Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreated Wastewater
Abstract
1. Introduction
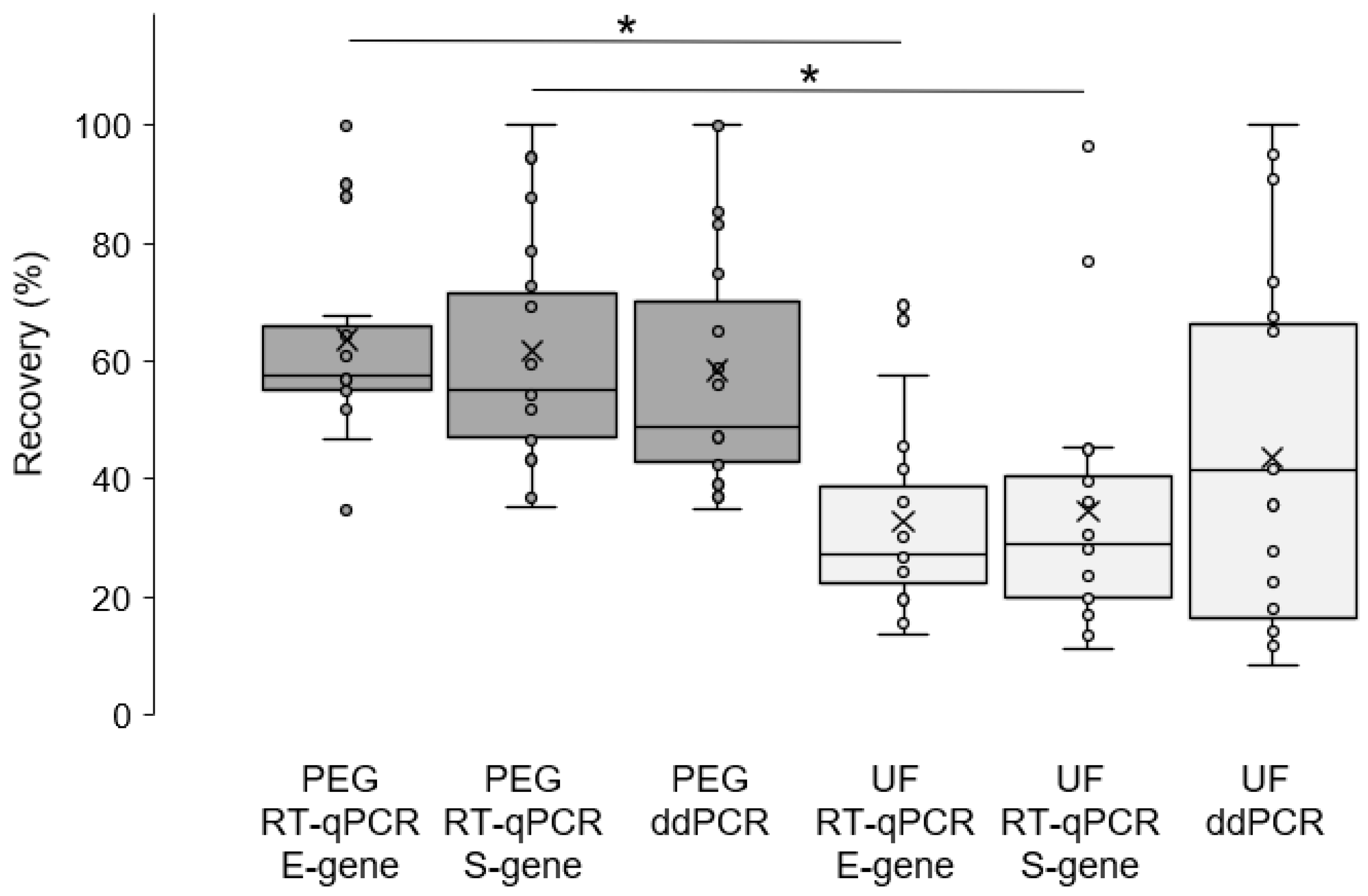
2. Results and Discussion
3. Material and Methods
3.1. Wastewater Samples
3.2. Spiking of Wastewater with SARS-CoV-2 Positive Clinical Specimens
3.3. Virus Concentration
3.4. RNA Extraction
3.5. RT-qPCR
3.6. Droplet Digital (dd)PCR
3.7. Quantification of Virus Recovery
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Inf. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Cevik, M.; Bamford, C.G.G.; Ho, A. COVID-19 pandemic-a focused review for clinicians. Clin. Microbiol. Infect. 2020, 26, 842–847. [Google Scholar] [CrossRef]
- van Doorn, A.S.; Meijer, B.; Frampton, C.M.A.; Barclay, M.L.; de Boer, N.K.H. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment. Pharmacol. Ther. 2020, 52, 1276–1288. [Google Scholar] [CrossRef]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Chaubey, A.K.; Pittman, C.U., Jr.; Mlsna, T.; Mohan, D. Coronavirus (SARS-CoV-2) in the environment: Occurrence, persistence, analysis in aquatic systems and possible management. Sci. Total Environ. 2020, 142698. [Google Scholar] [CrossRef]
- Lu, D.; Huang, Z.; Luo, J.; Zhang, X.; Sha, S. Primary concentration-The critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: A mini-review. Sci. Total Environ. 2020, 747, 41245. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R.; et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Rusinol, M.; Martínez-Puchol, S.; Fores, E.; Itarte, M.; Girones, R.; Bofill-Mas, S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health 2020, 17, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Been, F.; Heijnen, L.; Petterson, S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: Opportunities and challenges. Curr. Opin. Environ. Sci. Health 2020, 17, 49–71. [Google Scholar] [CrossRef]
- Pérez-Cataluña, A.; Cuevas-Ferrando, E.; Randazzo, W.; Falcó, I.; Allende, A.; Sánchez, G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2020, 758, 143870. [Google Scholar] [CrossRef]
- Philo, S.E.; Keim, E.K.; Swanstrom, R.; Ong, A.Q.W.; Burnor, E.A.; Kossik, A.L.; Harrison, J.C.; Demeke, B.A.; Zhou, N.A.; Beck, N.K.; et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2020, 760, 144215. [Google Scholar] [CrossRef]
- Barril, P.A.; Pianciola, L.A.; Mazzeo, M.; Ousset, M.J.; Jaureguiberry, M.V.; Alessandrello, M.; Sánchez, G.; Oteiza, J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2020, 756, 144105. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Weber, F.A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany-Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751, 141750. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef]
- Amdiouni, H.; Maunula, L.; Hajjami, K.; Faouzi, A.; Soukri, A.; Nourlil, J. Recovery comparison of two virus concentration methods from wastewater using cell culture and real-time PCR. Curr. Microbiol. 2012, 65, 432–437. [Google Scholar] [CrossRef]
- Trottier, J.; Darques, R.; Ait Mouheb, N.; Partiot, E.; Bakhache, W.; Deffieu, M.S.; Gaudin, R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health 2020, 10, 100157. [Google Scholar] [CrossRef]
- Corpuz, M.V.A.; Buonerba, A.; Vigliotta, G.; Zarra, T.; Ballesteros, F., Jr.; Campiglia, P.; Belgiorno, V.; Korshin, G.; Naddeo, V. Viruses in wastewater: Occurrence, abundance and detection methods. Sci. Total Environ. 2020, 745, 140910. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 2020, 5, e00614–e00620. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumke, R.; de la Cruz Barron, M.; Oertel, R.; Helm, B.; Kallies, R.; Berendonk, T.U.; Dalpke, A. Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreated Wastewater. Pathogens 2021, 10, 195. https://doi.org/10.3390/pathogens10020195
Dumke R, de la Cruz Barron M, Oertel R, Helm B, Kallies R, Berendonk TU, Dalpke A. Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreated Wastewater. Pathogens. 2021; 10(2):195. https://doi.org/10.3390/pathogens10020195
Chicago/Turabian StyleDumke, Roger, Magali de la Cruz Barron, Reinhard Oertel, Björn Helm, Rene Kallies, Thomas U. Berendonk, and Alexander Dalpke. 2021. "Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreated Wastewater" Pathogens 10, no. 2: 195. https://doi.org/10.3390/pathogens10020195
APA StyleDumke, R., de la Cruz Barron, M., Oertel, R., Helm, B., Kallies, R., Berendonk, T. U., & Dalpke, A. (2021). Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreated Wastewater. Pathogens, 10(2), 195. https://doi.org/10.3390/pathogens10020195





