Clinical Parasitology and Parasitome Maps as Old and New Tools to Improve Clinical Microbiomics
Abstract
:1. Introduction
2. Role and Relationship amongst Gut Microbiota Citizens
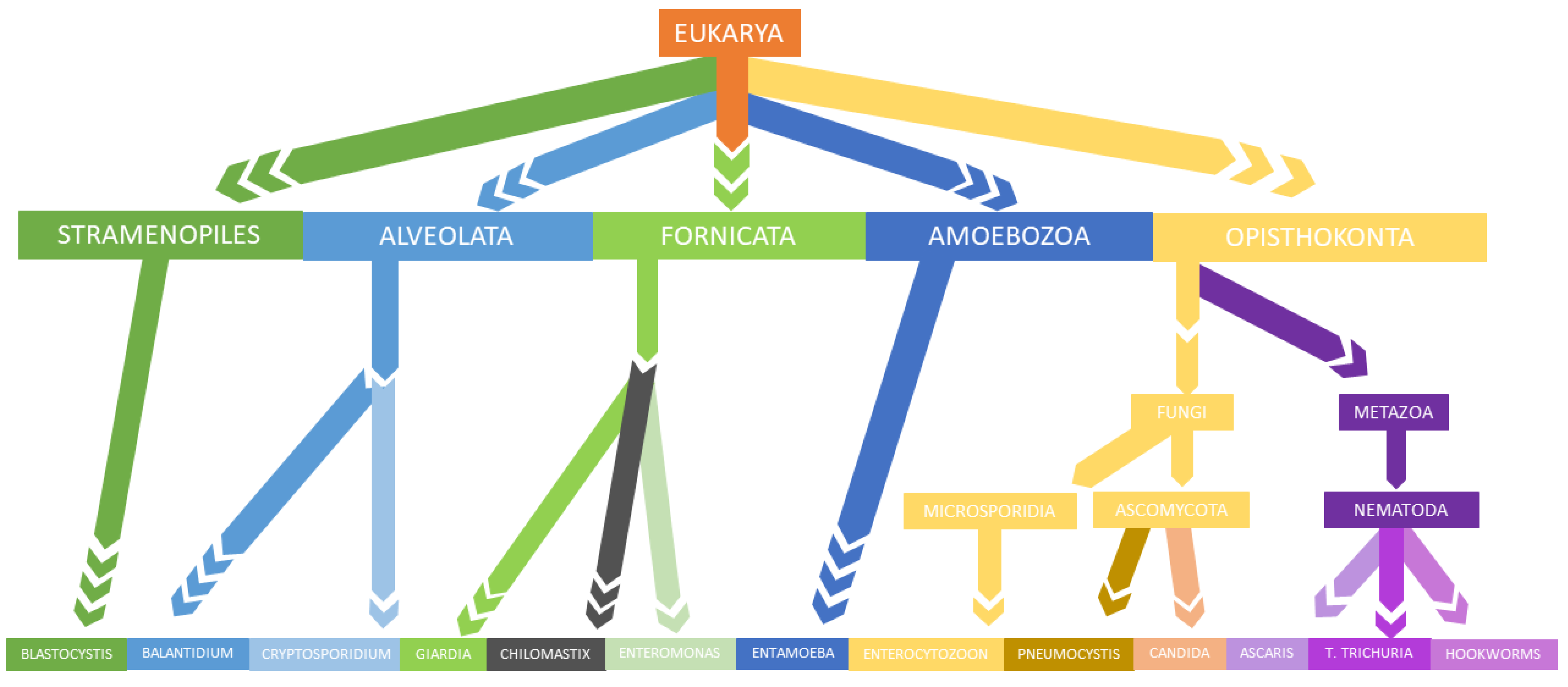
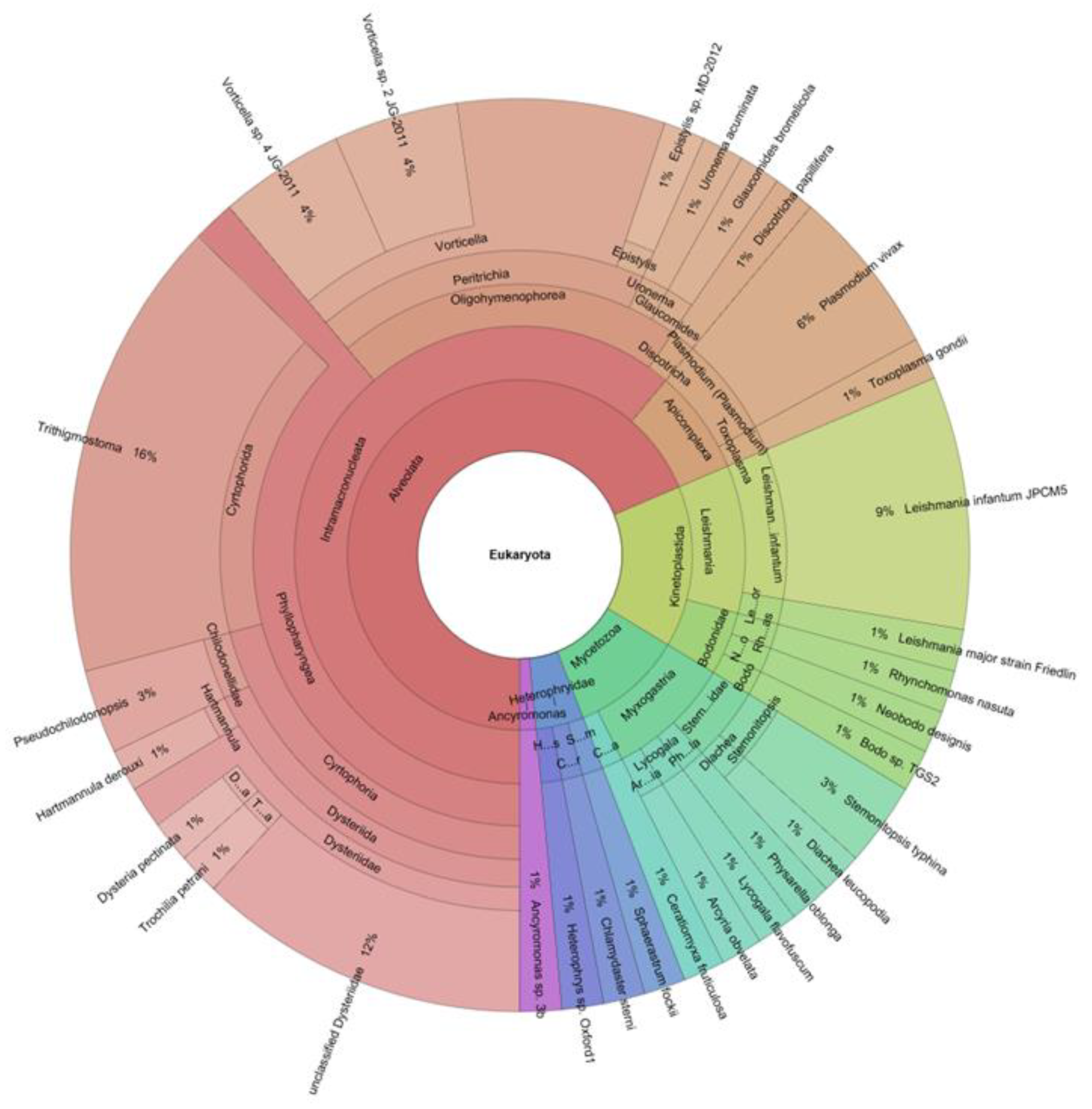
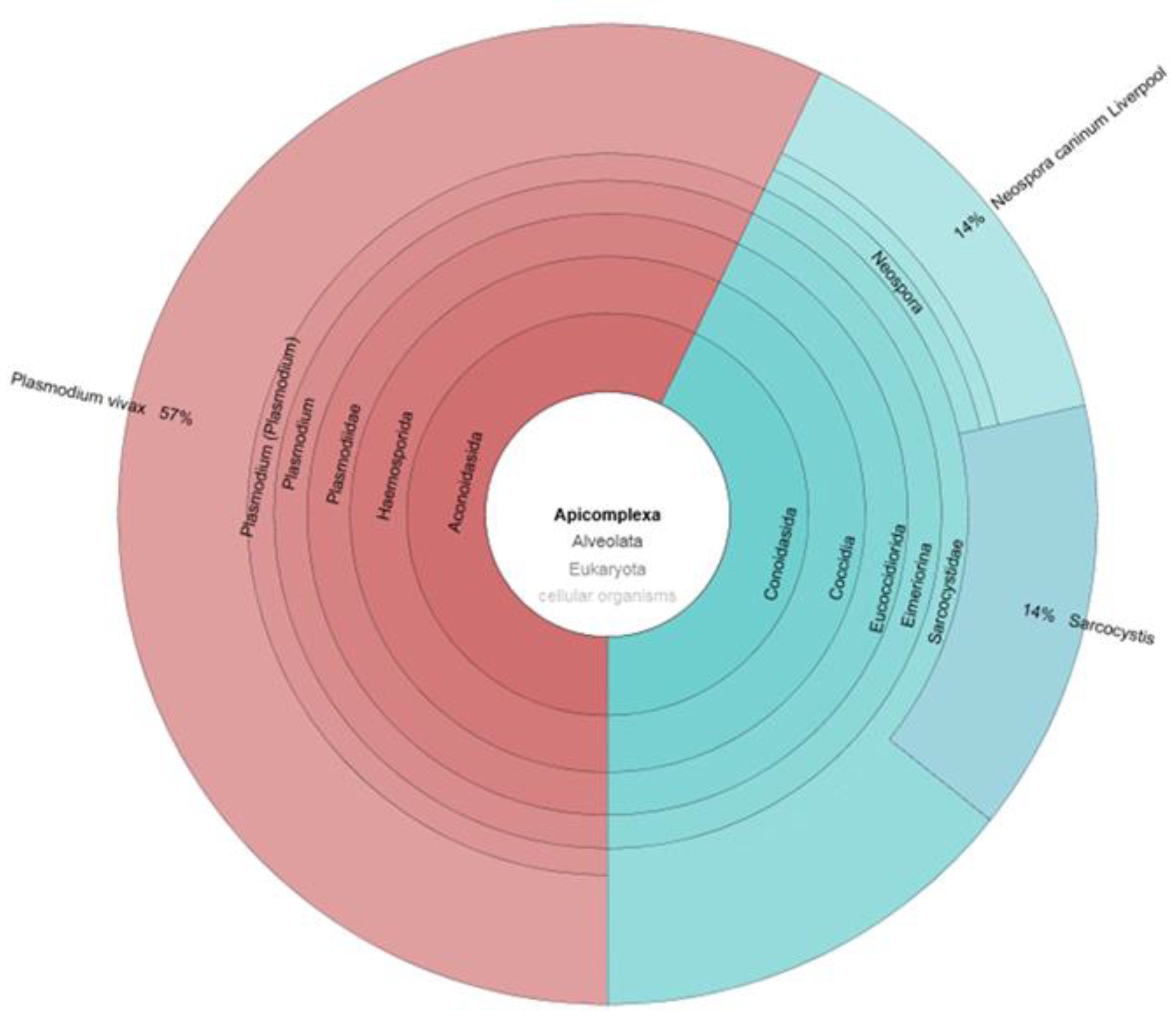
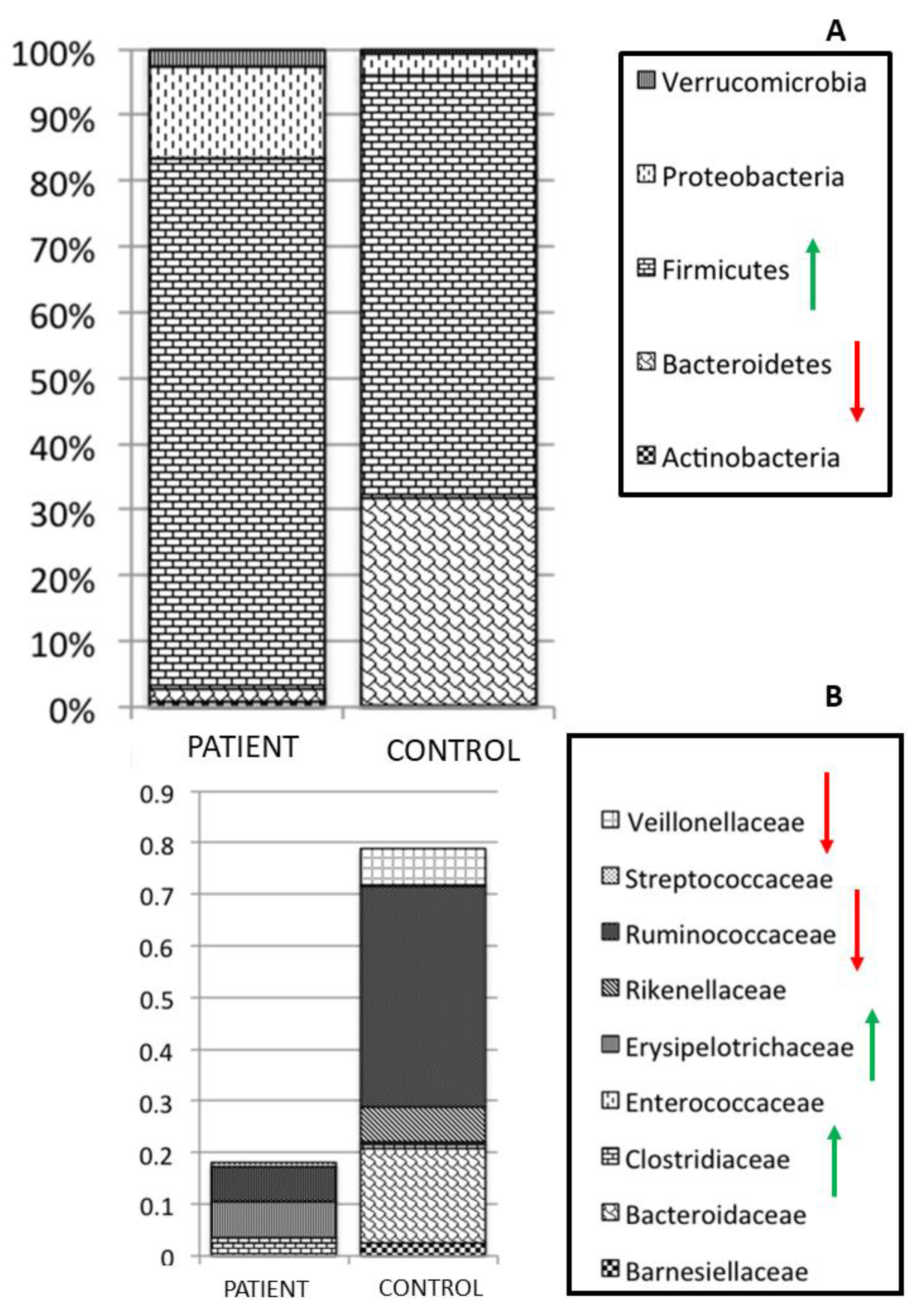
3. Parasites and Gut Microbiota Profiling
| Reference | Parasite | Type of Infection | Type of Study | Type of Sequencing | Gut Microbiota Composition |
|---|---|---|---|---|---|
| [67] | Blastocystis | Natural | Human study | 16S rDNA sequencing | Increase in Clostridia, Mollicutes, Clostridiales, Ruminococcaceae and Prevotellaceae. Decrease in Bacilli, Lactobacillales, Enterococcaceae, Streptococcaceae, Lactobacillaceae and Enterobacteriaceae. |
| [68] | Blastocystis | Natural | Human study | 16S rDNA sequencing | No significant change. |
| [69] | Blastocystis | Mouse infected with Blastocystis ST3 | Murine model | 16S rDNA sequencing | Increase in Bilophila and Butyricimonas in the Blastocystis-colonized group. Decrease in Defluviitaleaceae. |
| [65] | Blastocystis | Natural | Human study | Shotgun metagenomics | Increase in Firmicutes and Clostridiales. Decrease in Bacteroides. |
| [61] | Entamoeba | Natural | Human study | 16S rDNA sequencing | Increase in Bacteroidales, Mollicutes, Christensenellaceae, Elusimicrobiaceae, Ruminococcaceae, Paraprevotellaceae, Treponema, Parabacteroides, Streptococcus, Butyrivibrio, Oscillospira, Desulfovibrio and Ruminococcus bromii. Decrease in Prevotella, Prevotella copri. |
| [62] | Entamoeba | Natural | Human study | 16S rDNA sequencing | Positive correlation between Bifidobacterium vs. B. fragilis, and Prevotella vs. Bacteroides. Negative correlation between Bifidobacterium vs. Bacteroides. |
| [63] | Entamoeba | Culture of E. histolytica | Culture study | 16S rDNA sequencing | Increase Lactobacillaceae, Clostridiaceae, Erysipelotrichaceae, and Bifidobacteriaceae. |
| [58] | Schistosoma | Infected with S. mansoni cercariae (Sm-exp) | Murine model | 16S rDNA sequencing | The authors supposed that susceptibility to Schistosoma infection in mice is partially dependent on the composition of the host baseline microbiota. |
| [59] | Necator americanus | Percutaneous infection with third-stage larvae N. americanus | Longitudinal study | 16S rDNA sequencing | Increase in Tenericutes, Mollicutes and Parabacteroides. |
| [60] | Strongiloides stercoralis | Natural | Case report | 16S rDNA sequencing | Increase in Bifidobacterium, Blautia, Ruminococcus, Bacteroides, Corynebacterium, Colinsella, Streptococcus, Coprococcus, and Oscillospora. Decrease in Staphylococcus, Lactobacillus, and Pediococcus. |
4. New Molecular Approaches in Translational and Clinical Parasitology
5. Gut Microbiota Profiling as Tools to Restore and Modulate Gut Microbiota
6. Materials and Methods
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Bancroft, A.J.; Hayes, K.S.; Grencis, R.K. Life on the Edge: The Balance between Macrofauna, Microflora and Host Immunity. Trends Parasitol. 2012, 28, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becattini, S.; Littmann, E.R.; Carter, R.A.; Kim, S.G.; Morjaria, S.M.; Ling, L.; Gyaltshen, Y.; Fontana, E.; Taur, Y.; Leiner, I.M.; et al. Commensal Microbes Provide First Line Defense against Listeria Monocytogenes Infection. J. Exp. Med. 2017, 214, 1973–1989. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Tam, N.M.; Jogova, M.; Robertson, M.L.; Li, Y.; Lupp, C.; Finlay, B.B. Antibiotic-Induced Perturbations of the Intestinal Microbiota Alter Host Susceptibility to Enteric Infection. Infect. Immun. 2008, 76, 4726–4736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity 2012, 37, 158–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molloy, M.J.; Bouladoux, N.; Belkaid, Y. Intestinal Microbiota: Shaping Local and Systemic Immune Responses. Semin. Immunol. 2012, 24, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, K.L.; Targan, S.R.; Elson, C.O. Microbiota Activation and Regulation of Innate and Adaptive Immunity. Immunol. Rev. 2014, 260, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Putignani, L.; Del Chierico, F.; Vernocchi, P.; Cicala, M.; Cucchiara, S.; Dallapiccola, B. Dysbiotrack Study Group Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition. Inflamm. Bowel Dis. 2016, 22, 487–504. [Google Scholar] [CrossRef] [Green Version]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes Ratio of the Human Microbiota Changes with Age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Putignani, L.; Del Chierico, F.; Petrucca, A.; Vernocchi, P.; Dallapiccola, B. The Human Gut Microbiota: A Dynamic Interplay with the Host from Birth to Senescence Settled during Childhood. Pediatr. Res. 2014, 76, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Del Chierico, F.; Vernocchi, P.; Bonizzi, L.; Carsetti, R.; Castellazzi, A.M.; Dallapiccola, B.; de Vos, W.; Guerzoni, M.E.; Manco, M.; Marseglia, G.L.; et al. Early-Life Gut Microbiota under Physiological and Pathological Conditions: The Central Role of Combined Meta-Omics-Based Approaches. J. Proteom. 2012, 75, 4580–4587. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-Free Recipients Reveal Host Habitat Selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putignani, L.; Dallapiccola, B. Foodomics as Part of the Host-Microbiota-Exposome Interplay. J. Proteom. 2016, 147, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [Green Version]
- Bobardt, S.D.; Dillman, A.R.; Nair, M.G. The Two Faces of Nematode Infection: Virulence and Immunomodulatory Molecules From Nematode Parasites of Mammals, Insects and Plants. Front. Microbiol. 2020, 11, 577846. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Horvat, J.C.; Beagley, K.W.; Hansbro, P.M. Immunological Decision-Making: How Does the Immune System Decide to Mount a Helper T-Cell Response? Immunology 2008, 123, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Lindon, J.C.; Wilson, I.D. The Challenges of Modeling Mammalian Biocomplexity. Nat. Biotechnol. 2004, 22, 1268–1274. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced Diversity of Faecal Microbiota in Crohn’s Disease Revealed by a Metagenomic Approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High Prevalence of Adherent-Invasive Escherichia Coli Associated with Ileal Mucosa in Crohn’s Disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal Flora in Inflammatory Bowel Disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Parfrey, L.W.; Walters, W.A.; Knight, R. Microbial Eukaryotes in the Human Microbiome: Ecology, Evolution, and Future Directions. Front. Microbiol. 2011, 2, 153. [Google Scholar] [CrossRef] [Green Version]
- Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitson, J.P.; Maizels, R.M. Vaccination against Helminth Parasite Infections. Expert Rev. Vaccines 2014, 13, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Putignani, L. Epidemiology of Human Cryptosporidiosis. Cryptosporidium Parasite Dis. 2014, 43–79. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Virgin, H.W. Kingdom-Agnostic Metagenomics and the Importance of Complete Characterization of Enteric Microbial Communities. Gastroenterology 2014, 146, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, L.; Koziol, A.; Marcos, S.; Botnen, A.B.; Aizpurua, O.; Gopalakrishnan, S.; Limborg, M.T.; Gilbert, M.T.P.; Alberdi, A. Holo-Omics: Integrated Host-Microbiota Multi-Omics for Basic and Applied Biological Research. iScience 2020, 23, 101414. [Google Scholar] [CrossRef]
- Putignani, L.; Gasbarrini, A.; Dallapiccola, B. Potential of Multiomics Technology in Precision Medicine. Curr. Opin. Gastroenterol. 2019, 35, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Matijašić, M.; Meštrović, T.; Paljetak, H.Č.; Perić, M.; Barešić, A.; Verbanac, D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 2020, 21, 2668. [Google Scholar] [CrossRef] [Green Version]
- Zuo, T.; Wong, S.H.; Lam, K.; Lui, R.; Cheung, K.; Tang, W.; Ching, J.Y.L.; Chan, P.K.S.; Chan, M.C.W.; Wu, J.C.Y.; et al. Bacteriophage Transfer during Faecal Microbiota Transplantation in Clostridium Difficile Infection Is Associated with Treatment Outcome. Gut 2018, 67, 634–643. [Google Scholar] [CrossRef] [Green Version]
- Dalmasso, M.; Hill, C.; Ross, R.P. Exploiting Gut Bacteriophages for Human Health. Trends Microbiol. 2014, 22, 399–405. [Google Scholar] [CrossRef]
- Jonas, O.; Seifman, R. Do We Need a Global Virome Project? Lancet Glob. Health 2019, 7, e1314–e1316. [Google Scholar] [CrossRef] [Green Version]
- Minot, S.; Bryson, A.; Chehoud, C.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Rapid Evolution of the Human Gut Virome. Proc. Natl. Acad. Sci. USA 2013, 110, 12450–12455. [Google Scholar] [CrossRef] [Green Version]
- Townsend, E.M.; Kelly, L.; Muscatt, G.; Box, J.D.; Hargraves, N.; Lilley, D.; Jameson, E. The Human Gut Phageome: Origins and Roles in the Human Gut Microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 643214. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Maurice, C.F. Ménage à Trois in the Human Gut: Interactions between Host, Bacteria and Phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Noverr, M.C. The Emerging World of the Fungal Microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [Green Version]
- Beheshti-Maal, A.; Shahrokh, S.; Ansari, S.; Mirsamadi, E.S.; Yadegar, A.; Mirjalali, H.; Zali, M.R. Gut Mycobiome: The Probable Determinative Role of Fungi in IBD Patients. Mycoses 2021, 64, 468–476. [Google Scholar] [CrossRef]
- Ianiro, G.; Iorio, A.; Porcari, S.; Masucci, L.; Perno, C.F.; Gasbarrini, A.; Putignani, L.; Cam-marota, G. How the Gut Parasitome Affects Human Health.
- Prommi, A.; Prombutara, P.; Watthanakulpanich, D.; Adisakwattana, P.; Kusolsuk, T.; Yoonuan, T.; Poodeepiyasawat, A.; Homsuwan, N.; Prummongkol, S.; Tanita, M.; et al. Intestinal Parasites in Rural Communities in Nan Province, Thailand: Changes in Bacterial Gut Microbiota Associated with Minute Intestinal Fluke Infection. Parasitology 2020, 147, 972–984. [Google Scholar] [CrossRef]
- Farthing, M.J. Giardiasis. Gastroenterol. Clin. N. Am. 1996, 25, 493–515. [Google Scholar] [CrossRef]
- Cryptosporidium–ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/B978012818731900080X?via%3Dihub (accessed on 21 September 2021).
- Putignani, L.; Menichella, D. Global Distribution, Public Health and Clinical Impact of the Protozoan Pathogen Cryptosporidium. Interdiscip. Perspect. Infect. Dis. 2010, 2010, 753512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caradonna, T.; Marangi, M.; Del Chierico, F.; Ferrari, N.; Reddel, S.; Bracaglia, G.; Normanno, G.; Putignani, L.; Giangaspero, A. Detection and Prevalence of Protozoan Parasites in Ready-to-Eat Packaged Salads on Sale in Italy. Food Microbiol. 2017, 67, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Formenti, F.; Cortés, A.; Brindley, P.J.; Cantacessi, C.; Rinaldi, G. A Bug’s Life: Delving into the Challenges of Helminth Microbiome Studies. PLoS Negl. Trop. Dis. 2020, 14, e0008446. [Google Scholar] [CrossRef]
- White, E.C.; Houlden, A.; Bancroft, A.J.; Hayes, K.S.; Goldrick, M.; Grencis, R.K.; Roberts, I.S. Manipulation of Host and Parasite Microbiotas: Survival Strategies during Chronic Nematode Infection. Sci. Adv. 2018, 4, eaap7399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzscheiter, M.; Layland, L.E.; Loffredo-Verde, E.; Mair, K.; Vogelmann, R.; Langer, R.; Wagner, H.; Prazeres da Costa, C. Lack of Host Gut Microbiota Alters Immune Responses and Intestinal Granuloma Formation during Schistosomiasis. Clin. Exp. Immunol. 2014, 175, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Yason, J.A.; Liang, Y.R.; Png, C.W.; Zhang, Y.; Tan, K.S.W. Interactions between a Pathogenic Blastocystis Subtype and Gut Microbiota: In Vitro and in Vivo Studies. Microbiome 2019, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro-Londono, M.A.; Bedoya-Urrego, K.; Garcia-Montoya, G.M.; Galvan-Diaz, A.L.; Alzate, J.F. Intestinal Parasitic Infection Alters Bacterial Gut Microbiota in Children. PeerJ 2019, 7, e6200. [Google Scholar] [CrossRef] [Green Version]
- Alzate, J.F.; Toro-Londoño, M.; Cabarcas, F.; Garcia-Montoya, G.; Galvan-Diaz, A. Contrasting Microbiota Profiles Observed in Children Carrying Either Blastocystis Spp. or the Commensal Amoebas Entamoeba Coli or Endolimax Nana. Sci. Rep. 2020, 10, 15354. [Google Scholar] [CrossRef] [PubMed]
- Rosa, B.A.; Supali, T.; Gankpala, L.; Djuardi, Y.; Sartono, E.; Zhou, Y.; Fischer, K.; Martin, J.; Tyagi, R.; Bolay, F.K.; et al. Differential Human Gut Microbiome Assemblages during Soil-Transmitted Helminth Infections in Indonesia and Liberia. Microbiome 2018, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Giacomin, P.; Zakrzewski, M.; Jenkins, T.P.; Su, X.; Al-Hallaf, R.; Croese, J.; de Vries, S.; Grant, A.; Mitreva, M.; Loukas, A.; et al. Changes in Duodenal Tissue-Associated Microbiota Following Hookworm Infection and Consecutive Gluten Challenges in Humans with Coeliac Disease. Sci. Rep. 2016, 6, 36797. [Google Scholar] [CrossRef]
- Cortés, A.; Clare, S.; Costain, A.; Almeida, A.; McCarthy, C.; Harcourt, K.; Brandt, C.; Tolley, C.; Rooney, J.; Berriman, M.; et al. Baseline Gut Microbiota Composition Is Associated With Schistosoma Mansoni Infection Burden in Rodent Models. Front. Immunol. 2020, 11, 593838. [Google Scholar] [CrossRef]
- Jenkins, T.P.; Pritchard, D.I.; Tanasescu, R.; Telford, G.; Papaiakovou, M.; Scotti, R.; Cortés, A.; Constantinescu, C.S.; Cantacessi, C. Experimental Infection with the Hookworm, Necator Americanus, Is Associated with Stable Gut Microbial Diversity in Human Volunteers with Relapsing Multiple Sclerosis. BMC Biol. 2021, 19, 74. [Google Scholar] [CrossRef]
- Pane, S.; Sacco, A.; Iorio, A.; Romani, L.; Putignani, L. Strongyloides Stercoralis Infestation in a Child: How a Nematode Can Affect Gut Microbiota. Int. J. Mol. Sci. 2021, 22, 2131. [Google Scholar] [CrossRef]
- Morton, E.R.; Lynch, J.; Froment, A.; Lafosse, S.; Heyer, E.; Przeworski, M.; Blekhman, R.; Ségurel, L. Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence. PLoS Genet. 2015, 11, e1005658. [Google Scholar] [CrossRef] [Green Version]
- Iebba, V.; Santangelo, F.; Totino, V.; Pantanella, F.; Monsia, A.; Di Cristanziano, V.; Di Cave, D.; Schippa, S.; Berrilli, F.; D’Alfonso, R. Gut Microbiota Related to Giardia Duodenalis, Entamoeba Spp. and Blastocystis Hominis Infections in Humans from Côte d’Ivoire. J. Infect. Dev. Ctries 2016, 10, 1035–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, L.R.; Verma, A.K.; Paul, J.; Bhattacharya, A. Phagocytosis of Gut Bacteria by Entamoeba Histolytica. Front. Cell. Infect. Microbiol. 2019, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Midha, A.; Schlosser, J.; Hartmann, S. Reciprocal Interactions between Nematodes and Their Microbial Environments. Front. Cell. Infect. Microbiol. 2017, 7, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beghini, F.; Pasolli, E.; Truong, T.D.; Putignani, L.; Cacciò, S.M.; Segata, N. Large-Scale Comparative Metagenomics of Blastocystis, a Common Member of the Human Gut Microbiome. ISME J. 2017, 11, 2848–2863. [Google Scholar] [CrossRef]
- Andersen, L.O.; Stensvold, C.R. Blastocystis in Health and Disease: Are We Moving from a Clinical to a Public Health Perspective? J. Clin. Microbiol. 2016, 54, 524–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audebert, C.; Even, G.; Cian, A.; Blastocystis Investigation Group; Loywick, A.; Merlin, S.; Viscogliosi, E.; Chabé, M. Colonization with the Enteric Protozoa Blastocystis Is Associated with Increased Diversity of Human Gut Bacterial Microbiota. Sci. Rep. 2016, 6, 25255. [Google Scholar] [CrossRef]
- Nagel, R.; Traub, R.J.; Allcock, R.J.N.; Kwan, M.M.S.; Bielefeldt-Ohmann, H. Comparison of Faecal Microbiota in Blastocystis-Positive and Blastocystis-Negative Irritable Bowel Syndrome Patients. Microbiome 2016, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billy, V.; Lhotská, Z.; Jirků, M.; Kadlecová, O.; Frgelecová, L.; Parfrey, L.W.; Pomajbíková, K.J. Blastocystis Colonization Alters the Gut Microbiome and, in Some Cases, Promotes Faster Recovery From Induced Colitis. Front. Microbiol. 2021, 12, 641483. [Google Scholar] [CrossRef]
- Stensvold, C.R.; van der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Stensvold, C.R.; Rajilić-Stojanović, M.; Heilig, H.G.H.J.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The Microbial Eukaryote Blastocystis Is a Prevalent and Diverse Member of the Healthy Human Gut Microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, P.D.; Stensvold, C.R.; Cotter, P.D. Development and Application of a Blastocystis Subtype-Specific PCR Assay Reveals That Mixed-Subtype Infections Are Common in a Healthy Human Population. Appl. Environ. Microbiol. 2015, 81, 4071–4076. [Google Scholar] [CrossRef] [Green Version]
- Holtman, G.A.; Kranenberg, J.J.; Blanker, M.H.; Ott, A.; Lisman-van Leeuwen, Y.; Berger, M.Y. Dientamoeba Fragilis Colonization Is Not Associated with Gastrointestinal Symptoms in Children at Primary Care Level. Fam. Pract. 2017, 34, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, A.M.; Stensvold, C.R.; Mirsepasi, H.; Engberg, J.; Friis-Møller, A.; Porsbo, L.J.; Hammerum, A.M.; Nordgaard-Lassen, I.; Nielsen, H.V.; Krogfelt, K.A. Active Ulcerative Colitis Associated with Low Prevalence of Blastocystis and Dientamoeba Fragilis Infection. Scand. J. Gastroenterol. 2013, 48, 638–639. [Google Scholar] [CrossRef]
- Coskun, A.; Malatyali, E.; Ertabaklar, H.; Yasar, M.B.; Karaoglu, A.O.; Ertug, S. Blastocystis in Ulcerative Colitis Patients: Genetic Diversity and Analysis of Laboratory Findings. Asian Pac. J. Trop. Med. 2016, 9, 916–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogsgaard, L.R.; Lee, O.; Johannesen, T.B.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. Characteristics of the Bacterial Microbiome in Association with Common Intestinal Parasites in Irritable Bowel Syndrome. Clin. Transl. Gastroenterol. 2018, 9, 161. [Google Scholar] [CrossRef]
- Berrilli, F.; Di Cave, D.; Cavallero, S.; D’Amelio, S. Interactions between Parasites and Microbial Communities in the Human Gut. Front. Cell. Infect. Microbiol. 2012, 2, 141. [Google Scholar] [CrossRef] [Green Version]
- Dheilly, N.M.; Martínez Martínez, J.; Rosario, K.; Brindley, P.J.; Fichorova, R.N.; Kaye, J.Z.; Kohl, K.D.; Knoll, L.J.; Lukeš, J.; Perkins, S.L.; et al. Parasite Microbiome Project: Grand Challenges. PLoS Pathog. 2019, 15, e1008028. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical Metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Chuvochina, M.; Oren, A.; Parks, D.H.; Soo, R.M. Prokaryotic Taxonomy and Nomenclature in the Age of Big Sequence Data. ISME J. 2021, 15, 1879–1892. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Ravel, J. The Vocabulary of Microbiome Research: A Proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef] [Green Version]
- Chistoserdovai, L. Functional Metagenomics: Recent Advances and Future Challenges. Biotechnol. Genet. Eng. Rev. 2010, 26, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.Y.; Lee, T.K.; Sul, W.J. Metagenomic Analysis of Chicken Gut Microbiota for Improving Metabolism and Health of Chickens—A Review. Asian-Australas J. Anim. Sci. 2015, 28, 1217–1225. [Google Scholar] [CrossRef] [Green Version]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A Review of Methods and Databases for Metagenomic Classification and Assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Campanaro, S.; Treu, L.; Kougias, P.G.; Zhu, X.; Angelidaki, I. Taxonomy of Anaerobic Digestion Microbiome Reveals Biases Associated with the Applied High Throughput Sequencing Strategies. Sci. Rep. 2018, 8, 1926. [Google Scholar] [CrossRef]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S RRNA Gene Sequencing of Mock Microbial Populations- Impact of DNA Extraction Method, Primer Choice and Sequencing Platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [Green Version]
- Colwell, R.K. III.1 Biodiversity: Concepts, Patterns, and Measurement; Princeton University Press: Princeton, NJ, USA, 2009; pp. 257–263. ISBN 978-1-4008-3302-3. [Google Scholar]
- Sala, C.; Vitali, S.; Giampieri, E.; do Valle, Ì.F.; Remondini, D.; Garagnani, P.; Bersanelli, M.; Mosca, E.; Milanesi, L.; Castellani, G. Stochastic Neutral Modelling of the Gut Microbiota’s Relative Species Abundance from next Generation Sequencing Data. BMC Bioinform. 2016, 17, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstock, G.M. Genomic Approaches to Studying the Human Microbiota. Nature 2012, 489, 250–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S RRNA and Shotgun Sequencing Data for the Taxonomic Characterization of the Gut Microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Del Chierico, F.; Abbatini, F.; Russo, A.; Quagliariello, A.; Reddel, S.; Capoccia, D.; Caccamo, R.; Ginanni Corradini, S.; Nobili, V.; De Peppo, F.; et al. Gut Microbiota Markers in Obese Adolescent and Adult Patients: Age-Dependent Differential Patterns. Front. Microbiol. 2018, 9, 1210. [Google Scholar] [CrossRef]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut Microbiota Profiling of Pediatric Nonalcoholic Fatty Liver Disease and Obese Patients Unveiled by an Integrated Meta-Omics-Based Approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef]
- Mortera, S.L.; Vernocchi, P.; Basadonne, I.; Zandonà, A.; Chierici, M.; Durighello, M.; Marzano, V.; Gardini, S.; Gasbarrini, A.; Urbani, A.; et al. A Metaproteomic-Based Gut Microbiota Profiling in Children Affected by Autism Spectrum Disorders. J. Proteom. 2021, 251, 104407. [Google Scholar] [CrossRef] [PubMed]
- Quagliariello, A.; Del Chierico, F.; Russo, A.; Reddel, S.; Conte, G.; Lopetuso, L.R.; Ianiro, G.; Dallapiccola, B.; Cardona, F.; Gasbarrini, A.; et al. Gut Microbiota Profiling and Gut-Brain Crosstalk in Children Affected by Pediatric Acute-Onset Neuropsychiatric Syndrome and Pediatric Autoimmune Neuropsychiatric Disorders Associated With Streptococcal Infections. Front. Microbiol. 2018, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- Dobson, G.; Hickey, C.; Trinder, J. Clostridium Difficile Colitis Causing Toxic Megacolon, Severe Sepsis and Multiple Organ Dysfunction Syndrome. Intensive Care Med. 2003, 29, 1030. [Google Scholar] [CrossRef]
- Mylonakis, E.; Ryan, E.T.; Calderwood, S.B. Clostridium Difficile—Associated Diarrhea: A Review. Arch. Intern. Med. 2001, 161, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium Difficile Infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased Diversity of the Fecal Microbiome in Recurrent Clostridium Difficile-Associated Diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Harstall, C.; Louie, T.; Veldhuyzen van Zanten, S.; Dieleman, L.A. Systematic Review: Faecal Transplantation for the Treatment of Clostridium Difficile-Associated Disease. Aliment. Pharmacol. Ther. 2012, 35, 865–875. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilić-Stojanović, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European Consensus Conference on Faecal Microbiota Transplantation in Clinical Practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Kelly, C.R.; Mullish, B.H.; Allegretti, J.R.; Kassam, Z.; Putignani, L.; Fischer, M.; Keller, J.J.; Costello, S.P.; et al. International Consensus Conference on Stool Banking for Faecal Microbiota Transplantation in Clinical Practice. Gut 2019, 68, 2111–2121. [Google Scholar] [CrossRef] [Green Version]
- Keller, J.J.; Ooijevaar, R.E.; Hvas, C.L.; Terveer, E.M.; Lieberknecht, S.C.; Högenauer, C.; Arkkila, P.; Sokol, H.; Gridnyev, O.; Mégraud, F.; et al. A Standardised Model for Stool Banking for Faecal Microbiota Transplantation: A Consensus Report from a Multidisciplinary UEG Working Group. United Eur. Gastroenterol. J. 2021, 9, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [Green Version]
- Marzano, V.; Mancinelli, L.; Bracaglia, G.; Del Chierico, F.; Vernocchi, P.; Di Girolamo, F.; Garrone, S.; Tchidjou Kuekou, H.; D’Argenio, P.; Dallapiccola, B.; et al. “Omic” Investigations of Protozoa and Worms for a Deeper Understanding of the Human Gut “Parasitome”. PLoS Negl. Trop. Dis. 2017, 11, e0005916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherz, V.; Greub, G.; Bertelli, C. Building up a Clinical Microbiota Profiling: A Quality Framework Proposal. Crit. Rev. Microbiol. 2021, 1–20. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pane, S.; Ristori, M.V.; Gardini, S.; Russo, A.; Del Chierico, F.; Putignani, L. Clinical Parasitology and Parasitome Maps as Old and New Tools to Improve Clinical Microbiomics. Pathogens 2021, 10, 1550. https://doi.org/10.3390/pathogens10121550
Pane S, Ristori MV, Gardini S, Russo A, Del Chierico F, Putignani L. Clinical Parasitology and Parasitome Maps as Old and New Tools to Improve Clinical Microbiomics. Pathogens. 2021; 10(12):1550. https://doi.org/10.3390/pathogens10121550
Chicago/Turabian StylePane, Stefania, Maria Vittoria Ristori, Simone Gardini, Alessandra Russo, Federica Del Chierico, and Lorenza Putignani. 2021. "Clinical Parasitology and Parasitome Maps as Old and New Tools to Improve Clinical Microbiomics" Pathogens 10, no. 12: 1550. https://doi.org/10.3390/pathogens10121550
APA StylePane, S., Ristori, M. V., Gardini, S., Russo, A., Del Chierico, F., & Putignani, L. (2021). Clinical Parasitology and Parasitome Maps as Old and New Tools to Improve Clinical Microbiomics. Pathogens, 10(12), 1550. https://doi.org/10.3390/pathogens10121550







