–17.56] |
| Bacteroidacae | 4.95 [1.79–8.10] | 5.08 [1.28–8.89] |
| Microbacteriaceae | 1.37 [0.19–2.55] | 6.00 [2.23–9.76] |
| Enterobacteriaceae | 2.12 [0–4.85] | 4.19 [0.23–8.16] |
| Bacillaceae | 4.13 [0.95–7.30] | 1.88 [1.08–2.69] |
| Sporomusaceae | 2.00 [0.64–3.07] | 1.42 [0.56–2.28] |
| Nocardioidaceae | 0.39 [0.13–0.66] | 1.35 [0–2.86] |
| Hungateiclostridiaceae | 2.54 [1.12–3.95] | 0.36 [0.24–0.49] |
| Clostridiaceae | 1.70 [1.04–2.36] | 1.02 [0.58–1.46] |
| Staphylococcaceae | 1.47 [0–3.64] | 1.02 [0.24–1.80] |
| Rhizobiaceae | 1.01 [0–2.62] | 1.35 [0.62–2.09] |
| Enterococcaceae | 1.52 [0–3.77] | 0.70 [0–1.43] |
| Propionibacteriaceae | 0.58 [0.02–1.13] | 1.52 [0–3.22] |
| Morganellaceae | 1.91 [0–5.98] | 0.05 [0.01–0.10] |
| Erwiniaceae | 0.28 [0.01–0.54] | 1.75 [0–3.72] |
| Sphingomonadaceae | 0.44 [0–1.14] | 1.56 [0.6–2.52] |
| Bradyrhizobiaceae | 0.68 [0.19–1.17] | 1.22 [0.53–1.91] |
| Relative Abundance Median | Mean Rank | Mann-Whitney U | p-Value | |||
|---|---|---|---|---|---|---|
| Control n = 14 | EPN-Resistant n = 13 | Control n = 14 | EPN-Resistant n = 13 | |||
| Lachnoclostridium | 2.028 | 0.956 | 8.92 | 5.07 | 46 | 0.029 |
| Anaerotignum | 1.850 | 0.264 | 9.59 | 4.01 | 28 | 0.001 |
| Tyzzerella | 1.256 | 0.085 | 9.48 | 4.51 | 31 | 0.004 |
| Paludicola | 0.492 | 0.169 | 8.81 | 5.19 | 49 | 0.044 |
| Ruminiclostridium | 0.325 | 0.051 | 9.19 | 4.81 | 39 | 0.010 |
| Mesorhizobium | 0.019 | 0.061 | 5.46 | 8.54 | 42.5 | 0.019 |
| Galbitalea | 0.001 | 0.139 | 5.22 | 8.78 | 36 | 0.007 |
| Conyzicola | 0.002 | 0.056 | 4.96 | 9.04 | 29 | 0.002 |
| Mycolicibacterium | 0.054 | 0.360 | 5.37 | 8.63 | 40 | 0.013 |
| Aeromicrobium | 0.007 | 0.232 | 5.29 | 8.70 | 38 | 0.009 |
| Herbiconiux | 0.003 | 0.162 | 5.04 | 8.96 | 31 | 0.003 |
| Cellulomonas | 0.003 | 0.223 | 5.20 | 8.80 | 35.5 | 0.005 |
| Friedmanniella | 0.001 | 0.164 | 4.59 | 9.41 | 19 | 0.0003 |
| Methylobacterium | 0.006 | 0.076 | 5.61 | 8.39 | 46.5 | 0.030 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajnaga, E.; Skowronek, M.; Kalwasińska, A.; Kazimierczak, W.; Ferenc, K.; Lis, M.; Wiater, A. Nanopore-Sequencing Characterization of the Gut Microbiota of Melolontha melolontha Larvae: Contribution to Protection against Entomopathogenic Nematodes? Pathogens 2021, 10, 396. https://doi.org/10.3390/pathogens10040396
Sajnaga E, Skowronek M, Kalwasińska A, Kazimierczak W, Ferenc K, Lis M, Wiater A. Nanopore-Sequencing Characterization of the Gut Microbiota of Melolontha melolontha Larvae: Contribution to Protection against Entomopathogenic Nematodes? Pathogens. 2021; 10(4):396. https://doi.org/10.3390/pathogens10040396
Chicago/Turabian StyleSajnaga, Ewa, Marcin Skowronek, Agnieszka Kalwasińska, Waldemar Kazimierczak, Karolina Ferenc, Magdalena Lis, and Adrian Wiater. 2021. "Nanopore-Sequencing Characterization of the Gut Microbiota of Melolontha melolontha Larvae: Contribution to Protection against Entomopathogenic Nematodes?" Pathogens 10, no. 4: 396. https://doi.org/10.3390/pathogens10040396
APA StyleSajnaga, E., Skowronek, M., Kalwasińska, A., Kazimierczak, W., Ferenc, K., Lis, M., & Wiater, A. (2021). Nanopore-Sequencing Characterization of the Gut Microbiota of Melolontha melolontha Larvae: Contribution to Protection against Entomopathogenic Nematodes? Pathogens, 10(4), 396. https://doi.org/10.3390/pathogens10040396